Selective Oral Decontamination of the Esophagus to Reduce Microbial Burden in Patients Undergoing Esophagectomy for Esophageal Cancer (SODA)—First Results from a Proof-of-Principle Study
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
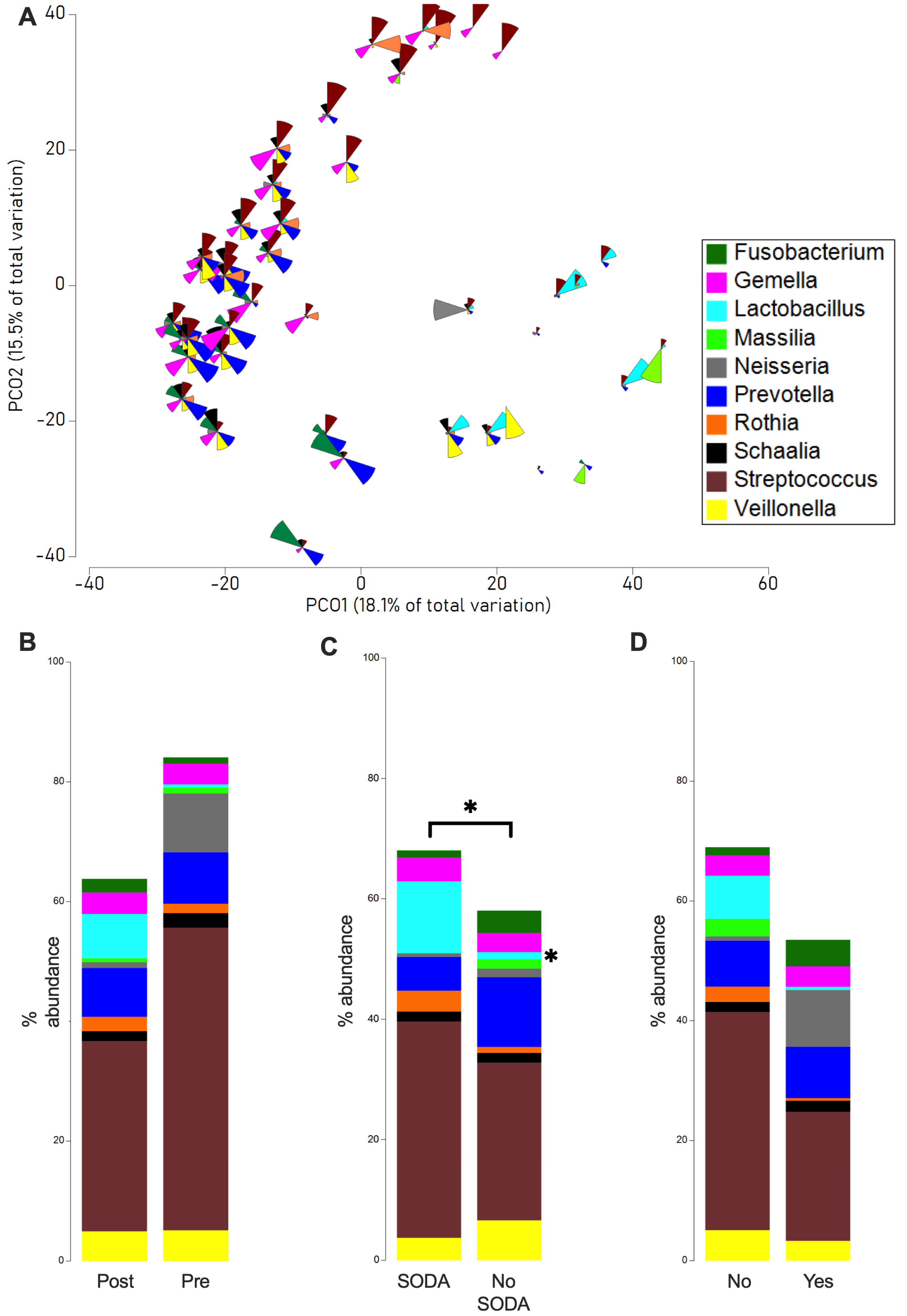
2.2. Microbiome Analysis
3. Discussion
4. Materials and Methods
4.1. Patient Enrolment
4.2. Treatment
4.3. Microbiome Analysis: DNA Extraction, Sequencing, and Bioinformatics
4.4. Statistical Analysis
4.5. Trial Registration
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Porschen, R.; Fischbach, W.; Gockel, I.; Hollerbach, S.; Hölscher, A.; Jansen, P.L.; Miehlke, S.; Pech, O.; Stahl, M.; Thuss-Patience, P.; et al. S3-Leitlinie—Diagnostik und Therapie der Plattenepithelkarzinome und Adenokarzinome des Ösophagus. Z. Fur Gastroenterol. 2019, 57, 336–418. [Google Scholar] [CrossRef] [PubMed]
- Schlottmann, F.; Strassle, P.D.; Patti, M.G. Transhiatal vs. Transthoracic Esophagectomy: A NSQIP Analysis of Postoperative Outcomes and Risk Factors for Morbidity. J. Gastrointest. Surg. 2017, 21, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Low, D.E.; Kuppusamy, M.K.; Alderson, D.; Cecconello, I.; Chang, A.C.; Darling, G.; Davies, A.; D’Journo, X.B.; Gisbertz, S.S.; Griffin, S.M.; et al. Benchmarking Complications Associated with Esophagectomy. Ann. Surg. 2019, 269, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Markar, S.; Gronnier, C.; Duhamel, A.; Bigourdan, J.-M.; Badic, B.; du Rieu, M.C.; Lefevre, J.H.; Turner, K.; Luc, G.; Mariette, C. Pattern of Postoperative Mortality After Esophageal Cancer Resection According to Center Volume: Results from a Large European Multicenter Study. Ann. Surg. Oncol. 2015, 22, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.F.; Atkins, B.Z.; Tong, B.C.; Harpole, D.H.; D’Amico, T.A.; Onaitis, M.W. A comprehensive evaluation for aspiration after esophagectomy reduces the incidence of postoperative pneumonia. J. Thorac. Cardiovasc. Surg. 2010, 140, 1266–1271. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Goense, L.; van Dijk, W.A.; Govaert, J.A.; van Rossum, P.S.; Ruurda, J.P.; van Hillegersberg, R. Hospital costs of complications after esophagectomy for cancer. Eur. J. Surg. Oncol. 2017, 43, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Fabbi, M.; Hagens, E.R.C.; van Berge Henegouwen, M.I.; Gisbertz, S.S. Anastomotic leakage after esophagectomy for esophageal cancer: Definitions, diagnostics, and treatment. Dis. Esophagus 2021, 34, doaa039. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hagens, E.R.C.; Reijntjes, M.A.; Anderegg, M.C.J.; Eshuis, W.J.; van Berge Henegouwen, M.I.; Gisbertz, S.S. Risk Factors and Consequences of Anastomotic Leakage After Esophagectomy for Cancer. Ann. Thorac. Surg. 2021, 112, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, U.; Baiocchi, G.L.; Celotti, A.; Parise, P.; Cossu, A.; Bonavina, L.; Bernardi, D.; de Manzoni, G.; Weindelmayer, J.; Verlato, G.; et al. Incidence and treatment of mediastinal leakage after esophagectomy: Insights from the multicenter study on mediastinal leaks. World J. Gastroenterol. 2019, 25, 356–366. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Derogar, M.; Orsini, N.; Sadr-Azodi, O.; Lagergren, P. Influence of major postoperative complications on health-related quality of life among long-term survivors of esophageal cancer surgery. J. Clin. Oncol. 2012, 30, 1615–1619. [Google Scholar] [CrossRef] [PubMed]
- Kofoed, S.C.; Calatayud, D.; Jensen, L.S.; Helgstrand, F.; Achiam, M.P.; De Heer, P.; Svendsen, L.B.; Esophageal, D. Intrathoracic anastomotic leakage after gastroesophageal cancer resection is associated with increased risk of recurrence. J. Thorac. Cardiovasc. Surg. 2015, 150, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Guyton, K.; Alverdy, J.C. The gut microbiota and gastrointestinal surgery. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Vilchez-Vargas, R.; Skieceviciene, J.; Lehr, K.; Varkalaite, G.; Thon, C.; Urba, M.; Morkūnas, E.; Kucinskas, L.; Bauraite, K.; Schanze, D.; et al. Gut microbial similarity in twins is driven by shared environment and aging. EBioMedicine 2022, 79, 104011. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fransen, L.F.C.; A Verhoeven, R.H.; Janssen, T.H.J.B.; van Det, M.J.; Gisbertz, S.S.; van Hillegersberg, R.; Klarenbeek, B.; A Kouwenhoven, E.; Nieuwenhuijzen, G.A.P.; Rosman, C.; et al. The association between postoperative complications and long-term survival after esophagectomy: A multicenter cohort study. Dis. Esophagus 2023, 36, doac086. [Google Scholar] [CrossRef] [PubMed]
- Kooij, C.D.; de Jongh, C.; Kingma, B.F.; Henegouwen, M.I.v.B.; Gisbertz, S.S.; Chao, Y.-K.; Chiu, P.W.; Rouanet, P.; Mourregot, A.; Immanuel, A.; et al. The Current State of Robot-Assisted Minimally Invasive Esophagectomy (RAMIE): Outcomes from the Upper GI International Robotic Association (UGIRA) Esophageal Registry. Ann. Surg. Oncol. 2025, 32, 823–833. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kingma, B.F.; Grimminger, P.P.; van der Sluis, P.C.; van Det, M.J.; Kouwenhoven, E.A.; Chao, Y.-K.; Tsai, C.-Y.; Fuchs, H.F.; Bruns, C.J.; Sarkaria, I.S.; et al. Worldwide Techniques and Outcomes in Robot-assisted Minimally Invasive Esophagectomy (RAMIE): Results From the Multicenter International Registry. Ann Surg. 2022, 276, e386–e392. [Google Scholar] [CrossRef] [PubMed]
- van der Sluis, P.C.; Babic, B.; Uzun, E.; Tagkalos, E.; Berlth, F.; Hadzijusufovic, E.; Lang, H.; Gockel, I.; van Hillegersberg, R.; Grimminger, P.P. Robot-assisted and conventional minimally invasive esophagectomy are associated with better postoperative results compared to hybrid and open transthoracic esophagectomy. Eur. J. Surg. Oncol. 2022, 48, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Roos, D.; Dijksman, L.M.; Tijssen, J.G.; Gouma, D.J.; Gerhards, M.F.; Oudemans-van Straaten, H.M. Systematic review of perioperative selective decontamination of the digestive tract in elective gastrointestinal surgery. Br. J. Surg. 2013, 100, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Scholten, J.; Reuvers, J.R.D.; Stockmann, H.B.A.C.; van Stralen, K.J.; van Egmond, M.; Bonjer, H.J.; Kazemier, G.; Abis, G.S.A.; Oosterling, S.J.; Acherman, Y.; et al. Selective Decontamination with Oral Antibiotics in Colorectal Surgery: 90-day Reintervention Rates and Long-Term Oncological Follow-Up. J. Gastrointest. Surg. 2023, 27, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Janssen, R.; Van Workum, F.; Baranov, N.; Blok, H.; Oever, J.T.; Kolwijck, E.; Tostmann, A.; Rosman, C.; Schouten, J. Selective Decontamination of the Digestive Tract to Prevent Postoperative Pneumonia and Anastomotic Leakage after Esophagectomy: A Retrospective Cohort Study. Antibiotics 2021, 10, 43. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Merboth, F.; Hasanovic, J.; Stange, D.; Distler, M.; Kaden, S.; Weitz, J.; Welsch, T. Strategiewechsel zur minimal-invasiven Ösophagektomie—Ergebnisse an einem zertifizierten Zentrum. Die Chir. 2022, 93, 694–701. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Näf, F.; Warschkow, R.; Kolb, W.; Zünd, M.; Lange, J.; Steffen, T. Selective decontamination of the gastrointestinal tract in patients undergoing esophageal resection. BMC Surg. 2010, 10, 36. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van Kasteren, M.E.; Gyssens, I.C.; Kullberg, B.J.; Bruining, H.A.; Stobberingh, E.E.; Goris, R.J. [Optimizing antibiotics policy in the Netherlands. V. SWAB guidelines for perioperative antibiotic prophylaxis. Foundation Antibiotics Policy Team]. Ned. Tijdschr. Geneeskd. 2000, 144, 2049–2055. [Google Scholar] [PubMed]
- de Jonge, E. Effects of selective decontamination of digestive tract on mortality and antibiotic resistance in the intensive-care unit. Curr. Opin. Crit. Care 2005, 11, 144–149. [Google Scholar] [CrossRef] [PubMed]
- de Smet, A.M.; Kluytmans, J.A.; Cooper, B.S.; Mascini, E.M.; Benus, R.F.; van der Werf, T.S.; van der Hoeven, J.G.; Pickkers, P.; Bogaers-Hofman, D.; van der Meer, N.J.; et al. Decontamination of the digestive tract and oropharynx in ICU patients. N. Engl. J. Med. 2009, 360, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Oostdijk, E.A.; de Wit, G.A.; Bakker, M.; de Smet, A.M.; Bonten, M.J. Selective decontamination of the digestive tract and selective oropharyngeal decontamination in intensive care unit patients: A cost-effectiveness analysis. BMJ Open 2013, 3, e002529. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tetteroo, G.W.; Wagenvoort, J.H.; Castelein, A.; Tilanus, H.W.; Ince, C.; Bruining, H.A. Selective decontamination to reduce gram-negative colonisation and infections after oesophageal resection. Lancet 1990, 335, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Erichsen, T.J.; Vonberg, R.-P.; Lenzen, H.; Negm, A.A.; Helfritz, F.A.; Emmanouilidis, N.; Manns, M.P.; Wedemeyer, J.; Suerbaum, S.; Lankisch, T.O. Microbiological analysis of fluids in postsurgical gastroesophageal intrathoracic leaks obtained by endoscopy: A new way to optimize antibiotic therapy. Digestion 2015, 91, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Ledda, A.L.; Tarantino, I.; Schiefer, S.; Ronellenfitsch, U.; Rebelo, A.; Sekulla, C.; Nienhüser, H.; Michalski, C.; Schmied, B.; Kleeff, J.; et al. Patterns of infectious complications and their implication on health system costs after esophagectomy for esophageal cancer: Real-world data from three European centers. Langenbeck’s Arch Surg. 2025, 410, 138. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lehr, K.; Lange, U.G.; Hipler, N.M.; Vilchez-Vargas, R.; Hoffmeister, A.; Feisthammel, J.; Buchloh, D.; Schanze, D.; Zenker, M.; Gockel, I.; et al. Prediction of anastomotic insufficiency based on the mucosal microbiome prior to colorectal surgery: A proof-of-principle study. Sci. Rep. 2024, 14, 15335. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guccione, C.; Yadlapati, R.; Shah, S.; Knight, R.; Curtius, K. Challenges in Determining the Role of Microbiome Evolution in Barrett’s Esophagus and Progression to Esophageal Adenocarcinoma. Microorganisms 2021, 9, 2003. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shijimaya, T.; Tahara, T.; Yamazaki, J.; Kobayashi, S.; Matsumoto, Y.; Nakamura, N.; Takahashi, Y.; Tomiyama, T.; Fukui, T.; Shibata, T.; et al. Microbiome of esophageal endoscopic wash samples is associated with resident flora in the esophagus and incidence of cancer. Sci. Rep. 2024, 14, 19525. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yano, Y.; Etemadi, A.; Abnet, C.C. Microbiome and Cancers of the Esophagus: A Review. Microorganisms 2021, 9, 1764. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- World Medical Association Declaration of Helsinki. Ethical principles for medical research involving human subjects. Bull. World Health Organ. 2001, 79, 373–374. [Google Scholar] [PubMed] [PubMed Central]
- Grimminger, P.P.; van der Horst, S.; Ruurda, J.P.; van Det, M.; Morel, P.; van Hillegersberg, R. Surgical robotics for esophageal cancer. Ann. N. Y. Acad. Sci. 2018, 1434, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Camarinha-Silva, A.; Jáuregui, R.; Chaves-Moreno, D.; Oxley, A.P.; Schaumburg, F.; Becker, K.; Wos-Oxley, M.L.; Pieper, D.H. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ. Microbiol. 2014, 16, 2939–2952. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maidak, B.L.; Olsen, G.J.; Larsen, N.; Overbeek, R.; McCaughey, M.J.; Woese, C.R. The RDP (Ribosomal Database Project). Nucleic Acids Res. 1997, 25, 109–111. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| Patient Characteristics | % | % | p-Value | ||
|---|---|---|---|---|---|
| SODA (n = 13) | No SODA (n = 9) | ||||
| Age (years, mean, IQR) | 62 (54–74) | 68 (39–83) | 0.135 | ||
| Gender | 0.667 | ||||
| Male | 12 | 92.3 | 8 | 88.9 | |
| Female | 1 | 7.7 | 1 | 11.1 | |
| ASA Score | 0.942 | ||||
| II | 2 | 15.4 | 2 | 22.2 | |
| III | 11 | 84.6 | 7 | 77.8 | |
| BMI (kg/m2, mean, IQR) | 25.3 (17.0–36.3) | 25.4 (22.1–30.1) | 0.93 | ||
| Reflux disease | 0.27 | ||||
| Yes | 3 | 23.1 | 4 | 44.4 | |
| No | 10 | 76.9 | 5 | 55.6 | |
| Barrett dysplasia | 0.35 | ||||
| Yes | 5 | 38.5 | 1 | 11.1 | |
| No | 8 | 61.5 | 8 | 88.9 | |
| Smoking | 0.032 | ||||
| Yes | 10 | 76.9 | 3 | 33.3 | |
| No | 3 | 23.1 | 6 | 66.7 | |
| Alcohol consumption | 0.497 | ||||
| Yes | 6 | 46.2 | 2 | 22.2 | |
| No | 7 | 53.8 | 7 | 77.8 | |
| Histology | 0.398 | ||||
| Squamous-cell carcinoma | 5 | 38.5 | 2 | 22.2 | |
| Adenocarcinoma | 8 | 61.5 | 7 | 77.8 | |
| UICC Stage | 0.791 | ||||
| II | 3 | 23.1 | 2 | 22.2 | |
| IIIa | 3 | 23.1 | 4 | 44.4 | |
| IIIb | 7 | 53.8 | 3 | 33.3 | |
| Tumor location | 0.599 | ||||
| Mid | 1 | 7.7 | 1 | 11.1 | |
| Distal | 12 | 92.3 | 8 | 88.9 | |
| Neoadjuvant therapy | 0.87 | ||||
| Yes | 10 | 76.9 | 1 | 11.1 | |
| No | 3 | 23.1 | 8 | 88.9 | |
| Duration of surgery in minutes (mean, IQR) | 411 (260–532) | 422 (373–480) | 0.803 | ||
| No. harvested lymph nodes (IQR) | 25 (11–38) | 23 (13–28) | 0.728 | ||
| Pneumonia | 0.035 | ||||
| Yes | 0 | 0.0 | 3 | 33.3 | |
| No | 13 | 100.0 | 6 | 66.7 | |
| Anastomotic leakage | 0.13 | ||||
| Yes | 1 | 7.7 | 2 | 22.2 | |
| No | 12 | 92.3 | 7 | 77.8 | |
| Reoperation | 0.436 | ||||
| Yes | 2 | 15.4 | 1 | 11.1 | |
| No | 11 | 84.6 | 8 | 88.9 | |
| Re-intubation | 0.09 | ||||
| Yes | 1 | 7.7 | 2 | 22.2 | |
| No | 12 | 92.3 | 7 | 77.8 | |
| Length of hospital stay in days (mean, IQR) | 45 (9–92) | 58 (15–85) | 0.127 | ||
| Length of ICU stay in days (mean, IQR) | 6 (1–83) | 12 (1–53) | 0.119 | ||
| ICU readmission | 1 | 7.7 | 2 | 22.2 | 0.291 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klose, J.; Lehr, K.; Ronellenfitsch, U.; Klose, M.A.; Ebert, D.; Rebelo, A.; Link, A.; Kleeff, J. Selective Oral Decontamination of the Esophagus to Reduce Microbial Burden in Patients Undergoing Esophagectomy for Esophageal Cancer (SODA)—First Results from a Proof-of-Principle Study. Antibiotics 2025, 14, 1033. https://doi.org/10.3390/antibiotics14101033
Klose J, Lehr K, Ronellenfitsch U, Klose MA, Ebert D, Rebelo A, Link A, Kleeff J. Selective Oral Decontamination of the Esophagus to Reduce Microbial Burden in Patients Undergoing Esophagectomy for Esophageal Cancer (SODA)—First Results from a Proof-of-Principle Study. Antibiotics. 2025; 14(10):1033. https://doi.org/10.3390/antibiotics14101033
Chicago/Turabian StyleKlose, Johannes, Konrad Lehr, Ulrich Ronellenfitsch, Michelle A. Klose, Daniel Ebert, Artur Rebelo, Alexander Link, and Jörg Kleeff. 2025. "Selective Oral Decontamination of the Esophagus to Reduce Microbial Burden in Patients Undergoing Esophagectomy for Esophageal Cancer (SODA)—First Results from a Proof-of-Principle Study" Antibiotics 14, no. 10: 1033. https://doi.org/10.3390/antibiotics14101033
APA StyleKlose, J., Lehr, K., Ronellenfitsch, U., Klose, M. A., Ebert, D., Rebelo, A., Link, A., & Kleeff, J. (2025). Selective Oral Decontamination of the Esophagus to Reduce Microbial Burden in Patients Undergoing Esophagectomy for Esophageal Cancer (SODA)—First Results from a Proof-of-Principle Study. Antibiotics, 14(10), 1033. https://doi.org/10.3390/antibiotics14101033









