Quorum Sensing Inhibition by Sponge-Associated Bacillus Species: Suppressing Pseudomonas aeruginosa Virulence Factors
Abstract
1. Introduction
2. Results
2.1. Identification of QS Inhibitory Sponge-Associated Bacteria Using Biosensor Sandwich Assays
2.2. 16S rRNA Gene Identification of Selected QS Inhibitory Isolates
2.3. Amplification and Sequencing of the aiiA Lactonase Gene
2.4. Characterization of Sponge-Associated Bacillus Species Extracts
2.4.1. Fourier Transform Infrared Analysis
2.4.2. Gas Chromatography-Mass Spectrometry Characterization
2.4.3. Isolate-Specific Metabolite Profiles
2.5. Sponge-Associated Bacterial Extracts Display QSI Activity Against C. violaceum ATCC 12472
2.6. Antimicrobial Testing of Sponge-Associated Bacillus Species Extracts Against Pseudomonas Aeruginosa ATCC 27853
2.7. Assessing P. aeruginosa ATCC 27853 Virulence Factor Inhibition
2.7.1. P. aeruginosa Pyocyanin Production Inhibition by Bacillus Species Extracts
2.7.2. P. aeruginosa Pyoverdine Production Inhibition by Bacillus Species Extracts
2.7.3. P. aeruginosa Elastase Production Inhibition by Bacillus Species Extracts
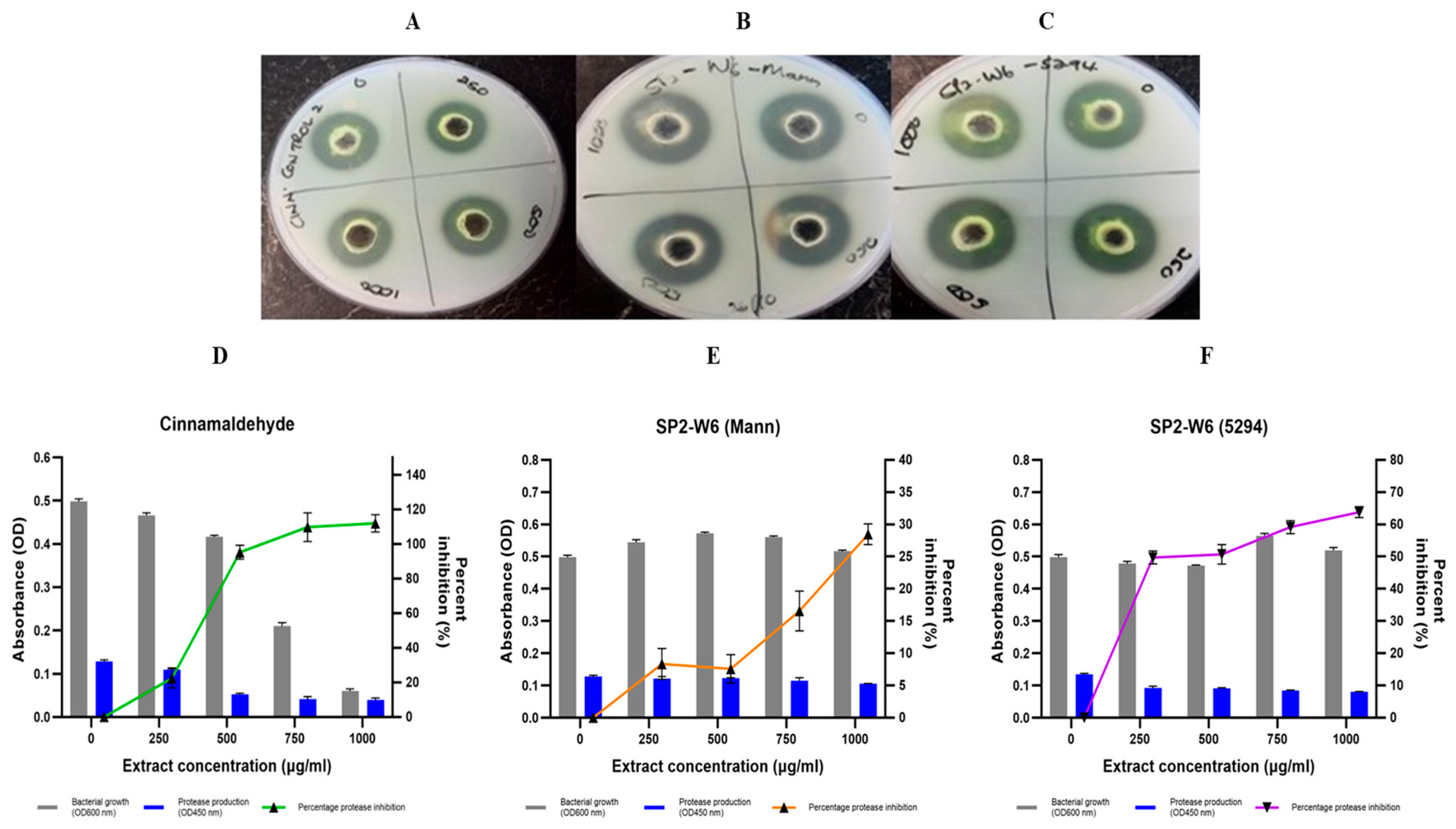
2.7.4. P. aeruginosa Protease Production Inhibition by Bacillus Species Extracts
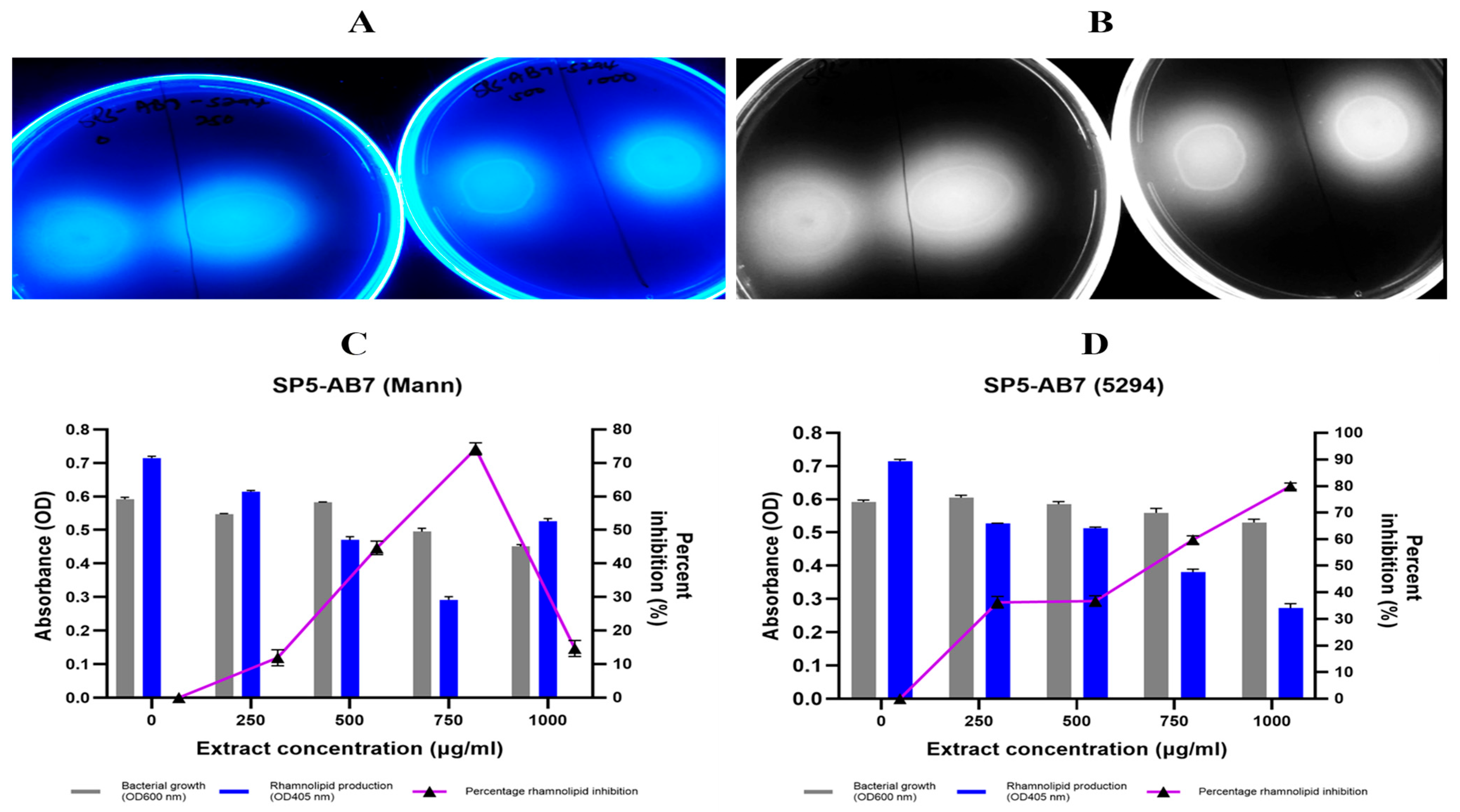
2.7.5. P. aeruginosa Rhamnolipid Production Inhibition by Bacillus Species Extracts
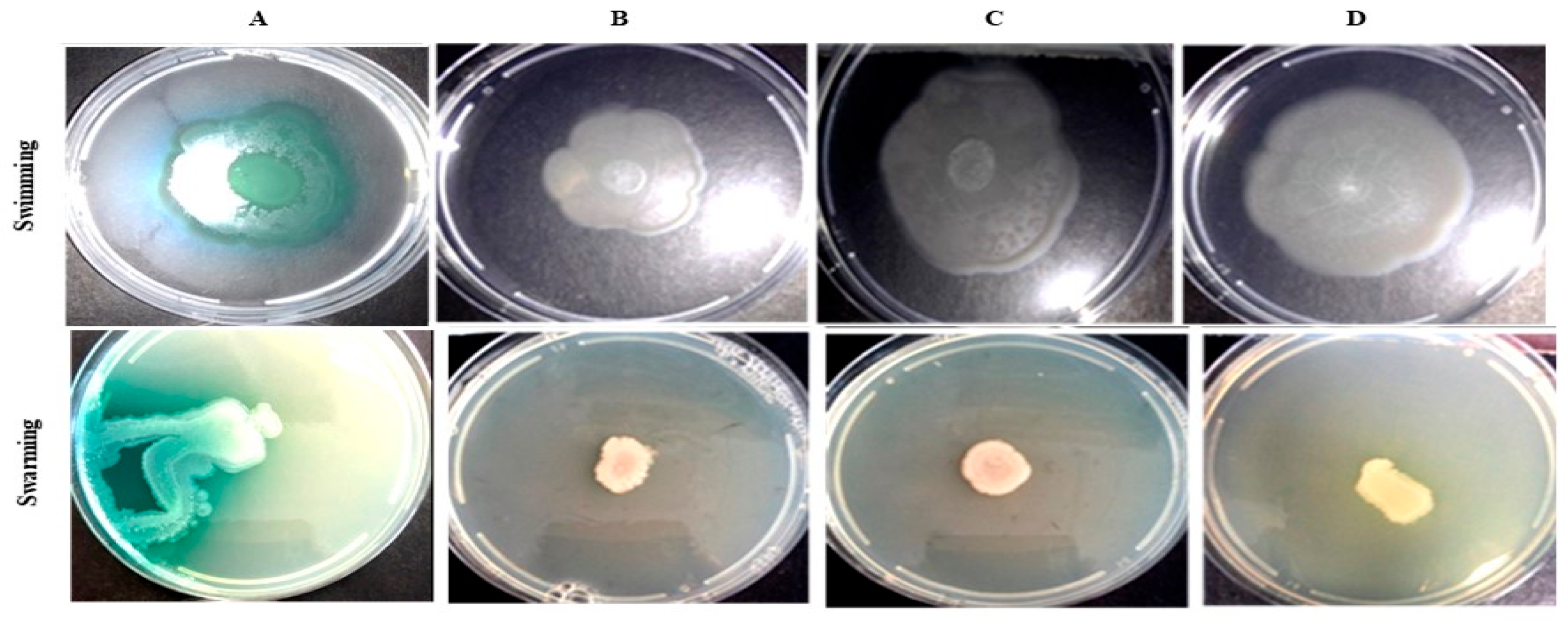
2.7.6. P. aeruginosa Swimming Motility Inhibition by Bacillus Species Extracts
2.7.7. P. aeruginosa Swarming Motility Inhibition by Bacillus Species Extracts
2.7.8. P. aeruginosa Initial Adhesion Inhibition by Bacillus Species Extracts
2.7.9. P. aeruginosa Mature Biofilm Inhibition by Bacillus Species Extracts
3. Discussion
4. Materials and Methods
4.1. Isolation of Sponge-Associated Bacteria
4.2. Quorum Sensing Inhibition Screening Using Chromobacterium violaceum
4.3. 16S rRNA Gene-Based Identification of Bacterial Isolates
4.4. PCR Amplification of aiiA Lactonase Gene
4.5. Chemical Characterization of Ethyl Acetate Culture Extracts
4.5.1. Extract Preparation
4.5.2. Fourier Transform Infrared (FTIR) Spectroscopy
4.5.3. Gas Chromatography-Mass Spectrometry (GC-MS)
4.6. Qualitative QSI Agar Well-Overlay Screening
4.7. Antimicrobial Testing of Bacterial Extracts Using the Agar-Well Diffusion Method
4.8. Screening of Extracts Against P. aeruginosa Virulence Factor Production
4.8.1. Inhibition of Pyocyanin Production
4.8.2. Inhibition of Pyoverdine Production
4.8.3. Inhibition of LasB Production
4.8.4. Qualitative Inhibition of Casein Hydrolysis
4.8.5. Quantitative Inhibition of Casein Hydrolysis
4.8.6. Qualitative Inhibition of Rhamnolipid Production
4.8.7. Quantitative Inhibition of Rhamnolipid Production
4.8.8. Inhibition of Swimming and Swarming Motility
4.8.9. Inhibition of Biofilm Production
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muimhneacháin, E.Ó.; Reen, F.J.; O’ Gara, F.; McGlacken, G.P. Analogues of Pseudomonas aeruginosa signalling molecules to tackle infections. Org. Biomol. Chem. 2018, 16, 169–179. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Soukarieh, F.; Williams, P.; Stocks, M.J.; Cámara, M. Pseudomonas aeruginosa quorum sensing systems as drug discovery targets: Current position and future perspectives. J. Med. Chem. 2018, 61, 10385–10402. [Google Scholar] [CrossRef] [PubMed]
- Hibbert, T.M.; Whiteley, M.; Renshaw, S.A.; Neill, D.R.; Fothergill, J.L. Emerging strategies to target virulence in Pseudomonas aeruginosa respiratory infections. Crit. Rev. Microbiol. 2024, 50, 1037–1052. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Krishnan, T.; Wang, H.; Chen, Y.; Yin, Y.-F.; Chong, Y.-M.; Tan, L.-Y.; Chong, T.M.; Chan, K.-G. Non-antibiotic quorum sensing inhibitors acting against N-acyl homoserine lactone synthase as druggable target. Sci. Rep. 2014, 4, 7245. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef]
- Schütz, C.; Empting, M. Targeting the Pseudomonas quinolone signal quorum sensing system for the discovery of novel anti-infective pathoblockers. J. Org. Chem. 2018, 14, 2627–2645. [Google Scholar] [CrossRef] [PubMed]
- Rather, M.A.; Saha, D.; Bhuyan, S.; Jha, A.N.; Mandal, M. Quorum quenching: A drug discovery approach against Pseudomonas aeruginosa. Microbiol. Res. 2022, 264, 127173. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Sendra, E.; Fernández-Muñoz, A.; Zamorano, L.; Oliver, A.; Horcajada, J.P.; Juan, C.; Gómez-Zorrilla, S. Impact of multidrug resistance on the virulence and fitness of Pseudomonas aeruginosa: A microbiological and clinical perspective. Infection 2024, 52, 1235–1268. [Google Scholar] [CrossRef]
- Chen, J.; Wang, B.; Lu, Y.; Guo, Y.; Sun, J.; Wei, B.; Zhang, H.; Wang, H. Quorum sensing inhibitors from marine microorganisms and their synthetic derivatives. Mar. Drugs 2019, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Dessaux, Y.; Llamas, I. Saline environments as a source of potential quorum sensing disruptors to control bacterial infections: A review. Mar. Drugs 2019, 17, 191. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Sukhikh, S.; Babich, O. Study of marine microorganism metabolites: New resources for bioactive natural products. Front. Microbiol. 2024, 14, 1285902. [Google Scholar] [CrossRef]
- Kiran, G.S.; Sekar, S.; Ramasamy, P.; Thinesh, T.; Hassan, S.; Lipton, A.N.; Ninawe, A.S.; Selvin, J. Marine sponge microbial association: Towards disclosing unique symbiotic interactions. Mar. Environ. Res. 2018, 140, 169–179. [Google Scholar] [CrossRef]
- Amelia, T.S.M.; Suaberon, F.A.C.; Vad, J.; Fahmi, A.D.M.; Saludes, J.P.; Bhubalan, K. Recent advances of marine sponge-associated microorganisms as a source of commercially viable natural products. Mar. Biotechnol. 2022, 24, 492–512. [Google Scholar] [CrossRef]
- Skindersoe, M.E.; Ettinger-Epstein, P.; Rasmussen, T.B.; Bjarnsholt, T.; de Nys, R.; Givskov, M. Quorum sensing antagonism from marine organisms. Mar. Biotechnol. 2008, 10, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Annapoorani, A.; Jabbar, A.K.; Musthafa, S.K.; Pandian, S.K.; Ravi, A.V. Inhibition of quorum sensing mediated virulence factors production in urinary pathogen Serratia marcescens PS1 by marine sponges. Indian J. Microbiol. 2012, 52, 160–166. [Google Scholar] [CrossRef]
- Pejin, B.; Talevska, A.; Ciric, A.; Glamoclija, J.; Nikolic, M.; Talevski, T.; Sokovic, M. Anti-quorum sensing activity of selected sponge extracts: A case study of Pseudomonas aeruginosa. Nat. Prod. Res. 2014, 28, 2330–2333. [Google Scholar] [CrossRef]
- Mai, T.; Tintillier, F.; Lucasson, A.; Moriou, C.; Bonno, E.; Petek, S.; Magré, K.; Al Mourabit, A.; Saulnier, D.; Debitus, C. Quorum sensing inhibitors from Leucetta chagosensis Dendy, 1863. Lett. Appl. Microbiol. 2015, 61, 311–317. [Google Scholar] [CrossRef]
- Quintana, J.; Brango-Vanegas, J.; Costa, G.M.; Castellanos, L.; Arévalo, C.; Duque, C. Marine organisms as source of extracts to disrupt bacterial communication: Bioguided isolation and identification of quorum sensing inhibitors from Ircinia felix. Rev. Bras. Farmacogn. 2015, 25, 199–207. [Google Scholar] [CrossRef]
- Saurav, K.; Bar-Shalom, R.; Haber, M.; Burgsdorf, I.; Oliviero, G.; Costantino, V.; Morgenstern, D.; Steindler, L. In search of alternative antibiotic drugs: Quorum-quenching activity in sponges and their bacterial isolates. Front. Microbiol. 2016, 7, 416. [Google Scholar] [CrossRef]
- Costantino, V.; Della Sala, G.; Saurav, K.; Teta, R.; Bar-Shalom, R.; Mangoni, A.; Steindler, L. Plakofuranolactone as a quorum quenching agent from the Indonesian sponge Plakortis cf. lita. Mar. Drugs 2017, 15, 59. [Google Scholar] [CrossRef]
- Batista, D.; Costa, R.; Carvalho, A.P.; Batista, W.R.; Rua, C.P.J.; de Oliveira, L.; Leomil, L.; Fróes, A.M.; Thompson, F.L.; Coutinho, R.; et al. Environmental conditions affect activity and associated microorganisms of marine sponges. Mar. Environ. Res. 2018, 142, 59–68. [Google Scholar] [CrossRef]
- Alam, P.; Alqahtani, A.S.; Mabood Husain, F.; Tabish Rehman, M.; Alajmi, M.F.; Noman, O.M.; El Gamal, A.A.; Al-Massarani, S.M.; Shavez Khan, M. Siphonocholin isolated from red sea sponge Siphonochalina siphonella attenuates quorum sensing controlled virulence and biofilm formation. Saudi Pharm. J. 2020, 28, 1383–1391. [Google Scholar] [CrossRef]
- Kovalerchik, D.; Singh, R.P.; Schlesinger, P.; Mahajni, A.; Shefer, S.; Fridman, M.; Ilan, M.; Carmeli, S. Bromopyrrole alkaloids of the sponge Agelas oroides collected near the Israeli Mediterranean coastline. J. Nat. Prod. 2020, 83, 374–384. [Google Scholar] [CrossRef]
- Tintillier, F.; Moriou, C.; Petek, S.; Fauchon, M.; Hellio, C.; Saulnier, D.; Ekins, M.; Hooper, J.N.A.; Al-Mourabit, A.; Debitus, C. Quorum sensing inhibitory and antifouling activities of new bromotyrosine metabolites from the polynesian sponge Pseudoceratina n. sp. Mar. Drugs 2020, 18, 272. [Google Scholar] [CrossRef] [PubMed]
- Saurav, K.; Borbone, N.; Burgsdorf, I.; Teta, R.; Caso, A.; Bar-Shalom, R.; Esposito, G.; Britstein, M.; Steindler, L.; Costantino, V. Identification of quorum sensing activators and inhibitors in the marine sponge Sarcotragus spinosulus. Mar. Drugs 2020, 18, 127. [Google Scholar] [CrossRef] [PubMed]
- Oluwabusola, E.T.; Katermeran, N.P.; Poh, W.H.; Goh, T.M.B.; Tan, L.T.; Diyaolu, O.; Tabudravu, J.; Ebel, R.; Rice, S.A.; Jaspars, M. Inhibition of the quorum sensing system, elastase production and biofilm formation in Pseudomonas aeruginosa by Psammaplin A and Bisaprasin. Molecules 2022, 27, 1721. [Google Scholar] [CrossRef]
- Liang, J.; She, J.; Fu, J.; Wang, J.; Ye, Y.; Yang, B.; Liu, Y.; Zhou, X.; Tao, H. Advances in natural products from the marine-sponge-associated microorganisms with antimicrobial activity in the last decade. Mar. Drugs 2023, 21, 236. [Google Scholar] [CrossRef] [PubMed]
- Amran, R.H.; Jamal, M.T.; Bowrji, S.; Sayegh, F.; Santanumurti, M.B.; Satheesh, S. Mini review: Antimicrobial compounds produced by bacteria associated with marine invertebrates. Folia Microbiol. 2024, 70, 271–292. [Google Scholar] [CrossRef]
- Indraningrat, A.; Smidt, H.; Sipkema, D. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Bibi, F.; Faheem, M.; Azhar, E.I.; Yasir, M.; Alvi, S.A.; Kamal, M.A.; Ullah, I.; Naseer, M.I. Bacteria from marine sponges: A source of new drugs. Curr. Drug Metab. 2017, 18, 11–15. [Google Scholar] [CrossRef]
- Laport, M.S. Isolating bacteria from sponges: Why and how? Curr. Pharm. Biotechnol. 2017, 18, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Freitas-Silva, J.; Silva-Oliveira, T.; Muricy, G.; Laport, M.S. Bacillus strains associated to Homoscleromorpha sponges are highly active against multidrug resistant bacteria. Curr. Microbiol. 2020, 77, 807–815. [Google Scholar] [CrossRef]
- Xiao, S.; Chen, N.; Chai, Z.; Zhou, M.; Xiao, C.; Zhao, S.; Yang, X. Secondary metabolites from marine-derived Bacillus: A comprehensive review of origins, structures, and bioactivities. Mar. Drugs 2022, 20, 567. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, T.; Wang, J.; Wang, Y.; Zhang, X.-H. The mechanisms and applications of quorum sensing (QS) and quorum quenching (QQ). J. Ocean Univ. China 2019, 18, 1427–1442. [Google Scholar] [CrossRef]
- Phelan, R.W.; O’Halloran, J.A.; Kennedy, J.; Morrissey, J.P.; Dobson, A.D.W.; O’Gara, F.; Barbosa, T.M. Diversity and bioactive potential of endospore-forming bacteria cultured from the marine sponge Haliclona simulans. J. Appl. Microbiol. 2012, 112, 65–78. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.; Reen, F.; Parages, M.; McCarthy, R.; Dobson, A.; O’Gara, F. Disruption of N-acyl-homoserine-lactone-specific signalling and virulence in clinical pathogens by marine sponge bacteria. Microb. Biotechnol. 2019, 12, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.F.M.; Goh, H.C.; Lim, S.C.; Pang, L.M.; Chin, J.S.F.; Tan, K.S.; Liang, Z.X.; Yang, L.; Glukhov, E.; Gerwick, W.H.; et al. Integrated genomic and metabolomic approach to the discovery of potential anti-quorum sensing natural products from microbes associated with marine samples from Singapore. Mar. Drugs 2019, 17, 72. [Google Scholar] [CrossRef]
- Singh, A.A.; Singh, A.K.; Nerurkar, A. Bacteria associated with marine macroorganisms as potential source of quorum-sensing antagonists. J. Basic Microbiol. 2020, 60, 799–808. [Google Scholar] [CrossRef]
- Singh, A.A.; Singh, A.K.; Nerurkar, A. Disrupting the quorum sensing mediated virulence in soft rot causing Pectobacterium carotovorum by marine sponge associated Bacillus sp. OA10. World J. Microbiol. Biotechnol. 2021, 37, 5. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Simões, M. Quorum sensing inhibition by marine bacteria. Mar. Drugs 2019, 17, 427. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Juarez, I.; López-Jácome, L.; Soberón-Chávez, G.; Tomás, M.; Castañeda-Tamez, P.; Hernández-Bárragan, I.; Cruz-Muñiz, M.; Maeda, T.; Wood, T.; García-Contreras, R. Exploiting quorum sensing inhibition for the control of Pseudomonas aeruginosa and Acinetobacter baumannii biofilms. Curr. Top. Med. Chem. 2017, 17, 1915–1927. [Google Scholar] [CrossRef]
- Nithya, C.; Aravindraja, C.; Pandian, S.K. Bacillus pumilus of Palk Bay origin inhibits quorum-sensing-mediated virulence factors in Gram-negative bacteria. Res. Microbiol. 2010, 161, 293–304. [Google Scholar] [CrossRef]
- Nithya, C.; Begum, M.F.; Pandian, S.K. Marine bacterial isolates inhibit biofilm formation and disrupt mature biofilms of Pseudomonas aeruginosa PAO1. Appl. Microbiol. Biotechnol. 2010, 88, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Musthafa, K.; Saroja, V.; Pandian, S.; Ravi, A. Antipathogenic potential of marine Bacillus sp. SS4 on N-acyl-homoserine-lactone-mediated virulence factors production in Pseudomonas aeruginosa (PAO1). J. Biosci. 2011, 36, 55–67. [Google Scholar] [CrossRef]
- Hall, S.; McDermott, C.; Anoopkumar-Dukie, S.; McFarland, A.; Forbes, A.; Perkins, A.; Davey, A.; Chess-Williams, R.; Kiefel, M.; Arora, D.; et al. Cellular effects of pyocyanin, a secreted virulence factor of Pseudomonas aeruginosa. Toxins 2016, 8, 236. [Google Scholar] [CrossRef]
- Mani, A.; Hameed, S.; Ramalingam, S.; Narayanan, M. Assessment of quorum quenching activity of Bacillus species against Pseudomonas aeruginosa MTCC 2297. Glob. J. Pharmacol. 2012, 6, 118–125. [Google Scholar] [CrossRef]
- Gellatly, S.; Robert, H. Pseudomonas aeruginosa: New insights into pathogenesis and host defences. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Guendouze, A.; Plener, L.; Bzdrenga, J.; Jacquet, P.; Rémy, B.; Elias, M.; Lavigne, J.; Daude, D.; Chabrière, E. Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front. Microbiol. 2017, 8, 227. [Google Scholar] [CrossRef]
- Granato, E.; Harrison, F.; Kümmerli, R.; Ross-Gillespie, A. Do bacterial “virulence factors” always increase virulence? A meta-analysis of pyoverdine production in Pseudomonas aeruginosa as a test case. Front. Microbiol. 2016, 7, 1952. [Google Scholar] [CrossRef] [PubMed]
- Bandara, H.; Lam, O.; Jin, L.; Samaranayake, L. Microbial chemical signalling: A current perspective. Crit. Rev. Microbiol. 2012, 3, 217–249. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Yoon, S. Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J. Microbiol. Biotechnol. 2017, 27, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Steindler, L.; Bertani, I.; De Sordi, L.; Schwager, S.; Eberl, L.; Venturi, V. LasI/R and RhlI/R quorum sensing in a strain of Pseudomonas aeruginosa beneficial to plants. Appl. Environ. Microbiol. 2009, 75, 5131–5140. [Google Scholar] [CrossRef]
- Morales-Soto, N.; Anyan, M.; Mattingly, A.; Madukoma, C.; Harvey, C.; Alber, M.; Déziel, E.; Kearns, D.; Shrout, J. Preparation, imaging and quantification of bacterial surface motility assays. J. Vis. Exp. 2012, 98, e52338. [Google Scholar] [CrossRef]
- Rekha, P.; Vasavi, H.; Vipin, C.; Saptami, K.; Arun, A. A medicinal herb Cassia alata attenuates quorum sensing in Chromobacterium violaceum and Pseudomonas aeruginosa. Lett. Appl. Microbiol. 2016, 64, 231–238. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J. Biofilm dispersion in Pseudomonas aeruginosa. J. Microbiol. 2016, 54, 71–85. [Google Scholar] [CrossRef]
- Moradali, F.; Ghods, S.; Rehm, H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Sayem, S.; Manzo, E.; Ciavatta, L.; Tramice, A.; Cordone, A.; Zanfardino, A.; De Felice, M.; Varcamonti, M. Anti-biofilm activity of an exopolysaccharide from a sponge-associated strain of Bacillus licheniformis. Microb. Cell Fact. 2011, 10, 74–85. [Google Scholar] [CrossRef]
- Holm, A.; Vikstrom, E. Quorum sensing communication between bacteria and human cells: Signals, targets, and functions. Front. Plant Sci. 2014, 5, 309. [Google Scholar] [CrossRef] [PubMed]
- Boopathi, S.; Vashisth, R.; Manoharan, P.; Kandasamy, R.; Sivakumar, N. Stigmatellin Y—An anti-biofilm compound from Bacillus subtilis BR4 possibly interferes in PQS-PqsR mediated quorum sensing system in Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2017, 27, 2113–2118. [Google Scholar] [CrossRef]
- Fong, J.; Zhang, C.; Yang, R.; Boo, Z.Z.; Tan, S.K.; Nielsen, T.E.; Givskov, M.; Liu, X.W.; Bin, W.; Su, H.; et al. Combination therapy strategy of quorum quenching enzyme and quorum sensing inhibitor in suppressing multiple quorum sensing pathways of P. aeruginosa. Sci. Rep. 2018, 8, 1155. [Google Scholar] [CrossRef]
- Algburi, A.; Zehm, S.; Netrebov, V.; Bren, A.; Chistyakov, V.; Chikindas, M. Subtilosin prevents biofilm formation by inhibiting bacterial quorum sensing. Probiotics Antimicrob. Proteins 2017, 9, 81–90. [Google Scholar] [CrossRef]
- Shah, S.D.; Patel, H.; Saiyad, S.M.; Bajpai, B. Effect of a phthalate derivative purified from Bacillus zhangzhouensis SK4 on quorum sensing regulated virulence factors of Pseudomonas aeruginosa. Microb. Pathog. 2024, 191, 106664. [Google Scholar] [CrossRef] [PubMed]
- Teasdale, M.E.; Donovan, K.A.; Forschner-Dancause, S.R.; Rowley, D.C. Gram-positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Mar. Biotechnol. 2011, 13, 722–732. [Google Scholar] [CrossRef]
- Patel, M.; Siddiqui, A.J.; Ashraf, S.A.; Surti, M.; Awadelkareem, A.M.; Snoussi, M.; Hamadou, W.S.; Bardakci, F.; Jamal, A.; Jahan, S.; et al. Lactiplantibacillus plantarum-derived biosurfactant attenuates quorum sensing-mediated virulence and biofilm formation in Pseudomonas aeruginosa and Chromobacterium violaceum. Microorganisms 2022, 10, 1026. [Google Scholar] [CrossRef]
- Almasoud, A.; Hettiarachchy, N.; Rayaprolu, S.; Babu, D.; Kwon, Y.M.; Mauromoustakos, A. Inhibitory effects of lactic and malic organic acids on autoinducer type 2 (AI-2) quorum sensing of Escherichia coli O157: H7 and Salmonella Typhimurium. LWT Food Sci. Technol. 2016, 66, 560–564. [Google Scholar] [CrossRef]
- Bao, X.; Bové, M.; Coenye, T. Organic acids and their salts potentiate the activity of selected antibiotics against Pseudomonas aeruginosa biofilms grown in a synthetic cystic fibrosis sputum medium. Antimicrob. Agents Chemother. 2022, 66, e01875-21. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.; Kumari, S.; Goel, G.; Azmi, W. Synergistic combination of malic acid with sodium hypochlorite impairs biofilm of Cronobacter sakazakii. LWT 2022, 155, 112902. [Google Scholar] [CrossRef]
- Musthafa, K.S.; Sivamaruthi, B.S.; Pandian, S.K.; Ravi, A.V. Quorum sensing inhibition in Pseudomonas aeruginosa PAO1 by antagonistic compound phenylacetic acid. Curr. Microbiol. 2012, 65, 475–480. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Dobretsov, S.; Al-Fori, M.; Gunasekera, S.P.; Sudesh, K.; Paul, V.J. Quorum-sensing inhibitory compounds from extremophilic microorganisms isolated from a hypersaline cyanobacterial mat. J. Ind. Microbiol. Biotechnol. 2013, 40, 759–772. [Google Scholar] [CrossRef]
- Kachhadia, R.; Kapadia, C.; Singh, S.; Gandhi, K.; Jajda, H.; Alfarraj, S.; Ansari, M.J.; Danish, S.; Datta, R. Quorum sensing inhibitory and quenching activity of Bacillus cereus RC1 extracts on soft rot-causing bacteria Lelliottia amnigena. ACS Omega 2022, 7, 25291–25308. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, I.; Jamal, M.T.; Pugazhendi, A.; Dhavamani, J.; Al-Shaeri, M.; Al-Maaqar, S.; Satheesh, S. Antibacterial and antibiofilm activity of extracts from sponge-associated bacterial endophytes. Prep. Biochem. Biotechnol. 2023, 53, 1143–1153. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, I.; Jamal, M.T.; Pugazhendi, A.; Dhavamani, J.; Satheesh, S. Antibiofilm activity of secondary metabolites from bacterial endophytes of Red Sea soft corals. Int. Biodeterior. Biodegrad. 2022, 173, 105462. [Google Scholar] [CrossRef]
- Gamal El-Din, M.I.; Youssef, F.S.; Altyar, A.E.; Ashour, M.L. GC/MS analyses of the essential oils obtained from different Jatropha species, their discrimination using chemometric analysis and assessment of their antibacterial and anti-biofilm activities. Plants 2022, 11, 1268. [Google Scholar] [CrossRef]
- Galdiero, E.; Ricciardelli, A.; D’Angelo, C.; De Alteriis, E.; Maione, A.; Albarano, L.; Casillo, A.; Corsaro, M.M.; Tutino, M.L.; Parrilli, E. Pentadecanoic acid against Candida albicans-Klebsiella pneumoniae biofilm: Towards the development of an anti-biofilm coating to prevent polymicrobial infections. Res. Microbiol. 2021, 172, 103880. [Google Scholar] [CrossRef]
- Crawford, D.L.; Lynch, J.M.; Whipps, M.J.; Ousley, M.A. Isolation and characterization of actinomycete antagonists of a fungal root pathogen. Appl. Environ. Microbiol. 1993, 59, 3899–3905. [Google Scholar] [CrossRef]
- Flemming, L.; Rawlings, D.; Chenia, H. Phenotypic and molecular characterisation of fish-borne Flavobacterium johnsoniae-like isolates from aquaculture systems in South Africa. Res. Microbiol. 2007, 158, 18–30. [Google Scholar] [CrossRef]
- Qin, S.; Li, J.; Chen, H.; Zhao, G.-Z.; Zhu, W.-Y.; Jiang, C.-L.; Xu, L.-H.; Li, W.-J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China Appl. Environ. Microbiol. 2009, 75, 61–76. [Google Scholar] [CrossRef]
- Wink, J. Methods for the Taxonomic Description of the Actinobacteria. Compendium of Actinobacteria. Available online: http://www.dsmz.de/fileadmin/Bereiche/Microbiology/Dateien/Bacterial_Nomenclature_uptodate/Actinomethods.pdf (accessed on 15 February 2016).
- Chu, W.; Lu, F.; Zhu, W.; Kang, C. Isolation and characterization of new potential probiotic bacteria based on quorum-sensing system. J. Appl. Microbiol. 2011, 110, 202–208. [Google Scholar] [CrossRef]
- McClean, K.; Winson, M.; Fish, L.; Taylor, A.; Chhabra, S.; Camara, M.; Daykin, M.; Lamb, J.; Swift, S.; Bycroft, B.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef]
- Morohoshi, T.; Kato, M.; Fukamachi, K.; Kato, N.; Ikeda, T. N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC12472. FEMS Microbiol. Lett. 2008, 279, 124–130. [Google Scholar] [CrossRef]
- Vaneechoutte, M.; Claeys, G.; Steyaert, S.; De Baere, T.; Peleman, R.; Verschraegen, G. Isolation of Moraxella canis from an ulcerated metastatic lymph node. J. Clin. Microbiol. 2000, 38, 3870–3871. [Google Scholar] [CrossRef]
- Coram, N.; Rawlings, D. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptospirillum ferriphilum sp. nov dominates South African commercial biooxidation tanks that operate at 40 °C. Appl. Environ. Microbiol. 2002, 68, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Huma, N.; Shankar, P.; Kushwah, J.; Bhushan, A.; Joshi, J.; Mukherjee, T.; Purohit, J.; Kalia, V. Diversity and polymorphism in AHL-lactonase gene (aiiA) of Bacillus. J. Microbiol. Biotechnol. 2011, 21, 1001–1011. [Google Scholar] [CrossRef]
- Pan, J.; Haung, T.; Yao, F.; Haung, Z.; Powell, C.A.; Qiu, S.; Guan, X. Expression and characterization of aiiA gene from Bacillus subtilis BS-1. Microbiol. Res. 2008, 163, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Baltes, M.; Dubois, J.; Hanocq, M. Ethyl acetate extraction procedure and isocratic high-performance liquid chromatographic assay for testosterone metabolites in cell microsomes. J. Chromatogr. B Biomed. Sci. Appl. 1998, 706, 201–207. [Google Scholar] [CrossRef]
- McLean, R.J.C.; Pierson, L.S.; Fuqua, C. A simple screening protocol for the identification of quorum signal antagonists. J. Microbiol. Methods 2004, 58, 351–360. [Google Scholar] [CrossRef]
- Chenia, H.Y. Anti-quorum sensing potential of crude Kigelia africana fruit extracts. Sensors 2013, 13, 2802–2817. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.A.; Abdelwahed, N.A. Application of statistical experimental design for optimization of silver nanoparticles biosynthesis by a nanofactory Streptomyces viridochromogenes. J. Microbiol. 2014, 52, 53–63. [Google Scholar] [CrossRef]
- Essar, D.; Eberly, E.; Hadero, A.; Crawfords, I. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef]
- Huerta, V.; Mihalik, K.; Crixell, S.H.; Vattem, D.A. Herbs, spices and medicinal plants used in Hispanic traditional medicine can decrease quorum sensing dependent virulence in Pseudomonas aeruginosa. Int. J. Appl. Res. Nat. Prod. 2008, 1, 9–15. [Google Scholar]
- George, M.; Pierce, G.; Gabriel, M.; Morris, C.; Ahearn, D. Effects of quorum sensing molecules of Pseudomonas aeruginosa on organism growth, elastase B production, and primary adhesion to hydrogel contact lenses. Eye Contact Lens 2005, 31, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Zacaria, J.; Delamare, A.; Costa, S.O.P.; Echeverrigaray, S. Diversity of extracellular proteases among Aeromonas determined by zymogram analysis. J. Appl. Microbiol. 2010, 109, 212–219. [Google Scholar] [CrossRef]
- Nicodème, M.; Grill, J.; Humbert, G.; Gaillard, J. Extracellular protease activity of different Pseudomonas strains: Dependence of proteolytic activity on culture conditions. J. Appl. Microbiol. 2005, 99, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Senturk, S.; Ulusoy, S.; Bosgelmez-Tinaz, G.; Yagci, A. Quorum sensing and virulence of Pseudomonas aeruginosa during urinary tract infections. J. Infect. Dev. Ctries. 2012, 6, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Kappeli, O.; Fiechter, A.; Reiser, J. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J. Bacteriol. 1991, 173, 4214–4219. [Google Scholar] [CrossRef]
- Rashid, M.H.; Rambaugh, K.; Passador, L.; Davies, D.G.; Hamood, A.N.; Iglewski, B.H.; Kornberg, A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 9636–9641. [Google Scholar] [CrossRef]
- Packiavathy, I.A.S.V.; Agilandeswari, P.; Syed Musthafa, K.; Pandian, S.K.; Ravi, A.V. Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl eugenol against Gram negative bacterial pathogens. Food Res. Int. 2012, 45, 85–92. [Google Scholar] [CrossRef]
- Basson, A.; Flemming, L.; Chenia, H. Evaluation of adherence, hydrophobicity, aggregation and biofilm development of Flavobacterium johnsoniae-like isolates. Microb. Ecol. 2008, 55, 1–14. [Google Scholar] [CrossRef]
- Pitt, B.; Hamilton, M.; Zelver, N.; Stewart, P. A microtiter-plate screening method for biofilm disinfection and removal. J. Microbiol. Methods 2003, 54, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.S. Investigating the Quorum-Sensing Inhibitory and Potential HIV Activity of Indigenous South African Sponge-Associated Bacterial Extracts. Master’s Thesis, University of KwaZulu-Natal, Durban, South Africa, 2018. [Google Scholar]
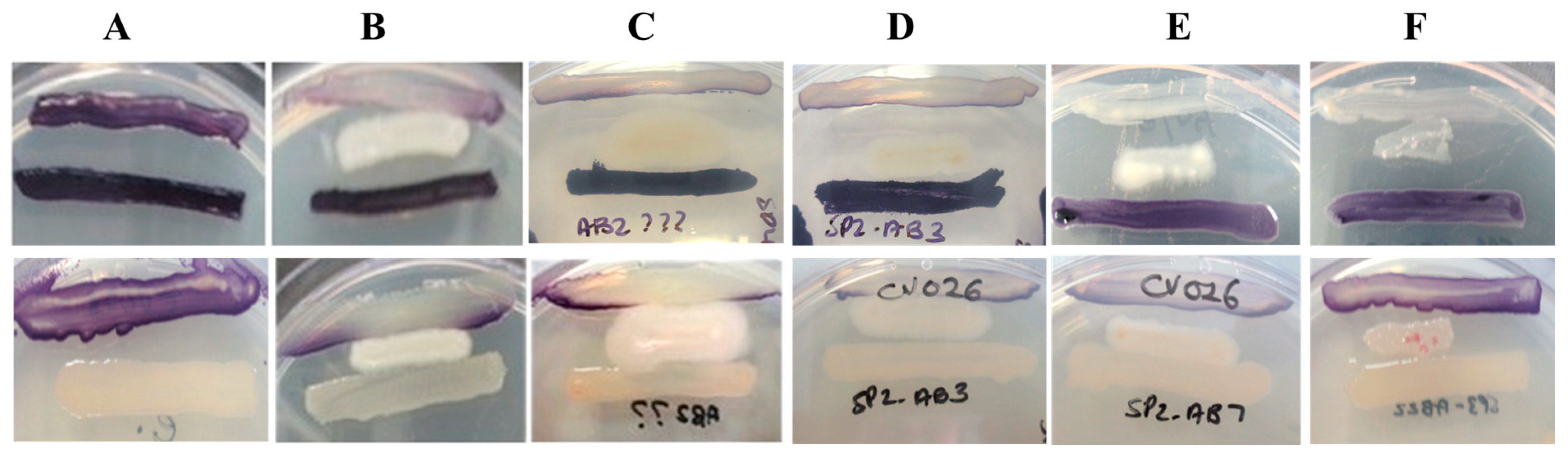
| Isolate Code | CV026 (Short-Chain AHLs) | VIR07 (Long-Chain AHLs) |
|---|---|---|
| Bacillus cereus ATCC 14579 | 4 | 4 |
| SP-AB2 (Bacillus sp.) | 4 | 4 |
| SP1-AB4 (Bacillus sp.) | 4 | 3 |
| SP1-V4 | 4 | 2 |
| SP2-AB1 | 3 | 0 |
| SP2-AB2 | 1 | 3 |
| SP2-AB3 | 4 | 4 |
| SP2-AB6 | 4 | 3 |
| SP2-AB7 (Bacillus sp.) | 3 | +4 |
| SP2-W3 | 1 | 2 |
| SP2-W6 (Bacillus sp.) | 2 | 2 |
| SP3-AB22 | 1 | +4 |
| SP4-AB2 | 3 | 4 |
| SP4-AB6 | 2 | 4 |
| SP5-AB7 (Bacillus sp.) | 1 | 4 |
| SP6-AB5 | 3 | 3 |
| Isolate Code | Nearest Type Strain (Accession Number) | Percentage Identity * | Sub-Group | Genbank Accession Number |
|---|---|---|---|---|
| SP-AB2 | Bacillus thuringiensis (NR_114581.1) | 99.85% | B. cereus | MH013306.1 |
| SP1-AB4 | Bacillus cereus (NR 115714.1) | 99.93% | B. cereus | MH013307.1 |
| SP2-AB7 | Bacillus mobilis (NR_157731.1) | 99.79% | B. cereus | MH013312.1 |
| SP2-W6 | Bacillus pumilus (NR_043242.1) | 99.05% | B. pumilus | MH013314.1 |
| SP5-AB7 | Bacillus wiedmannii (NR_152692.1) | 99.81% | B. cereus | MH013318.1 |
| Extract Code | Percentage Pyocyanin Inhibition | Percentage Pyoverdine Inhibition | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 250 µg/mL ** | 500 µg/mL ** | 750 µg/mL ** | 1000 µg/mL ** | 250 µg/mL ** | 500 µg/mL ** | 750 µg/mL ** | 1000 µg/mL ** | |||||||||
| Mann * | 5294 * | Mann ** | 5294 * | Mann * | 5294 ** | Mann ** | 5294 ** | Mann * | 5294 * | Mann ** | 5294 * | Mann * | 5294 ** | Mann ** | 5294 ** | |
| SP-AB2 | 17.80 | 17.99 | 23.02 | 26.58 | 29.57 | 28.24 | 32.63 | 36.21 | 12.05 | 9.47 | 26.19 | 14.61 | 23.13 | 18.52 | 38.43 | 26.89 |
| SP1-AB4 | 25.66 | 30.09 | 32.72 | 29.05 | 24.75 | 30.60 | 37.57 | 32.37 | 30.15 | 31.33 | 31.43 | 28.26 | 31.32 | 29.80 | 29.37 | 23.96 |
| SP2-AB7 | 23.10 | 19.83 | 28.31 | 17.58 | 25.09 | 16.81 | 31.54 | 19.70 | 10.55 | 23.82 | 6.39 | 19.02 | 21.45 | 18.87 | 18.02 | 30.92 |
| SP2-W6 | −0.73 # | 3.37 | 16.67 | 3.59 | 15.93 | 5.85 | 8.06 | 36.76 | 7.15 | 2.51 | 4.64 | 4.77 | 9.80 | 5.63 | 27.91 | 7.55 |
| SP5-AB7 | 30.46 | 18.44 | 35.33 | 21.40 | 39.36 | 23.69 | 32.69 | 30.97 | 8.72 | 31.56 | 2.69 | 34.86 | 3.90 | 33.40 | 8.17 | 29.33 |
| Cinnamaldehyde | 9.79 | 12.89 | 37.34 $ | 79.53 $ | −41.39 | −57.07 | −205.22 $ | −932.37 $ | ||||||||
| 10% DMSO | 17.43 | −6.13 # | 10.79 | 0.59 | 13.33 | 15.30 | 12.65 | 18.09 | ||||||||
| Extract Code | Percentage Elastase Inhibition | Percentage Protease Inhibition (Azocasein Assay) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 250 µg/mL * | 500 µg/mL ** | 1000 µg/mL ** | 250 µg/mL ** | 500 µg/mL ** | 750 µg/mL ** | 1000 µg/mL ** | ||||||||
| Mann ** | 5294 | Mann * | 5294 | Mann * | 5294 * | Mann | 5294 * | Mann | 5294 * | Mann * | 5294 ** | Mann * | 5294 ** | |
| SP-AB2 | 17.85 | −4.17 # | 26.04 | −0.93 # | 25.66 | 2.35 | 13.85 | 38.55 | 4.96 | 34.47 | 7.08 | 40.72 | 9.19 | 53.51 |
| SP1-AB4 | 16.63 | −3.5 # | 20.74 | 25.16 | 50.32 | 11.39 | 38.89 | 12.47 | 43.69 | 16.35 | 50.07 | 16.01 | 41.02 | 15.66 |
| SP2-AB7 | 30.92 | 21.63 | 38.75 | 40.17 | 51.76 | 46.01 | −1.88 # | 23.06 | 31.67 | 39.75 | 30.08 | 42.11 | 42.36 | 59.93 |
| SP2-W6 | 14.95 | −8.20 # | 43.08 | −1.26 # | 5.09 | 34.41 | 8.33 | 49.73 | 7.59 | 50.69 | 16.56 | 59.17 | 28.47 | 63.81 |
| SP5-AB7 | 15.18 | 12.43 | 19.40 | 35.46 | 71.32 | 43.86 | 35.05 | 9.30 | 31.28 | 15.06 | 34.36 | 43.22 | 38.77 | 46.90 |
| Cinnamaldehyde | 38.30 | 42.67 | 67.45 $ | 22.38 | 95.23 | 109.72 $ | 111.91 $ | |||||||
| 10% DMSO | 7.51 | 7.28 | 12.81 | 3.80 | 6.33 | 10.13 | 12.66 | |||||||
| Extract Code | Percentage Rhamnolipid Inhibition (Orcinol Assay) | |||||||
|---|---|---|---|---|---|---|---|---|
| 250 µg/mL | 500 µg/mL * | 750 µg/mL ** | 1000 µg/mL ** | |||||
| Mann | 5294 | Mann | 5294 ** | Mann | 5294 * | Mann | 5294 | |
| SP-AB2 | 5.54 | 11.92 | 14.18 | 44.65 | 21.08 | 74.22 | 21.30 | 14.69 |
| SP1-AB4 | 12.22 | 45.17 | 17.41 | 50.30 | 17.44 | 54.68 | 9.02 | 56.81 |
| SP2-AB7 | 86.03 | 47.53 | 79.47 | 50.95 | 80.71 | 51.27 | 85.73 | 51.49 |
| SP2-W6 | −40.52 # | −14.99 # | −34.96 # | 14.17 | 3.14 | 4.23 | 21.80 | −24.67 # |
| SP5-AB7 | 89.31 | 36.25 | 93.88 | 36.72 | 93.07 | 59.74 | 88.44 | 79.96 |
| Furanone | 26.96 | 23.90 | 24.37 | 28.90 | ||||
| 10% DMSO | 26.90 | 28.39 | 25.75 | 20.88 | ||||
| Isolate Code | Percentage Virulence Factor Inhibition at 1000 µg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pyocyanin ** | Pyoverdine ** | Elastase LasB ** | Protease (Casein Agar) | Protease (Azocasein) ** | Rhamnolipid (CTAB agar) | Rhamnolipid (Orcinol) ** | Swimming ** | Swarming ** | |
| SP-AB2 (Mann) | 32.63 | 38.43 | 25.66 * | 10.50 * | 9.19 * | 54.73 | 21.30 | 6.72 * | 13.29 * |
| SP-AB2 (5294) | 36.21 | 26.89 | 2.35 | 21.44 | 53.51 | 39.64 * | 14.69 * | 37.35 | 42.41 * |
| SP1-AB4 (Mann) | 37.57 | 29.37 * | 50.32 | 30.39 | 41.02 * | 16.71 * | 9.02 * | 37.35 | 5.7 * |
| SP1-AB4 (5294) | 32.37 | 23.96 * | 11.39 * | 14.15 * | 15.66 * | 42.06 | 56.81 | 18.28 * | 51.9 * |
| SP2-AB7 (Mann) | 31.54 | 18.02 * | 51.76 | 18.46 * | 42.36 | 46.89 | 85.73 | 18.28 * | 8.86 * |
| SP2-AB7 (5294) | 19.70 | 30.92 | 46.01 | 17.96 | 59.93 | 39.64 * | 51.49 | 37.94 * | 29.11 * |
| SP2-W6 (Mann) | 8.06 * | 27.91 | 5.09 * | 24.42 * | 28.47 | 9.46 * | 21.80 | 37.94 * | 11.39 * |
| SP2-W6 (5294) | 36.76 | 7.55 | 34.41 | 22.93 * | 63.81 | 10.67 * | −24.67 * | 28.24 * | 45.57 |
| SP5-AB7 (Mann) | 32.69 * | 8.17 | 71.32 | 21.44 * | 38.77 | 67.41 * | 88.44 * | 28.24 * | 29.11 |
| SP5-AB7 (5294) | 30.97 | 29.33 * | 43.86 | 25.91 | 46.90 | 42.06 * | 79.96 | 50.59 * | 24.05 |
| Cinnamaldehyde | 79.53 $ | −932.37 $ | 67.45 $ | 54.26 $ | 111.91 $ | 54.13 | 28.90 # | 44.37 $ | −8.23 $ |
| 10% DMSO | 0.59 | 18.09 | 12.81 | 6.52 | 12.66 | −1.40 | 20.88 | 20.91 | −49.37 |
| Extract Code | Percentage Initial Adhesion Inhibition (%BFR) | Percentage Mature Biofilm Inhibition (%BFR) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.5 mg/mL | 1 mg/mL * | 5 mg/mL ** | 10 mg/mL ** | 0.5 mg/mL | 1 mg/mL * | 5 mg/mL ** | 10 mg/mL ** | |||||||||
| Mann | 5294 * | Mann | 5294 ** | Mann | 5294 | Mann | 5294 | Mann | 5294 ** | Mann * | 5294 ** | Mann | 5294 ** | Mann | 5294 ** | |
| SP-AB2 | −17.73 # | 3.50 | −17.92 # | 13.82 | 78.77 $ | 20.80 | 88.77 $ | 40.96 | 35.96 | 11.78 | 40.43 | 12.11 | 40.67 | 12.29 | 86.28 | 12.33 |
| SP1-AB4 | 2.84 | 6.97 | 31.13 | 16.54 | 58.89 | 69.39 | 73.58 | 81.41 $ | 12.04 | 11.69 | 12.25 | 14.53 | 12.34 | 15.34 | 13.26 | 17.82 |
| SP2-AB7 | 1.27 | 4.99 | 2.94 | 4.03 | 2.93 | 2.78 | 8.32 | 4.35 | 7.49 | 11.18 | 10.87 | 11.21 | 11.14 | 12.48 | 13.81 | 12.55 |
| SP2-W6 | −2.97 # | 11.09 | −3.16 # | 19.39 | −3.22 # | 41.11 | −2.36 # | 41.84 | 10.06 | 12.56 | 9.75 | 13.63 | 7.53 | 14.24 | 7.97 | 14.74 |
| SP5-AB7 | 21.26 | 19.54 | 31.43 | 17.59 | 66.51 | 14.47 | 71.70 $ | 1.13 | 8.57 | 11.26 | 9.86 | 11.71 | 10.20 | 12.20 | 15.71 | 12.54 |
| Cinnamaldehyde | −22.50 # | −21.12 # | 70.25 $ | 70.19 $ | 38.85 | 38.55 | 45.39 | 46.43 $ | ||||||||
| 10% DMSO | −23.36 # | −21.12 # | −18.47 # | −18.52 # | 14.63 | 15.48 | 18.16 | 21.03 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacobs, C.S.; Naicker, R.; Chenia, H.Y. Quorum Sensing Inhibition by Sponge-Associated Bacillus Species: Suppressing Pseudomonas aeruginosa Virulence Factors. Antibiotics 2025, 14, 1035. https://doi.org/10.3390/antibiotics14101035
Jacobs CS, Naicker R, Chenia HY. Quorum Sensing Inhibition by Sponge-Associated Bacillus Species: Suppressing Pseudomonas aeruginosa Virulence Factors. Antibiotics. 2025; 14(10):1035. https://doi.org/10.3390/antibiotics14101035
Chicago/Turabian StyleJacobs, Carrie Shelouise, Ryan Naicker, and Hafizah Yousuf Chenia. 2025. "Quorum Sensing Inhibition by Sponge-Associated Bacillus Species: Suppressing Pseudomonas aeruginosa Virulence Factors" Antibiotics 14, no. 10: 1035. https://doi.org/10.3390/antibiotics14101035
APA StyleJacobs, C. S., Naicker, R., & Chenia, H. Y. (2025). Quorum Sensing Inhibition by Sponge-Associated Bacillus Species: Suppressing Pseudomonas aeruginosa Virulence Factors. Antibiotics, 14(10), 1035. https://doi.org/10.3390/antibiotics14101035






