ISApl4, a New IS1595 Family Insertion Sequence Forming a Novel Pseudo-Compound Transposon That Confers Antimicrobial Multidrug Resistance in Actinobacillus pleuropneumoniae
Abstract
1. Introduction
2. Results
2.1. Identification of ISApl4
2.2. Analysis of the Tn7560 Sequence
2.3. Single Nucleotide Polymorphism (SNP) Analysis
2.4. Confirmation of ISApl4 Activity
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates and Antimicrobial Susceptibility Testing
4.2. Genome Analysis and Identification of ISApl4 Insertions
4.3. Generation of a Closed Genome of AP_123
4.4. SNP Analysis of the ISApl4-Containing Isolates
4.5. Analysis of ISApl4 Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| CLSI | Clinical and Laboratory Standards Institute |
| DR | Direct repeat |
| gDNA | Genomic DNA |
| ICE | Integrative and conjugative element |
| IR | Inverted repeat |
| IS | Insertion sequence |
| LPS | Lipopolysaccharide |
| MGE | Mobile genetic element |
| MDR | Multidrug resistance |
| MIC | Minimal inhibitory concentration |
| NAD | Nicotinamide adenine dinucleotide |
| PCR | Polymerase chain reaction |
| RT-PCR | Reverse transcription polymerase chain reaction |
| SNP | Single nucleotide polymorphism |
| TU | Translocatable unit |
| UK | United Kingdom |
| USA | United States of America |
References
- Michael, G.B.; Bossé, J.T.; Schwarz, S. Antimicrobial Resistance in Pasteurellaceae of Veterinary Origin. Microbiol. Spectr. 2018, 6, ARBA-0022-2017. [Google Scholar] [CrossRef]
- White, A.; Hughes, J.M. Critical Importance of a One Health Approach to Antimicrobial Resistance. EcoHealth 2019, 16, 404–409. [Google Scholar] [CrossRef]
- da Silva, G.C.; Gonçalves, O.S.; Rosa, J.N.; França, K.C.; Bossé, J.T.; Santana, M.F.; Langford, P.R.; Bazzolli, D.M.S. Mobile Genetic Elements Drive Antimicrobial Resistance Gene Spread in Pasteurellaceae Species. Front. Microbiol. 2021, 12, 773284. [Google Scholar] [CrossRef]
- Sassu, E.L.; Bossé, J.T.; Tobias, T.J.; Gottschalk, M.; Langford, P.R.; Hennig-Pauka, I. Update on Actinobacillus pleuropneumoniae-knowledge, gaps and challenges. Transbound. Emerg. Dis. 2018, 65 (Suppl. 1), 72–90. [Google Scholar] [CrossRef]
- Loera-Muro, A.; Angulo, C. New trends in innovative vaccine development against Actinobacillus pleuropneumoniae. Vet. Microbiol. 2018, 217, 66–75. [Google Scholar] [CrossRef]
- Bossé, J.T.; Li, Y.; Rogers, J.; Fernandez Crespo, R.; Li, Y.; Chaudhuri, R.R.; Holden, M.T.; Maskell, D.J.; Tucker, A.W.; Wren, B.W.; et al. Whole Genome Sequencing for Surveillance of Antimicrobial Resistance in Actinobacillus pleuropneumoniae. Front. Microbiol. 2017, 8, 311. [Google Scholar] [CrossRef] [PubMed]
- Vilaró, A.; Novell, E.; Enrique-Tarancón, V.; Balielles, J.; Vilalta, C.; Martinez, S.; Fraile Sauce, L.J. Antimicrobial Susceptibility Pattern of Porcine Respiratory Bacteria in Spain. Antibiotics 2020, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.M.; Bossé, J.T.; Stegger, M.; Li, Y.; Langford, P.R.; Kielland, C.; Klem, T.B.; Gulliksen, S.M.; Ranheim, B.; Grøntvedt, C.A.; et al. Comparative Genome Sequence Analysis of Actinobacillus pleuropneumoniae Serovar 8 Isolates From Norway, Denmark, and the United Kingdom Indicates Distinct Phylogenetic Lineages and Differences in Distribution of Antimicrobial Resistance Genes. Front. Microbiol. 2021, 12, 729637. [Google Scholar] [CrossRef] [PubMed]
- Holmer, I.; Salomonsen, C.M.; Jorsal, S.E.; Astrup, L.B.; Jensen, V.F.; Høg, B.B.; Pedersen, K. Antibiotic resistance in porcine pathogenic bacteria and relation to antibiotic usage. BMC Vet. Res. 2019, 15, 449. [Google Scholar] [CrossRef]
- Krüger-Haker, H.; Kostova, V.; Hanke, D.; Kaspar, H.; Fiedler, S.; Schwarz, S. Genetic basis of macrolide resistance in porcine Pasteurella multocida isolates from the German national resistance monitoring program GERM-Vet 2008–2021. J. Antimicrob. Chemother. 2024, 79, 2975–2979. [Google Scholar] [CrossRef]
- Jahnen, J.; Hanke, D.; Kadlec, K.; Schwarz, S.; Krüger-Haker, H. Antimicrobial Resistance in Pasteurella multocida Isolates from Bovine Mastitis Can Be Associated with Multidrug-Resistance-Mediating Integrative and Conjugative Elements (ICEs). Antibiotics 2025, 14, 153. [Google Scholar] [CrossRef]
- Kostova, V.; Hanke, D.; Kaspar, H.; Fiedler, S.; Schwarz, S.; Krüger-Haker, H. Macrolide resistance in Mannheimia haemolytica isolates associated with bovine respiratory disease from the German national resistance monitoring program GERM-Vet 2009 to 2020. Front. Microbiol. 2024, 15, 1356208. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef]
- Consuegra, J.; Gaffé, J.; Lenski, R.E.; Hindré, T.; Barrick, J.E.; Tenaillon, O.; Schneider, D. Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria. Nat. Commun. 2021, 12, 980. [Google Scholar] [CrossRef]
- Wu, Y.; Aandahl, R.Z.; Tanaka, M.M. Dynamics of bacterial insertion sequences: Can transposition bursts help the elements persist? BMC Evol. Biol. 2015, 15, 288. [Google Scholar] [CrossRef]
- Wagner, A. Cooperation is fleeting in the world of transposable elements. PLoS Comput. Biol. 2006, 2, e162. [Google Scholar] [CrossRef]
- Harmer, C.J.; Pong, C.H.; Hall, R.M. Structures bounded by directly-oriented members of the IS26 family are pseudo-compound transposons. Plasmid 2020, 111, 102530. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yang, W.; Schwarz, S.; Xu, Q.; Yang, Q.; Wang, L.; Liu, S.; Zhang, W. Characterization of the novel optrA-carrying pseudo-compound transposon Tn7363 and an Inc18 plasmid carrying cfr(D) in Vagococcus lutrae. J. Antimicrob. Chemother. 2022, 77, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhu, Y.; Schwarz, S.; Wang, L.; Liu, W.; Yang, W.; Liu, S.; Zhang, W. Integrative and conjugative elements in streptococci can act as vectors for plasmids and translocatable units integrated via IS1216E. Int. J. Antimicrob. Agents 2023, 61, 106793. [Google Scholar] [CrossRef] [PubMed]
- Tegetmeyer, H.E.; Jones, S.C.; Langford, P.R.; Baltes, N. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet. Microbiol. 2008, 128, 342–353. [Google Scholar] [CrossRef]
- Siguier, P.; Gagnevin, L.; Chandler, M. The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. Res. Microbiol. 2009, 160, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Varani, A.; Perochon, J.; Chandler, M. Exploring bacterial insertion sequences with ISfinder: Objectives, uses, and future developments. Methods Mol. Biol. 2012, 859, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Price, N.L.; Raivio, T.L. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 2009, 191, 1798–1815. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Wang, H.; Lv, Z.; Hu, Y.; Wang, H.; Shu, F.; Zhu, C.; Xue, T. The Two-Component System CpxRA Affects Antibiotic Susceptibility and Biofilm Formation in Avian Pathogenic Escherichia coli. Animals 2023, 13, 383. [Google Scholar] [CrossRef]
- Matter, L.B.; Ares, M.A.; Abundes-Gallegos, J.; Cedillo, M.L.; Yáñez, J.A.; Martínez-Laguna, Y.; De la Cruz, M.A.; Girón, J.A. The CpxRA stress response system regulates virulence features of avian pathogenic Escherichia coli. Environ. Microbiol. 2018, 20, 3363–3377. [Google Scholar] [CrossRef]
- Shetty, D.; Abrahante, J.E.; Chekabab, S.M.; Wu, X.; Korber, D.R.; Vidovic, S. Role of CpxR in Biofilm Development: Expression of Key Fimbrial, O-Antigen and Virulence Operons of Salmonella Enteritidis. Int. J. Mol. Sci. 2019, 20, 5146. [Google Scholar] [CrossRef]
- Schink, A.-K.; Hanke, D.; Semmler, T.; Brombach, J.; Bethe, A.; Lübke-Becker, A.; Teske, K.; Müller, K.E.; Schwarz, S. Novel multiresistance-mediating integrative and conjugative elements carrying unusual antimicrobial resistance genes in Mannheimia haemolytica and Pasteurella multocida. J. Antimicrob. Chemother. 2022, 77, 2033–2035. [Google Scholar] [CrossRef]
- Kyselková, M.; Chrudimský, T.; Husník, F.; Chroňáková, A.; Heuer, H.; Smalla, K.; Elhottová, D. Characterization of tet(Y)-carrying LowGC plasmids exogenously captured from cow manure at a conventional dairy farm. FEMS Microbiol. Ecol. 2016, 92, fiw075. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Schwarz, S. Occurrence and linkage of genes coding for resistance to sulfonamides, streptomycin and chloramphenicol in bacteria of the genera Pasteurella and Mannheimia. FEMS Microbiol. Lett. 2001, 205, 283–290. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; CLSI Supplement VET01S 7th ed; CLSI: Wayne, PA, USA, 2024. [Google Scholar]
- Kehrenberg, C.; Catry, B.; Haesebrouck, F.; de Kruif, A.; Schwarz, S. tet(L)-mediated tetracycline resistance in bovine Mannheimia and Pasteurella isolates. J. Antimicrob. Chemother. 2005, 56, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Bossé, J.T.; Chaudhuri, R.R.; Li, Y.; Leanse, L.G.; Fernandez Crespo, R.; Coupland, P.; Holden, M.T.; Bazzolli, D.M.; Maskell, D.J.; Tucker, A.W.; et al. Complete Genome Sequence of MIDG2331, a Genetically Tractable Serovar 8 Clinical Isolate of Actinobacillus pleuropneumoniae. Genome Announc. 2016, 4, e01667-15. [Google Scholar] [CrossRef]
- Cheng, Y.; Chen, Y.; Liu, Y.; Song, J.; Chen, Y.; Shan, T.; Xiao, Y.; Zhou, K. Detection of a new tet(X6)-encoding plasmid in Acinetobacter towneri. J. Glob. Antimicrob. Resist. 2021, 25, 132–136. [Google Scholar] [CrossRef]
- Alaimo, C.; Catrein, I.; Morf, L.; Marolda, C.L.; Callewaert, N.; Valvano, M.A.; Feldman, M.F.; Aebi, M. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 2006, 25, 967–976. [Google Scholar] [CrossRef]
- Marolda, C.L.; Tatar, L.D.; Alaimo, C.; Aebi, M.; Valvano, M.A. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide o antigen. J. Bacteriol. 2006, 188, 5124–5135. [Google Scholar] [CrossRef]
- Franco, A.V.; Liu, D.; Reeves, P.R. A Wzz (Cld) protein determines the chain length of K lipopolysaccharide in Escherichia coli O8 and O9 strains. J. Bacteriol. 1996, 178, 1903–1907. [Google Scholar] [CrossRef]
- Murray, G.L.; Attridge, S.R.; Morona, R. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 2003, 47, 1395–1406. [Google Scholar] [CrossRef]
- Kachlany, S.C.; Planet, P.J.; Bhattacharjee, M.K.; Kollia, E.; DeSalle, R.; Fine, D.H.; Figurski, D.H. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 2000, 182, 6169–6176. [Google Scholar] [CrossRef]
- Bhattacharjee, M.K.; Kachlany, S.C.; Fine, D.H.; Figurski, D.H. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 2001, 183, 5927–5936. [Google Scholar] [CrossRef] [PubMed]
- Au, K.G.; Cabrera, M.; Miller, J.H.; Modrich, P. Escherichia coli mutY gene product is required for specific A-G----C.G mismatch correction. Proc. Natl. Acad. Sci. USA 1988, 85, 9163–9166. [Google Scholar] [CrossRef] [PubMed]
- Russelburg, L.P.; O’Shea Murray, V.L.; Demir, M.; Knutsen, K.R.; Sehgal, S.L.; Cao, S.; David, S.S.; Horvath, M.P. Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY. ACS Chem. Biol. 2020, 15, 93–102. [Google Scholar] [CrossRef]
- Bossé, J.T.; Li, Y.; Cohen, L.M.; Stegger, M.; Angen, Ø.; Lacouture, S.; Gottschalk, M.; Lei, L.; Koene, M.; Kuhnert, P.; et al. Complete genome for Actinobacillus pleuropneumoniae serovar 8 reference strain 405: Comparative analysis with draft genomes for different laboratory stock cultures indicates little genetic variation. Microb. Genom. 2021, 7, 000687. [Google Scholar] [CrossRef] [PubMed]
- Snesrud, E.; Ong, A.C.; Corey, B.; Kwak, Y.I.; Clifford, R.; Gleeson, T.; Wood, S.; Whitman, T.J.; Lesho, E.P.; Hinkle, M.; et al. Analysis of Serial Isolates of mcr-1-Positive Escherichia coli Reveals a Highly Active ISApl1 Transposon. Antimicrob. Agents Chemother. 2017, 61, e00056-17. [Google Scholar] [CrossRef] [PubMed]
- Bossé, J.T.; Li, Y.; Walker, S.; Atherton, T.; Fernandez Crespo, R.; Williamson, S.M.; Rogers, J.; Chaudhuri, R.R.; Weinert, L.A.; Oshota, O.; et al. Identification of dfrA14 in two distinct plasmids conferring trimethoprim resistance in Actinobacillus pleuropneumoniae. J. Antimicrob. Chemother. 2015, 70, 2217–2222. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Hawkey, J.; Hamidian, M.; Wick, R.R.; Edwards, D.J.; Billman-Jacobe, H.; Hall, R.M.; Holt, K.E. ISMapper: Identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genom. 2015, 16, 667. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bossé, J.T.; Li, Y.; Fernandez Crespo, R.; Chaudhuri, R.R.; Rogers, J.; Holden, M.T.; Maskell, D.J.; Tucker, A.W.; Wren, B.W.; Rycroft, A.N.; et al. ICEApl1, an Integrative Conjugative Element Related to ICEHin1056, Identified in the Pig Pathogen Actinobacillus pleuropneumoniae. Front. Microbiol. 2016, 7, 810. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Fernandez Crespo, R.; Leanse, L.G.; Langford, P.R.; Bossé, J.T. Characterization of the Actinobacillus pleuropneumoniae SXT-related integrative and conjugative element ICEApl2 and analysis of the encoded FloR protein: Hydrophobic residues in transmembrane domains contribute dynamically to florfenicol and chloramphenicol efflux. J. Antimicrob. Chemother. 2018, 73, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Bossé, J.T.; Soares-Bazzolli, D.M.; Li, Y.; Wren, B.W.; Tucker, A.W.; Maskell, D.J.; Rycroft, A.N.; Langford, P.R. The generation of successive unmarked mutations and chromosomal insertion of heterologous genes in Actinobacillus pleuropneumoniae using natural transformation. PLoS ONE 2014, 9, e111252. [Google Scholar] [CrossRef] [PubMed]
- Maglennon, G.A.; Cook, B.S.; Deeney, A.S.; Bossé, J.T.; Peters, S.E.; Langford, P.R.; Maskell, D.J.; Tucker, A.W.; Wren, B.W.; Rycroft, A.N. Transposon mutagenesis in Mycoplasma hyopneumoniae using a novel mariner-based system for generating random mutations. Vet. Res. 2013, 44, 124. [Google Scholar] [CrossRef] [PubMed]
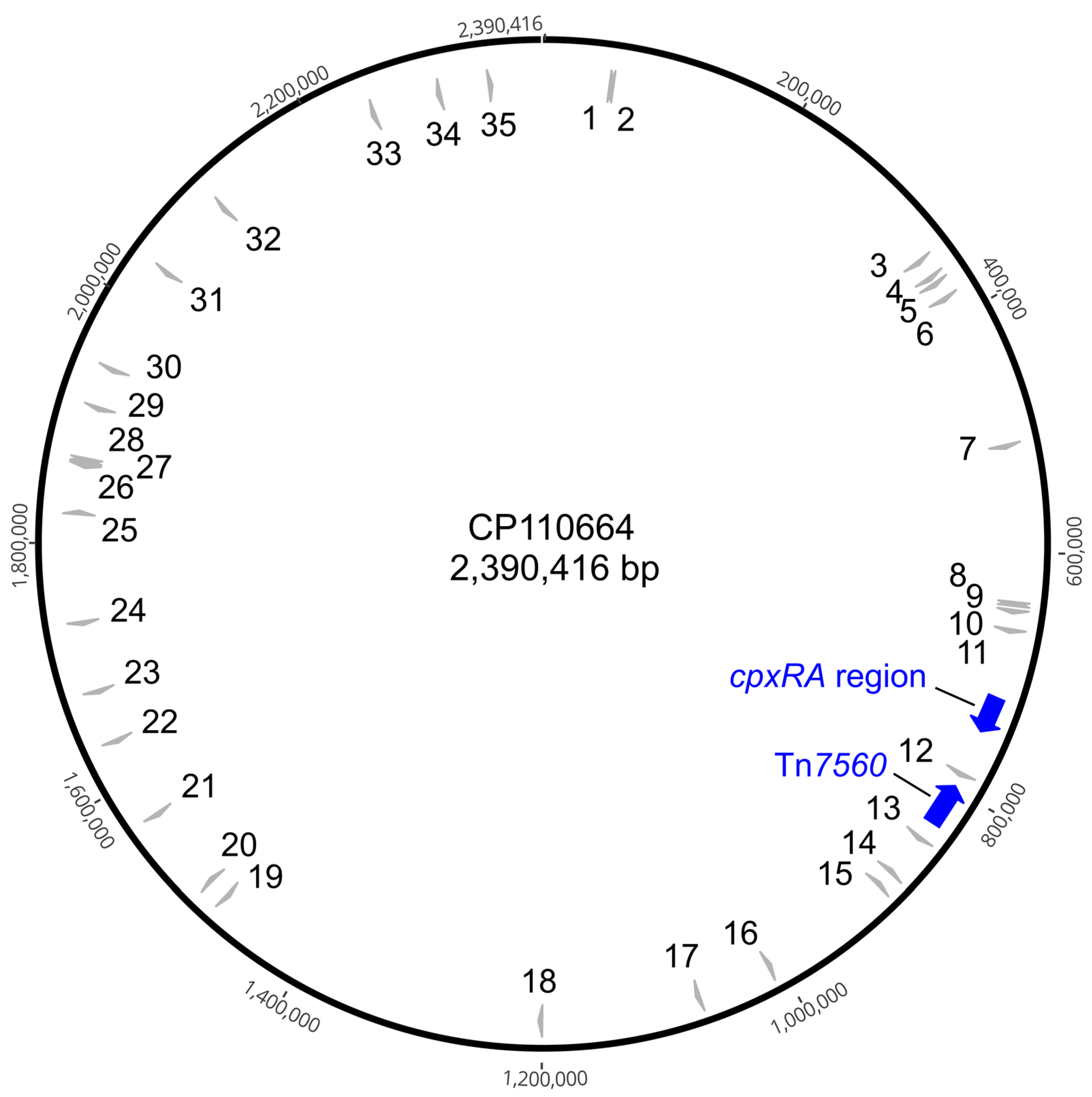
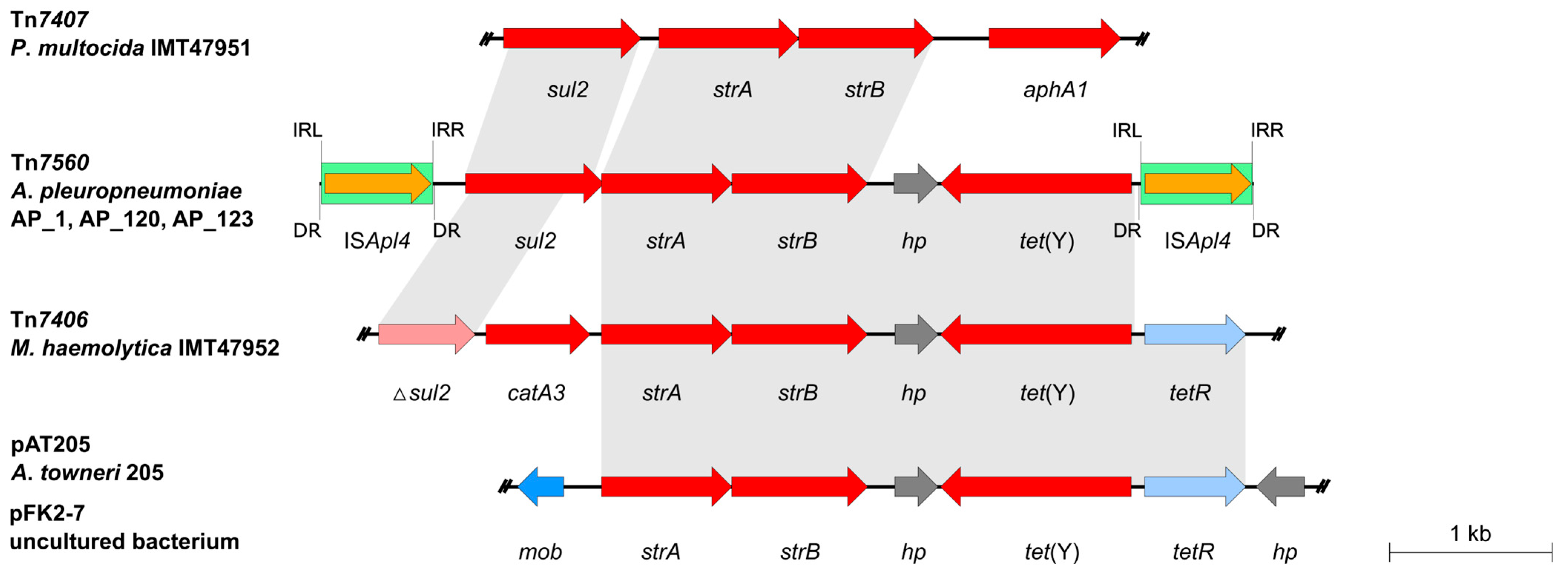

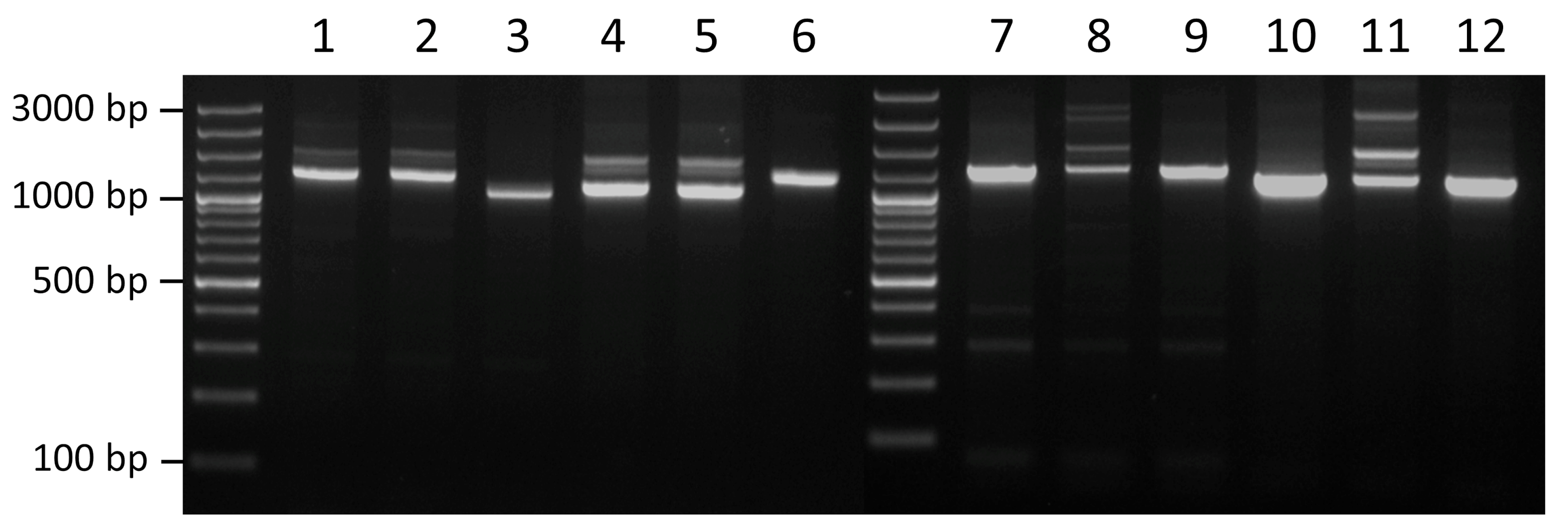
| ISApl4 Copy No. | 8-bp DR | Genomic Context |
|---|---|---|
| 1 | AAAAAAAA | non-coding region |
| 2 | AAAAAGAA | non-coding region |
| 3 | AAACAAAA | intragenic (DMT family transporter/EamA domain-containing protein) |
| 4 | AAACATAA | non-coding region |
| 5 | AAATTTAA | non-coding region |
| 6 | AAATTTTT | non-coding region |
| 7 | AAATTTTT | non-coding region |
| 8 | AACAATTA | intragenic (tadA; CpaF family protein/tight adherence protein A) |
| 9 | AACTAAAA | non-coding region |
| 10 | AAGTTTTA | non-coding region |
| 11 | AATAAATA | intragenic (sulfatase N-terminal domain-containing protein) |
| 12 | AATTAATT | intragenic (tetratricopeptide repeat protein) |
| 13 | ATAAAAAA | intragenic (hypothetical protein) |
| 14 | ATAATTTA | non-coding region |
| 15 | ATGATAAA | non-coding region |
| 16 | ATGATTTT | intragenic (udk; uridine kinase) |
| 17 | ATTAAATA | non-coding region |
| 18 | CAATTTAA | intragenic (hypothetical protein) |
| 19 | GATATTTT | intragenic (phoB; phosphate regulon transcriptional regulator) |
| 20 | TAAATAAA | intragenic (Tex family protein/predicted transcriptional accessory protein) |
| 21 | TAAATGAA | non-coding region |
| 22 | TAAATTAA | intragenic (manX; PTS mannose transporter subunit IIAB) |
| 23 | TAATAAAA | intragenic (LPS O-antigen chain length determinant protein, WzzB/FepE family) |
| 24 | TAATTTAA | intragenic (N-6 DNA methylase/class I SAM-dependent DNA methyltransferase) |
| 25 | TAATTTTT | intragenic (wzxE; lipid III flippase WzxE/lipopolysaccharide biosynthesis protein) |
| 26 | TATCTTAA | non-coding region |
| 27 | TTAAAAAT | non-coding region |
| 28 | TTAATAAA | non-coding region |
| 29 | TTAATAAA | non-coding region |
| 30 | TTAATTTA | non-coding region |
| 31 | TTAGTTTT | intragenic (mutY; A/G-specific adenine glycosylase) |
| 32 | TTATTAAA | intragenic (DUF1523 domain-containing protein) |
| 33 | TTTAATTA | intragenic (hypothetical protein) |
| 34 | TTTTAAGG | non-coding region |
| 35 | TTTTATAA | non-coding region |
| Isolate | Tetracycline MIC | Sulfisoxazole MIC | Streptomycin MIC |
|---|---|---|---|
| AP_1 | 16 | >512 | >256 |
| MIDG2331/AP_1 | 16 | >512 | >256 |
| AP_120 | 16 | >512 | >256 |
| MIDG2331/AP_120 | 16 | >512 | >256 |
| AP_123 | 8 | >512 | >256 |
| MIDG2331/AP_123 | 8 | >512 | >256 |
| MIDG2331 | 2 | 32 | 64 |
| Primer Name | Sequence 5′ to 3′ | Target/Purpose |
|---|---|---|
| ISApl4_tnp_for | TTGAAGTTACTGCTCGTTCGGC | ISApl4 tnp; 429-bp amplicon |
| ISApl4_tnp_rev | CGTTGATGTGATTATGGTCTTTTGC | |
| ISApl4_inv1 | TGAGAATTTCTGGTCGCAAGCC | ISApl4 circular form (718-bp amplicon) and transposon insertion site mapping |
| ISApl4_inv2 | GCCGTTGATGTGATTATGGTCTTTTG | |
| tetY_for | TATGGACGGCGGATTATTTTGCTG | tet(Y) gene; 530-bp amplicon |
| tetY_rev | GTGCCAATATCCCAATTCAAGCG | |
| sul2_for | TCAACATAACCTCGGACAGTTTCTC | sul2 gene; 212-bp amplicon |
| sul2_rev | GGGAATGCCATCTGCCTTGAGC | |
| strA_for | GATTTTGTTTTTCGACGTGGTGAC | strA-strB; 796-bp amplicon |
| strB_rev | GTGTCCGCAATGAGAACAGG | |
| oligo 254 | CGACTGGACCTGGA | Generation of linkers |
| oligo 256 | GATAAGCAGGGATCGGAACCTCCAGGTCCAGTCG | |
| oligo 258 | GATAAGCAGGGATCGGAACC | Linker-specific primer for linker PCR |
| tetY_5′_out | CATCAAGCGCGACAACAATGAG | Transposon-specific primer for linker PCR |
| sul2_up_out | TCTGGAAGCAAAGCAAGGAAAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bossé, J.T.; Li, Y.; Stegger, M.; Cohen, L.M.; Angen, Ø.; Overballe-Petersen, S.; Hanke, D.; Schwarz, S.; Langford, P.R.; Krüger-Haker, H. ISApl4, a New IS1595 Family Insertion Sequence Forming a Novel Pseudo-Compound Transposon That Confers Antimicrobial Multidrug Resistance in Actinobacillus pleuropneumoniae. Antibiotics 2025, 14, 1021. https://doi.org/10.3390/antibiotics14101021
Bossé JT, Li Y, Stegger M, Cohen LM, Angen Ø, Overballe-Petersen S, Hanke D, Schwarz S, Langford PR, Krüger-Haker H. ISApl4, a New IS1595 Family Insertion Sequence Forming a Novel Pseudo-Compound Transposon That Confers Antimicrobial Multidrug Resistance in Actinobacillus pleuropneumoniae. Antibiotics. 2025; 14(10):1021. https://doi.org/10.3390/antibiotics14101021
Chicago/Turabian StyleBossé, Janine T., Yanwen Li, Marc Stegger, Liza Miriam Cohen, Øystein Angen, Søren Overballe-Petersen, Dennis Hanke, Stefan Schwarz, Paul R. Langford, and Henrike Krüger-Haker. 2025. "ISApl4, a New IS1595 Family Insertion Sequence Forming a Novel Pseudo-Compound Transposon That Confers Antimicrobial Multidrug Resistance in Actinobacillus pleuropneumoniae" Antibiotics 14, no. 10: 1021. https://doi.org/10.3390/antibiotics14101021
APA StyleBossé, J. T., Li, Y., Stegger, M., Cohen, L. M., Angen, Ø., Overballe-Petersen, S., Hanke, D., Schwarz, S., Langford, P. R., & Krüger-Haker, H. (2025). ISApl4, a New IS1595 Family Insertion Sequence Forming a Novel Pseudo-Compound Transposon That Confers Antimicrobial Multidrug Resistance in Actinobacillus pleuropneumoniae. Antibiotics, 14(10), 1021. https://doi.org/10.3390/antibiotics14101021







