On-Site Inactivation for Disinfection of Antibiotic-Resistant Bacteria in Hospital Effluent by UV and UV-LED
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Disinfection of Wastewater by UV and UV-LED
2.3. Microbial Analysis
2.4. Quantitative PCR (qPCR) Analysis
2.5. Bacterial Community Structure Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Disinfection of the WWTP Wastewater and Hospital Effluent by UV and UV-LED Treatment
3.2. Inactivation Kinetics of AMRB and AMSB in the WWTP Wastewater and Hospital Effluent by UV and UV-LED Treatment
3.3. Removal of Antimicrobial-Resistance Genes in WWTP Wastewater and Hospital Effluent by UV and UV-LED Treatment
3.4. Bacterial Community Structure Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, N.A.; Vambol, V.; Vambol, S.; Bolibrukh, B.; Sillanpaa, M.; Changani, F.; Esrafili, A.; Yousefi, M. Hospital effluent guidelines and legislation scenario around the globe: A critical review. J. Environ. Chem. Eng. 2021, 9, 105874. [Google Scholar] [CrossRef]
- Cameron, A.; Esiovwa, R.; Connolly, J.; Hursthouse, A.; Henriquez, F. Antimicrobial resistance as a global health threat: The need to learn lessons from the COVID-19 pandemic. Glob. Policy 2022, 13, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, Y.; Li, L.; Liu, J.; Yan, X. Distribution, sources, and potential risks of antibiotic resistance genes in wastewater treatment plant: A review. Environ. Pollut. 2022, 310, 119870. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance 2014; World Health Organization: Geneva, Switzerland, 2014; pp. 1–232. [Google Scholar]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019; pp. 1–139. [Google Scholar]
- Jim, O.N. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014, 1–16. [Google Scholar]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- The Government of Japan. National Action Plan on Antimicrobial Resistance (AMR), 2016; The Government of Japan: Tokyo, Japan, 2016; pp. 1–69. [Google Scholar]
- The Government of Japan. National Action Plan on Antimicrobial Resistance (AMR), 2023; The Government of Japan: Tokyo, Japan, 2023; pp. 1–90. [Google Scholar]
- Al Salah, D.M.M.; Ngweme, G.N.; Laffite, A.; Otamonga, J.P.; Mulaji, C.; Poté, J. Hospital wastewaters: A reservoir and source of clinically relevant bacteria and antibiotic resistant genes dissemination in urban river under tropical conditions. Ecotoxicol. Environ. Saf. 2020, 200, 110767. [Google Scholar] [CrossRef] [PubMed]
- Hassoun-Kheir, N.; Stabholz, Y.; Kreft, J.U.; de la Cruz, R.; Romalde, J.L.; Nesme, J.; Sørensen, S.J.; Smets, B.F.; Graham, D.; Paul, M. Comparison of antibiotic-resistant bacteria and antibiotic resistance genes abundance in hospital and community wastewater: A systematic review. Sci. Total Environ. 2020, 743, 140804. [Google Scholar] [CrossRef] [PubMed]
- Sekizuka, T.; Tanaka, R.; Hashino, M.; Yatsu, K.; Kuroda, M. Comprehensive genome and plasmidome analysis of antimicrobial resistant bacteria in wastewater treatment plant effluent of Tokyo. Antibiotics 2022, 11, 1283. [Google Scholar] [CrossRef]
- Muraki, Y.; Kitamura, M.; Maeda, Y.; Kitahara, T.; Mori, T.; Ikeue, H.; Tsugita, M.; Tadano, K.; Takada, K.; Akamatsu, T.; et al. Nationwide surveillance of antimicrobial consumption and resistance to Pseudomonas aeruginosa isolates at 203 Japanese hospitals in 2010. Infection 2013, 41, 415–423. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Stefani, S.; Chung, D.R.; Lindsay, J.A.; Friedrich, A.W.; Kearns, A.M.; Westh, H.; MacKenzie, F.M. Meticillin-resistant Staphylococcus aureus (MRSA): Global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 2012, 39, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018, 31, e00018–e00020. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nature Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed]
- Shiozaki, Y. The action for the antimicobial resistance issue in Japan. Jpn. Assoc. Infect. Dis. 2018, 91, 915–923. [Google Scholar]
- Khan, M.T.; Shah, I.A.; Ihsanullah, I.; Naushad, M.; Ali, S.; Shah, S.H.A.; Mohammad, A.W. Hospital wastewater as a source of environmental contamination: An overview of management practices, environmental risks, and treatment processes. J. Water Proc. Eng. 2021, 41, 101990. [Google Scholar] [CrossRef]
- Verlicchi, P. Trends, new insights and perspectives in the treatment of hospital effluents. Curr. Opin. Environ. Sci. Health 2021, 19, 100217. [Google Scholar] [CrossRef] [PubMed]
- Pariente, M.I.; Segura, Y.; Álvarez-Torrellas, S.; Casas, J.A.; de Pedro, Z.M.; Diaz, E.; García, J.; López-Muñoz, M.J.; Marugán, J.; Mohedano, A.F.; et al. Critical review of technologies for the on-site treatment of hospital wastewater: From conventional to combined advanced processes. J. Environ. Manag. 2022, 320, 115769. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Vikesland, P.J.; Davis, B.C.; de Roda Husman, A.M. Seizing the moment: Now is the time for integrated global surveillance of antimicrobial resistance in wastewater environments. Curr. Opin. Microbiol. 2021, 64, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Sekizuka, T.; Itokawa, K.; Tanaka, R.; Hashino, M.; Yatsu, K.; Kuroda, M. Metagenomic analysis of urban wastewater treatment plant effluents in tokyo. Infect. Drug Resist. 2022, 15, 4763–4777. [Google Scholar] [CrossRef] [PubMed]
- Rekhate, C.V.; Srivastava, J.K. Recent advances in ozone-based advanced oxidation processes for treatment of wastewater- A review. Chem. Eng. J. Adv. 2020, 3, 100031. [Google Scholar] [CrossRef]
- Bhandari, G.; Chaudhary, P.; Gangola, S.; Gupta, S.; Gupta, A.; Rafatullah, M.; Chen, S. A review on hospital wastewater treatment technologies: Current management practices and future prospects. J. Water Proc. Eng. 2023, 56, 104516. [Google Scholar] [CrossRef]
- Adeoye, J.B.; Tan, Y.H.; Lau, S.Y.; Tan, Y.Y.; Chiong, T.; Mubarak, N.M.; Khalid, M. Advanced oxidation and biological integrated processes for pharmaceutical wastewater treatment: A review. J. Environ. Manag. 2024, 353, 120170. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.; Foster, T.; Shimki, N.T.; Willetts, J. Hospital wastewater (HWW) treatment in low- and middle-income countries: A systematic review of microbial treatment efficacy. Sci. Total Environ. 2024, 921, 170994. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, A.; Deivayanai, V.C.; Kumar, P.S.; Rangasamy, G.; Hemavathy, R.V.; Harshana, T.; Gayathri, N.; Alagumalai, K. A detailed review on advanced oxidation process in treatment of wastewater: Mechanism, challenges and future outlook. Chemosphere 2022, 308, 136524. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Shah, N.S.; Ali Khan, J.; Naushad, M.; Boczkaj, G.; Jamil, F.; Khan, S.; Li, L.; Murtaza, B.; Han, C. Pharmaceuticals wastewater treatment via different advanced oxidation processes: Reaction mechanism, operational factors, toxicities, and cost evaluation—A review. Sep. Purif. Technol. 2024, 347, 127458. [Google Scholar] [CrossRef]
- Li, X.; Cai, M.; Wang, L.; Niu, F.; Yang, D.; Zhang, G. Evaluation survey of microbial disinfection methods in UV-LED water treatment systems. Sci. Total Environ. 2019, 659, 1415–1427. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Hill, K.; Deere, D. Review of generic screening level assumptions for quantitative microbial risk assessment (qmra) for estimating public health risks from australian drinking water sources contaminated with Cryptosporidium by recreational activities. Water Res. 2022, 220, 118659. [Google Scholar] [CrossRef] [PubMed]
- Matafonova, G.; Batoev, V. Recent advances in application of UV light-emitting diodes for degrading organic pollutants in water through advanced oxidation processes: A review. Water Res. 2018, 132, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.H.; Suwan, P.; Koottatep, T.; Beck, S.E. Application of a novel, continuous-feeding ultraviolet light emitting diode (UV-LED) system to disinfect domestic wastewater for discharge or agricultural reuse. Water Res. 2019, 153, 53–62. [Google Scholar] [CrossRef]
- Song, K.; Mohseni, M.; Taghipour, F. Application of ultraviolet light-emitting diodes (UV-LEDs) for water disinfection: A review. Water Res. 2016, 94, 341–349. [Google Scholar] [CrossRef]
- Pelayo, D.; Rivero, M.J.; Santos, G.; Gómez, P.; Ortiz, I. Techno-economic evaluation of UV light technologies in water remediation. Sci. Total Environ. 2023, 868, 161376. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Q.; Wang, W.L.; Huo, Z.Y.; Lu, Y.; Hu, H.Y. Comparison of UV-LED and low pressure UV for water disinfection: Photoreactivation and dark repair of Escherichia coli. Water Res. 2017, 126, 134–143. [Google Scholar] [CrossRef]
- Nyangaresi, P.O.; Qin, Y.; Chen, G.; Zhang, B.; Lu, Y.; Shen, L. Comparison of UV-LED photolytic and UV-LED/TiO2 photocatalytic disinfection for escherichia coli in water. Catal. Today 2019, 335, 200–207. [Google Scholar] [CrossRef]
- Wan, Q.; Cao, R.; Wen, G.; Xu, X.; Xia, Y.; Wu, G.; Li, Y.; Wang, J.; Xu, H.; Lin, Y.; et al. Efficacy of UV-LED based advanced disinfection processes in the inactivation of waterborne fungal spores: Kinetics, photoreactivation, mechanism and energy requirements. Sci. Total Environ. 2022, 803, 150107. [Google Scholar] [CrossRef]
- Dulin, D.; Mill, T. Development and evaluation of sunlight actinometers. Environ. Sci. Technol. 1982, 16, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Loeb, S.; Kim, J.H. Led revolution: Fundamentals and prospects for UV disinfection applications. Environ. Sci. Water Res. Technol. 2017, 3, 188–202. [Google Scholar] [CrossRef]
- Ma, B.; Seyedi, S.; Wells, E.; McCarthy, D.; Crosbie, N.; Linden, K.G. Inactivation of biofilm-bound bacterial cells using irradiation across UVC wavelengths. Water Res. 2022, 217, 118379. [Google Scholar] [CrossRef]
- Bae, J.Y.; Kim, Y.; Kim, H.; Kim, Y.; Jin, J.; Bae, B.S. Ultraviolet light stable and transparent sol–gel methyl siloxane hybrid material for UV light-emitting diode (UV LED) encapsulant. ACS Appl. Mater. Interfaces 2015, 7, 1035–1039. [Google Scholar] [CrossRef]
- Gao, Z.C.; Lin, Y.L.; Xu, B.; Xia, Y.; Hu, C.Y.; Cao, T.C.; Zou, X.Y.; Gao, N.Y. Evaluating iopamidol degradation performance and potential dual-wavelength synergy by UV-LED irradiation and UV-LED/chlorine treatment. Chem. Eng. J. 2019, 360, 806–816. [Google Scholar] [CrossRef]
- Cai, A.; Deng, J.; Ye, C.; Zhu, T.; Ling, X.; Shen, S.; Guo, H.; Li, X. Highly efficient removal of deet by UV-LED irradiation in the presence of iron-containing coagulant. Chemosphere 2022, 286, 131613. [Google Scholar] [CrossRef]
- Song, K.; Taghipour, F.; Mohseni, M. Microorganisms inactivation by wavelength combinations of ultraviolet light-emitting diodes (UV-LEDs). Sci. Total Environ. 2019, 665, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.Y.; Lin, Y.L.; Xu, B.; Cao, T.C.; Tang, Y.L.; Pan, Y.; Gao, Z.C.; Gao, N.-Y. Enhanced inactivation of E. coli by pulsed UV-LED irradiation during water disinfection. Sci. Total Environ. 2019, 650, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Pousty, D.; Hofmann, R.; Gerchman, Y.; Mamane, H. Wavelength-dependent time–dose reciprocity and stress mechanism for UV-LED disinfection of Escherichia coli. J. Photochem. Photobiol. B 2021, 217, 112129. [Google Scholar] [CrossRef]
- Gerchman, Y.; Mamane, H.; Friedman, N.; Mandelboim, M. UV-LED disinfection of coronavirus: Wavelength effect. J. Photochem. Photobiol. B 2020, 212, 112044. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H.; Saito, A.; Kaneko, C.; Sugiyama, H.; Okabayashi, T.; Fujimoto, S. Rapid inactivation of SARS-CoV-2 variants by continuous and intermittent irradiation with a deep-ultraviolet light-emitting diode (DUV-LED) device. Pathogens 2021, 10, 754. [Google Scholar] [CrossRef]
- Verlicchi, P. Hospital Wastewaters: Characteristics, Management, Treatment and Environmental Risks; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–243. [Google Scholar]
- Khan, N.A.; Ahmed, S.; Farooqi, I.H.; Ali, I.; Vambol, V.; Changani, F.; Yousefi, M.; Vambol, S.; Khan, S.U.; Khan, A.H. Occurrence, sources and conventional treatment techniques for various antibiotics present in hospital wastewaters: A critical review. Trends Anal. Chem. 2020, 129, 115921. [Google Scholar] [CrossRef]
- Fatimazahra, S.; Latifa, M.; Laila, S.; Monsif, K. Review of hospital effluents: Special emphasis on characterization, impact, and treatment of pollutants and antibiotic resistance. Environ. Monit. Assess. 2023, 195, 393. [Google Scholar] [CrossRef] [PubMed]
- Haeusser, S.; Weber, M.; Mauer, C.; Linnemann, V.; Pfannstiel, A.; Pinnekamp, J.; Wintgens, T.; Klümper, C.; Beier, S. On-site treatment of hospital wastewater in a full-scale treatment plant in Germany: SARS-CoV-2 and treatment performance. Water Sci. Technol. 2023, 87, 1747–1763. [Google Scholar] [CrossRef]
- Gutierrez, M.; Mutavdžić Pavlović, D.; Stipaničev, D.; Repec, S.; Avolio, F.; Zanella, M.; Verlicchi, P. A thorough analysis of the occurrence, removal and environmental risks of organic micropollutants in a full-scale hybrid membrane bioreactor fed by hospital wastewater. Sci. Total Environ. 2024, 914, 169848. [Google Scholar] [CrossRef]
- Yu, S.Y.; Xie, Z.H.; Wu, X.; Zheng, Y.Z.; Shi, Y.; Xiong, Z.K.; Zhou, P.; Liu, Y.; He, C.-S.; Pan, Z.C.; et al. Review of advanced oxidation processes for treating hospital sewage to achieve decontamination and disinfection. Chin. Chem. Lett. 2024, 35, 108714. [Google Scholar] [CrossRef]
- Azuma, T.; Uchiyama, T.; Zhang, D.; Usui, M.; Hayashi, T. Distribution and characteristics of carbapenem-resistant and extended-spectrum β-lactamase (ESBL) producing Escherichia coli in hospital effluents, sewage treatment plants, and river water in an urban area of Japan. Sci. Total Environ. 2022, 839, 156232. [Google Scholar] [CrossRef]
- Japan Meteorological Agency. Weather Statistics. Available online: http://www.jma.go.jp/jma/index.html (accessed on 23 July 2024).
- Zheng, J.; Su, C.; Zhou, J.; Xu, L.; Qian, Y.; Chen, H. Effects and mechanisms of ultraviolet, chlorination, and ozone disinfection on antibiotic resistance genes in secondary effluents of municipal wastewater treatment plants. Chem. Eng. J. 2017, 317, 309–316. [Google Scholar] [CrossRef]
- Dunkin, N.; Weng, S.; Coulter, C.G.; Jacangelo, J.G.; Schwab, K.J. Impacts of virus processing on human norovirus GI and GII persistence during disinfection of municipal secondary wastewater effluent. Water Res. 2018, 134, 1–12. [Google Scholar] [CrossRef]
- Nyangaresi, P.O.; Qin, Y.; Chen, G.; Zhang, B.; Lu, Y.; Shen, L. Effects of single and combined UV-LEDs on inactivation and subsequent reactivation of E.coli in water disinfection. Water Res. 2018, 147, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzfathy, M.; Malayeri, A.H.; Mohseni, M.; Taghipour, F. UV-LED fluence determination by numerical method for microbial inactivation studies. J. Photochem. Photobiol. A 2020, 392, 112406. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency (EPA). Fate, Transport and Transformation: Test Guidelines OPPTS 835.2210; Direct Photolysis Rate in Water by Sunlight; U.S. Environmental Protection Agency (EPA): Washington, DC, USA, 1998; pp. 1–35. [Google Scholar]
- U.S. Environmental Protection Agency (EPA). Fate, Transport and Transformation: Test Guidelines OPPTS 835.2240; Photodegradation in Water; U.S. Environmental Protection Agency (EPA): Washington, DC, USA, 1998; pp. 1–4. [Google Scholar]
- OECD. Phototransformation of Chemicals in Water-Direct Photolysis, OECD Guideline for the Testing of Chemicals No. 316; OECD: Paris, France, 2008; pp. 1–53. [Google Scholar]
- Japan Sewage Works Association. Statistics of Sewerage; Japan Sewage Works Association: Tokyo, Japan, 2022. (In Japanese) [Google Scholar]
- Azuma, T.; Hayashi, T. Effects of natural sunlight on antimicrobial-resistant bacteria (AMRB) and antimicrobial-susceptible bacteria (AMSB) in wastewater and river water. Sci. Total Environ. 2021, 766, 142568. [Google Scholar] [CrossRef]
- Hijnen, W.A.M.; Beerendonk, E.F.; Medema, G.J. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: A review. Water Res. 2006, 40, 3–22. [Google Scholar] [CrossRef]
- He, H.; Zhou, P.; Shimabuku, K.K.; Fang, X.; Li, S.; Lee, Y.; Dodd, M.C. Degradation and deactivation of bacterial antibiotic resistance genes during exposure to free chlorine, monochloramine, chlorine dioxide, ozone, ultraviolet light, and hydroxyl radical. Environ. Sci. Technol. 2019, 53, 2013–2026. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Global Action Plan on Antimicrobial Resistance; World Health Organization (WHO): Geneva, Switzerland, 2015; pp. 1–19. [Google Scholar]
- World Health Organization (WHO). Antibiotic-Resistant “Priority Pathogens”—A Catalogue of 12 Families of Bacteria that Pose the Greatest Threat to Human Health. Available online: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ (accessed on 23 July 2024).
- Alekshun, M.N.; Levy, S.B. Molecular mechanisms of antibacterial multidrug resistance. Cell 2007, 128, 1037–1050. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat eskape pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Azuma, T.; Usui, M.; Hayashi, T. Inactivation of antibiotic-resistant bacteria in wastewater by ozone-based advanced water treatment processes. Antibiotics 2022, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- bioMérieux (France). Manufacturer’s Protocol for chromIDTM Chromogenic Media. Available online: http://www.biomerieux.fr/diagnostic-clinique/milieux-de-culture (accessed on 23 July 2024).
- Lamba, M.; Graham, D.W.; Ahammad, S.Z. Hospital wastewater releases of carbapenem-resistance pathogens and genes in urban India. Environ. Sci. Technol. 2017, 51, 13906–13912. [Google Scholar] [CrossRef] [PubMed]
- Glady-Croue, J.; Niu, X.Z.; Ramsay, J.P.; Watkin, E.; Murphy, R.J.T.; Croue, J.P. Survival of antibiotic resistant bacteria following artificial solar radiation of secondary wastewater effluent. Sci. Total Environ. 2018, 626, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Haller, L.; Chen, H.; Ng, C.; Le, T.H.; Koh, T.H.; Barkham, T.; Sobsey, M.; Gin, K.Y.H. Occurrence and characteristics of extended-spectrum β-lactamase- and carbapenemase- producing bacteria from hospital effluents in singapore. Sci. Total Environ. 2018, 615, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Azuma, T.; Otomo, K.; Kunitou, M.; Shimizu, M.; Hosomaru, K.; Mikata, S.; Ishida, M.; Hisamatsu, K.; Yunoki, A.; Mino, Y.; et al. Environmental fate of pharmaceutical compounds and antimicrobial-resistant bacteria in hospital effluents, and contributions to pollutant loads in the surface waters in Japan. Sci. Total Environ. 2019, 657, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Serna-Galvis, E.A.; Vélez-Peña, E.; Osorio-Vargas, P.; Jiménez, J.N.; Salazar-Ospina, L.; Guaca-González, Y.M.; Torres-Palma, R.A. Inactivation of carbapenem-resistant Klebsiella pneumoniae by photo-Fenton: Residual effect, gene evolution and modifications with citric acid and persulfate. Water Res. 2019, 161, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Sauter, D.; Stange, C.; Schumacher, V.; Tiehm, A.; Gnirss, R.; Wintgens, T. Impact of ozonation and biological post-treatment of municipal wastewater on microbiological quality parameters. Environ. Sci. Water Res. Technol. 2021, 7, 1643–1656. [Google Scholar] [CrossRef]
- Sib, E.; Voigt, A.M.; Wilbring, G.; Schreiber, C.; Faerber, H.A.; Skutlarek, D.; Parcina, M.; Mahn, R.; Wolf, D.; Brossart, P.; et al. Antibiotic resistant bacteria and resistance genes in biofilms in clinical wastewater networks. Int. J. Hyg. Environ. Health 2019, 222, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, C.; Zacharias, N.; Essert, S.M.; Wasser, F.; Müller, H.; Sib, E.; Precht, T.; Parcina, M.; Bierbaum, G.; Schmithausen, R.M.; et al. Clinically relevant antibiotic-resistant bacteria in aquatic environments—An optimized culture-based approach. Sci. Total Environ. 2021, 750, 142265. [Google Scholar] [CrossRef]
- Tsunoda, R.; Usui, M.; Tagaki, C.; Fukuda, A.; Boonla, C.; Anomasiri, W.; Sukpanyatham, N.; Akapelwa, M.L.; Nakajima, C.; Tamura, Y.; et al. Genetic characterization of coliform bacterial isolates from environmental water in Thailand. J. Infect. Chemother. 2021, 27, 722–728. [Google Scholar] [CrossRef]
- The eDNA Society. Environmental DNA Sampling and Experiment Manual Ver. 2.1; The eDNA Society: Otsu, Japan, 2021; pp. 1–93. [Google Scholar]
- Katada, S.; Fukuda, A.; Nakajima, C.; Suzuki, Y.; Azuma, T.; Takei, A.; Takada, H.; Okamoto, E.; Kato, T.; Tamura, Y.; et al. Aerobic composting and anaerobic digestion decrease the copy numbers of antibiotic-resistant genes and the levels of lactose-degrading Enterobacteriaceae in dairy farms in Hokkaido, Japan. Front. Microbiol. 2021, 12, 737420. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xing, S.; Chen, Y.; Wu, R.; Wu, Y.; Wang, Y.; Mi, J.; Liao, X. Profiles of bacteria/phage-comediated args in pig farm wastewater treatment plants in China: Association with mobile genetic elements, bacterial communities and environmental factors. J. Hazard. Mater. 2021, 404, 124149. [Google Scholar] [CrossRef] [PubMed]
- Saladin, M.; Cao, V.T.B.; Lambert, T.; Donay, J.L.; Herrmann, J.L.; Ould-Hocine, Z.; Verdet, C.; Delisle, F.; Philippon, A.; Arlet, G. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three parisian hospitals. FEMS Microbiol. Lett. 2002, 209, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Kojima, A.; Ishii, Y.; Ishihara, K.; Esaki, H.; Asai, T.; Oda, C.; Tamura, Y.; Takahashi, T.; Yamaguchi, K. Extended-spectrum-β-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: Report from the Japanese veterinary antimicrobial resistance monitoring program. Antimicrob. Age. Chemother. 2005, 49, 3533. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Miyanaga, K.; Tanji, Y. Persistence of antibiotic-resistant and -sensitive Proteus mirabilis strains in the digestive tract of the housefly (Musca domestica) and green bottle flies (Calliphoridae). Appl. Microbiol. Biotechnol. 2014, 98, 8357–8366. [Google Scholar] [CrossRef] [PubMed]
- Laffite, A.; Al Salah, D.M.M.; Slaveykova, V.I.; Otamonga, J.P.; Poté, J. Impact of anthropogenic activities on the occurrence and distribution of toxic metals, extending-spectra β-lactamases and carbapenem resistance in sub-Saharan African urban rivers. Sci. Total Environ. 2020, 727, 138129. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Chen, Q.L.; Chen, M.L.; Li, H.Z.; Liao, H.; Pu, Q.; Zhu, Y.G.; Cui, L. Temporal dynamics of antibiotic resistome in the plastisphere during microbial colonization. Environ. Sci. Technol. 2020, 54, 11322–11332. [Google Scholar] [CrossRef] [PubMed]
- Ávila, C.; García-Galán, M.J.; Borrego, C.M.; Rodríguez-Mozaz, S.; García, J.; Barceló, D. New insights on the combined removal of antibiotics and args in urban wastewater through the use of two configurations of vertical subsurface flow constructed wetlands. Sci. Total Environ. 2021, 755, 142554. [Google Scholar] [CrossRef]
- Li, S.; Yao, Q.; Liu, J.; Yu, Z.; Li, Y.; Jin, J.; Liu, X.; Wang, G. Liming mitigates the spread of antibiotic resistance genes in an acid black soil. Sci. Total Environ. 2022, 817, 152971. [Google Scholar] [CrossRef]
- Zhang, Y.; Pei, M.; Zhang, B.; He, Y.; Zhong, Y. Changes of antibiotic resistance genes and bacterial communities in the advanced biological wastewater treatment system under low selective pressure of tetracycline. Water Res. 2021, 207, 117834. [Google Scholar] [CrossRef]
- Ohore, O.E.; Wei, Y.; Wang, Y.; Nwankwegu, A.S.; Wang, Z. Tracking the influence of antibiotics, antibiotic resistomes, and salinity gradient in modulating microbial community assemblage of surface water and the ecological consequences. Chemosphere 2022, 305, 135428. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, H.; Wei, Y.; Chen, Z.; Chen, T.; Liang, Y.; Yin, J.; Yang, D.; Yang, Z.; Shi, D.; et al. Comprehensive insights into profiles and bacterial sources of intracellular and extracellular antibiotic resistance genes in groundwater. Environ. Pollut. 2022, 307, 119541. [Google Scholar] [CrossRef] [PubMed]
- Flach, C.F.; Hutinel, M.; Razavi, M.; Åhrén, C.; Larsson, D.G.J. Monitoring of hospital sewage shows both promise and limitations as an early-warning system for carbapenemase-producing Enterobacterales in a low-prevalence setting. Water Res. 2021, 200, 117261. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Liu, Y.; Wang, R.; Zhang, T.; Lu, W. Behaviors of antibiotic resistance genes (ARGs) and metal resistance genes (mrgs) during the pilot-scale biophysical drying treatment of sewage sludge: Reduction of args and enrichment of mrgs. Sci. Total Environ. 2022, 809, 152221. [Google Scholar] [CrossRef] [PubMed]
- Watts, G.S.; Youens-Clark, K.; Slepian, M.J.; Wolk, D.M.; Oshiro, M.M.; Metzger, G.S.; Dhingra, D.; Cranmer, L.D.; Hurwitz, B.L. 16s RNA gene sequencing on a benchtop sequencer: Accuracy for identification of clinically important bacteria. J. Appl. Microbiol. 2017, 123, 1584–1596. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16s rrna gene sequencing for species and strain-level microbiome analysis. Nature Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Deng, M.; Chen, J.; Gou, J.; Hou, J.; Li, D.; He, X. The effect of different carbon sources on water quality, microbial community and structure of biofloc systems. Aquaculture 2018, 482, 103–110. [Google Scholar] [CrossRef]
- Quartaroli, L.; Silva, C.M.; Silva, L.C.F.; Lima, H.S.; de Paula, S.O.; Dias, R.S.; Carvalho, K.B.; Souza, R.S.; Bassin, J.P.; da Silva, C.C. Effect of the gradual increase of salt on stability and microbial diversity of granular sludge and ammonia removal. J. Environ. Manag. 2019, 248, 109273. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Li, G.; Wang, C.; Jing, Y.; Zhu, Y.; Zhang, S.; Liu, Y. Community dynamics of prokaryotic and eukaryotic microbes in an estuary reservoir. Sci. Rep. 2014, 4, 6966. [Google Scholar] [CrossRef]
- Yu, P.; Sun, Y.; Huang, Z.; Zhu, F.; Sun, Y.; Jiang, L. The effects of ectomycorrhizal fungi on heavy metals’ transport in pinus massoniana and bacteria community in rhizosphere soil in mine tailing area. J. Hazard. Mater. 2020, 381, 121203. [Google Scholar] [CrossRef]
- Azuma, T.; Hayashi, T. Disinfection of antibiotic-resistant bacteria in sewage and hospital effluent by ozonation. Ozone Sci. Eng. 2021, 43, 413–426. [Google Scholar] [CrossRef]
- An, X.L.; Su, J.Q.; Li, B.; Ouyang, W.Y.; Zhao, Y.; Chen, Q.L.; Cui, L.; Chen, H.; Gillings, M.R.; Zhang, T.; et al. Tracking antibiotic resistome during wastewater treatment using high throughput quantitative pcr. Environ. Int. 2018, 117, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Ogwugwa, V.H.; Oyetibo, G.O.; Amund, O.O. Taxonomic profiling of bacteria and fungi in freshwater sewer receiving hospital wastewater. Environ. Res. 2021, 192, 110319. [Google Scholar] [CrossRef]
- Cheng, J.H.; Tang, X.Y.; Guan, Z.; Liu, C. Occurrence of antibiotic resistome in farmland soils near phosphorus chemical industrial area. Sci. Total Environ. 2021, 796, 149053. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Hui, X.; Yang, W.; Hua, Y.; Liu, H.; Dai, X. Implications for mitigation of antibiotic resistance: Differential response of intracellular and extracellular antibiotic resistance genes to sludge fermentation coupled with thermal hydrolysis. Water Res. 2022, 209, 117876. [Google Scholar] [CrossRef]
- Wasserstein, R.L.; Lazar, N.A. The ASA statement on p-values: Context, process, and purpose. Am. Stat. 2016, 70, 129–133. [Google Scholar] [CrossRef]
- Agathokleous, E. Environmental pollution impacts: Are p values over-valued? Sci. Total Environ. 2022, 850, 157807. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.M.; Kim, H.Y.; Park, W.; Kim, T.H.; Yu, S. A comparative study of disinfection efficiency and regrowth control of microorganism in secondary wastewater effluent using UV, ozone, and ionizing irradiation process. J. Hazard. Mater. 2015, 295, 201–208. [Google Scholar] [CrossRef]
- Ofori, I.; Maddila, S.; Lin, J.; Jonnalagadda, S.B. Ozone initiated inactivation of Escherichia coli and Staphylococcus aureus in water: Influence of selected organic solvents prevalent in wastewaters. Chemosphere 2018, 206, 43–50. [Google Scholar] [CrossRef]
- Torii, S.; Itamochi, M.; Katayama, H. Inactivation kinetics of waterborne virus by ozone determined by a continuous quench flow system. Water Res. 2020, 186, 116291. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Guidelines for Environmental Infection Control in Health-Care Facilities Recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013; pp. 1–241. [Google Scholar]
- Garcia, A.B.; Vinuela-Prieto, J.M.; Lopez-Gonzalez, L.; Candel, F.J. Correlation between resistance mechanisms in Staphylococcus aureus and cell wall and septum thickening. Infect. Drug Resist. 2017, 10, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Azuma, T.; Hayashi, T. On-site chlorination responsible for effective disinfection of wastewater from hospital. Sci. Total Environ. 2021, 776, 145951. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; He, H.; Dodd, M.C.; Lee, Y. Degradation kinetics of antibiotic resistance gene mecA of methicillin-resistant Staphylococcus aureus (MRSA) during water disinfection with chlorine, ozone, and ultraviolet light. Environ. Sci. Technol. 2021, 55, 2541–2552. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bu, L.; Wu, Y.; Sun, J.; Li, G.; Zhou, S. Disinfection profiles and mechanisms of E. coli, S. aureus, and B. subtilis in UV365/chlorine process: Inactivation, reactivation, and DBP formation. Sep. Purif. Technol. 2022, 287, 120584. [Google Scholar] [CrossRef]
- Torii, S.; David, S.C.; Larivé, O.; Cariti, F.; Kohn, T. Observed kinetics of enterovirus inactivation by free chlorine are host cell-dependent. Environ. Sci. Technol. 2023, 57, 18483–18490. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.; Ye, Y.; Chang, P.H.; Thirunarayanan, D.; Wigginton, K.R. Nucleic acid photolysis by UV254 and the impact of virus encapsidation. Environ. Sci. Technol. 2018, 52, 10408–10415. [Google Scholar] [CrossRef]
- Liu, X.; Hu, J.Y. Effect of DNA sizes and reactive oxygen species on degradation of sulphonamide resistance sul1 genes by combined UV/free chlorine processes. J. Hazard. Mater. 2020, 392, 122283. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Choi, Y.; Wu, S.J.; Fang, X.; Anderson, A.K.; Liou, S.Y.; Roberts, M.C.; Lee, Y.; Dodd, M.C. Application of nucleotide-based kinetic modeling approaches to predict antibiotic resistance gene degradation during UV- and chlorine-based wastewater disinfection processes: From bench- to full-scale. Environ. Sci. Technol. 2022, 56, 15141–15155. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.P.; Bowen, J.C.; Freeman, D.H.; Sharpless, C.M. Rapid and reproducible characterization of the wavelength dependence of aquatic photochemical reactions using light-emitting diodes. Environ. Sci. Technol. Lett. 2021, 8, 437–442. [Google Scholar] [CrossRef]
- Wang, M.; Ateia, M.; Awfa, D.; Yoshimura, C. Regrowth of bacteria after light-based disinfection—What we know and where we go from here. Chemosphere 2021, 268, 128850. [Google Scholar] [CrossRef]
- Lennon, J.T.; Muscarella, M.E.; Placella, S.A.; Lehmkuhl, B.K.; Zhou, J. How, when, and where relic DNA affects microbial diversity. mBio 2018, 9, e00618–e00637. [Google Scholar] [CrossRef] [PubMed]
- Burkert, A.; Douglas Thomas, A.; Waldrop Mark, P.; Mackelprang, R.; Atomi, H. Changes in the active, dead, and dormant microbial community structure across a pleistocene permafrost chronosequence. Appl. Environ. Microbiol. 2019, 85, e02646-18. [Google Scholar] [CrossRef]
- Morrison, C.; Atkinson, A.; Zamyadi, A.; Kibuye, F.; McKie, M.; Hogard, S.; Mollica, P.; Jasim, S.; Wert, E.C. Critical review and research needs of ozone applications related to virus inactivation: Potential implications for SARS-CoV-2. Ozone Sci. Eng. 2021, 43, 2–20. [Google Scholar] [CrossRef]
- Epelle, E.I.; Macfarlane, A.; Cusack, M.; Burns, A.; Okolie, J.A.; Mackay, W.; Rateb, M.; Yaseen, M. Ozone application in different industries: A review of recent developments. Chem. Eng. J. 2023, 454, 140188. [Google Scholar] [CrossRef] [PubMed]
- Loeb, B.L. Ozone: A valuable tool for addressing today’s environmental issues. A review of forty-five years of Ozone: Science & Engineering. Ozone Sci. Eng. 2024, 46, 2–25. [Google Scholar]
- Wang, R.; Ji, M.; Zhai, H.; Guo, Y.; Liu, Y. Occurrence of antibiotics and antibiotic resistance genes in wwtp effluent-receiving water bodies and reclaimed wastewater treatment plants. Sci. Total Environ. 2021, 796, 148919. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Feng, K.; Wang, Z.; Zhang, Y.; Yang, M.; Zhu, Y.G.; Virta, M.P.J.; Deng, Y. High-throughput single-cell technology reveals the contribution of horizontal gene transfer to typical antibiotic resistance gene dissemination in wastewater treatment plants. Environ. Sci. Technol. 2021, 55, 11824–11834. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.; Shin, H.; Hur, H.G.; Unno, T. Higher abundance of core antimicrobial resistant genes in effluent from wastewater treatment plants. Water Res. 2022, 208, 117882. [Google Scholar] [CrossRef]
- Huang, Y.H.; Liu, Y.; Du, P.-P.; Zeng, L.J.; Mo, C.H.; Li, Y.W.; Lü, H.; Cai, Q.Y. Occurrence and distribution of antibiotics and antibiotic resistant genes in water and sediments of urban rivers with black-odor water in Guangzhou, South China. Sci. Total Environ. 2019, 670, 170–180. [Google Scholar] [CrossRef]
- Cheng, X.; Xu, J.; Smith, G.; Zhang, Y. Metagenomic insights into dissemination of antibiotic resistance across bacterial genera in wastewater treatment. Chemosphere 2021, 271, 129563. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, L.; Chen, J.; Fan, X.; Xie, S.; Huang, J.; Yu, G. Antibiotic resistance genes and mobile genetic elements in a rural river in southeast China: Occurrence, seasonal variation and association with the antibiotics. Sci. Total Environ. 2021, 778, 146131. [Google Scholar] [CrossRef]
- Grenni, P. Antimicrobial resistance in rivers: A review of the genes detected and new challenges. Environ. Toxicol. Chem. 2022, 41, 687–714. [Google Scholar] [CrossRef]
- Stange, C.; Sidhu, J.P.S.; Toze, S.; Tiehm, A. Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment. Int. J. Hyg. Environ. Health 2019, 222, 541–548. [Google Scholar] [CrossRef]
- van Bruggen, A.H.C.; Goss, E.M.; Havelaar, A.; van Diepeningen, A.D.; Finckh, M.R.; Morris, J.G. One health—Cycling of diverse microbial communities as a connecting force for soil, plant, animal, human and ecosystem health. Sci. Total Environ. 2019, 664, 927–937. [Google Scholar] [CrossRef]
- Booton, R.D.; Meeyai, A.; Alhusein, N.; Buller, H.; Feil, E.; Lambert, H.; Mongkolsuk, S.; Pitchforth, E.; Reyher, K.K.; Sakcamduang, W.; et al. One health drivers of antibacterial resistance: Quantifying the relative impacts of human, animal and environmental use and transmission. One Health 2021, 12, 100220. [Google Scholar] [CrossRef]
- Auguet, O.; Pijuan, M.; Borrego, C.M.; Rodriguez-Mozaz, S.; Triadó-Margarit, X.; Giustina, S.V.D.; Gutierrez, O. Sewers as potential reservoirs of antibiotic resistance. Sci. Total Environ. 2017, 605–606, 1047–1054. [Google Scholar] [CrossRef]
- He, P.; Zhou, Y.; Shao, L.; Huang, J.; Yang, Z.; Lü, F. The discrepant mobility of antibiotic resistant genes: Evidence from their spatial distribution in sewage sludge flocs. Sci. Total Environ. 2019, 697, 134176. [Google Scholar] [CrossRef]
- Zhang, X.; Li, R. Variation and distribution of antibiotic resistance genes and their potential hosts in microbial electrolysis cells treating sewage sludge. Bioresour. Technol. 2020, 315, 123838. [Google Scholar] [CrossRef]
- Lee, J.; Jeon, J.H.; Shin, J.; Jang, H.M.; Kim, S.; Song, M.S.; Kim, Y.M. Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. Sci. Total Environ. 2017, 605–606, 906–914. [Google Scholar] [CrossRef]
- Narciso-da-Rocha, C.; Rocha, J.; Vaz-Moreira, I.; Lira, F.; Tamames, J.; Henriques, I.; Martinez, J.L.; Manaia, C.M. Bacterial lineages putatively associated with the dissemination of antibiotic resistance genes in a full-scale urban wastewater treatment plant. Environ. Int. 2018, 118, 179–188. [Google Scholar] [CrossRef]
- Gwenzi, W.; Musiyiwa, K.; Mangori, L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: A hotspot reservoir. J. Environ. Chem. Eng. 2020, 8, 102220. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Quantitative Microbial Risk Assessment, Application for Water Safety Management; World Health Organization (WHO): Geneva, Switzerland, 2016; pp. 1–187. [Google Scholar]
- Le Page, G.; Gunnarsson, L.; Snape, J.; Tyler, C.R. Integrating human and environmental health in antibiotic risk assessment: A critical analysis of protection goals, species sensitivity and antimicrobial resistance. Environ. Int. 2017, 109, 155–169. [Google Scholar] [CrossRef]
- Pepper, I.L.; Brooks, J.P.; Gerba, C.P. Antibiotic resistant bacteria in municipal wastes: Is there reason for concern? Environ. Sci. Technol. 2018, 52, 3949–3959. [Google Scholar] [CrossRef]
- Schages, L.; Wichern, F.; Kalscheuer, R.; Bockmühl, D. Winter is coming—Impact of temperature on the variation of beta-lactamase and mcr genes in a wastewater treatment plant. Sci. Total Environ. 2020, 712, 136499. [Google Scholar] [CrossRef]
- Schoen, M.E.; Jahne, M.A.; Garland, J.; Ramirez, L.; Lopatkin, A.J.; Hamilton, K.A. Quantitative microbial risk assessment of antimicrobial resistant and susceptible Staphylococcus aureus in reclaimed wastewaters. Environ. Sci. Technol. 2021, 55, 15246–15255. [Google Scholar] [CrossRef]
- Wang, L.; Ye, C.; Guo, L.; Chen, C.; Kong, X.; Chen, Y.; Shu, L.; Wang, P.; Yu, X.; Fang, J. Assessment of the UV/chlorine process in the disinfection of Pseudomonas aeruginosa: Efficiency and mechanism. Environ. Sci. Technol. 2021, 55, 9221–9230. [Google Scholar] [CrossRef]
- Iakovides, I.C.; Manoli, K.; Karaolia, P.; Michael-Kordatou, I.; Manaia, C.M.; Fatta-Kassinos, D. Reduction of antibiotic resistance determinants in urban wastewater by ozone: Emphasis on the impact of wastewater matrix towards the inactivation kinetics, toxicity and bacterial regrowth. J. Hazard. Mater. 2021, 420, 126527. [Google Scholar] [CrossRef]
- García-Espinoza, J.D.; Robles, I.; Durán-Moreno, A.; Godínez, L.A. Photo-assisted electrochemical advanced oxidation processes for the disinfection of aqueous solutions: A review. Chemosphere 2021, 274, 129957. [Google Scholar] [CrossRef]
- Li, H.; Dechesne, A.; He, Z.; Jensen, M.M.; Song, H.L.; Smets, B.F. Electrochemical disinfection may increase the spread of antibiotic resistance genes by promoting conjugal plasmid transfer. Sci. Total Environ. 2023, 858, 159846. [Google Scholar] [CrossRef]
- Kiejza, D.; Kotowska, U.; Polińska, W.; Karpińska, J. Peracids—New oxidants in advanced oxidation processes: The use of peracetic acid, peroxymonosulfate, and persulfate salts in the removal of organic micropollutants of emerging concern—A review. Sci. Total Environ. 2021, 790, 148195. [Google Scholar] [CrossRef]
- Sahulka, S.Q.; Bhattarai, B.; Bhattacharjee, A.S.; Tanner, W.; Mahar, R.B.; Goel, R. Differences in chlorine and peracetic acid disinfection kinetics of Enterococcus faecalis and Escherichia fergusonii and their susceptible strains based on gene expressions and genomics. Water Res. 2021, 203, 117480. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.W.; Wu, Y.H.; Chen, G.Q.; Wang, H.B.; Wang, Y.H.; Tong, X.; Bai, Y.; Xu, Y.Q.; Zhang, Z.-W.; Ikuno, N.; et al. Chlorine-resistant bacteria (CRB) in the reverse osmosis system for wastewater reclamation: Isolation, identification and membrane fouling mechanisms. Water Res. 2022, 209, 117966. [Google Scholar] [CrossRef]
- Wang, R.; Alamin, M.; Tsuji, S.; Hara-Yamamura, H.; Hata, A.; Zhao, B.; Ihara, M.; Honda, R. Removal performance of SARS-CoV-2 in wastewater treatment by membrane bioreactor, anaerobic-anoxic-oxic, and conventional activated sludge processes. Sci. Total Environ. 2022, 851, 158310. [Google Scholar] [CrossRef]
- Zhou, Z.; Tran, P.Q.; Kieft, K.; Anantharaman, K. Genome diversification in globally distributed novel marine proteobacteria is linked to environmental adaptation. ISME J. 2020, 14, 2060–2077. [Google Scholar] [CrossRef]
- Niestępski, S.; Harnisz, M.; Ciesielski, S.; Korzeniewska, E.; Osińska, A. Environmental fate of Bacteroidetes, with particular emphasis on Bacteroides fragilis group bacteria and their specific antibiotic resistance genes, in activated sludge wastewater treatment plants. J. Hazard. Mater. 2020, 394, 122544. [Google Scholar] [CrossRef]
- Wallace, M.J.; Jean, S.; Wallace, M.A.; Burnham, C.A.D.; Dantas, G. Comparative genomics of Bacteroides fragilis group isolates reveals species-dependent resistance mechanisms and validates clinical tools for resistance prediction. mBio 2022, 13, e03603-21. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Knopp, G.; Dötsch, A.; Wieland, A.; Schwartz, T. Ozone treatment of conditioned wastewater selects antibiotic resistance genes, opportunistic bacteria, and induce strong population shifts. Sci. Total Environ. 2016, 559, 103–112. [Google Scholar] [CrossRef]
- Czekalski, N.; Imminger, S.; Salhi, E.; Veljkovic, M.; Kleffel, K.; Drissner, D.; Hammes, F.; Bürgmann, H.; von Gunten, U. Inactivation of antibiotic resistant bacteria and resistance genes by ozone: From laboratory experiments to full-scale wastewater treatment. Environ. Sci. Technol. 2016, 50, 11862–11871. [Google Scholar] [CrossRef] [PubMed]
- Gomi, R.; Matsuda, T.; Yamamoto, M.; Chou, P.H.; Tanaka, M.; Ichiyama, S.; Yoneda, M.; Matsumura, Y. Characteristics of carbapenemase-producing Enterobacteriaceae in wastewater revealed by genomic analysis. Antimicrob. Age. Chemother. 2018, 62, e02501–e02517. [Google Scholar] [CrossRef]
- Sekizuka, T.; Yatsu, K.; Inamine, Y.; Segawa, T.; Nishio, M.; Kishi, N.; Kuroda, M. Complete genome sequence of a blakpc-2-positive Klebsiella pneumoniae strain isolated from the effluent of an urban sewage treatment plant in Japan. mSphere 2018, 3, e00314–e00318. [Google Scholar] [CrossRef]
- Sekizuka, T.; Inamine, Y.; Segawa, T.; Kuroda, M. Characterization of NDM-5- and CTX-M-55-coproducing Escherichia coli GSH8M-2 isolated from the effluent of a wastewater treatment plant in Tokyo Bay. Infect. Drug Resist. 2019, 12, 2243–2249. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.; Tacão, M.; Pureza, L.; Gonçalves, J.; Silva, A.; Cruz-Schneider, M.P.; Henriques, I. Occurrence of carbapenemase-producing Enterobacteriaceae in a portuguese river: blaNDM, blaKPC and blaGES among the detected genes. Environ. Pollut. 2020, 260, 113913. [Google Scholar] [CrossRef] [PubMed]
- Hiller, C.X.; Hübner, U.; Fajnorova, S.; Schwartz, T.; Drewes, J.E. Antibiotic microbial resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review. Sci. Total Environ. 2019, 685, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems –A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Piña, B.; Bayona, J.M.; Christou, A.; Fatta-Kassinos, D.; Guillon, E.; Lambropoulou, D.; Michael, C.; Polesel, F.; Sayen, S. On the contribution of reclaimed wastewater irrigation to the potential exposure of humans to antibiotics, antibiotic resistant bacteria and antibiotic resistance genes—NEREUS COST Action ES1403 position paper. J. Environ. Chem. Eng. 2020, 8, 102131. [Google Scholar] [CrossRef]
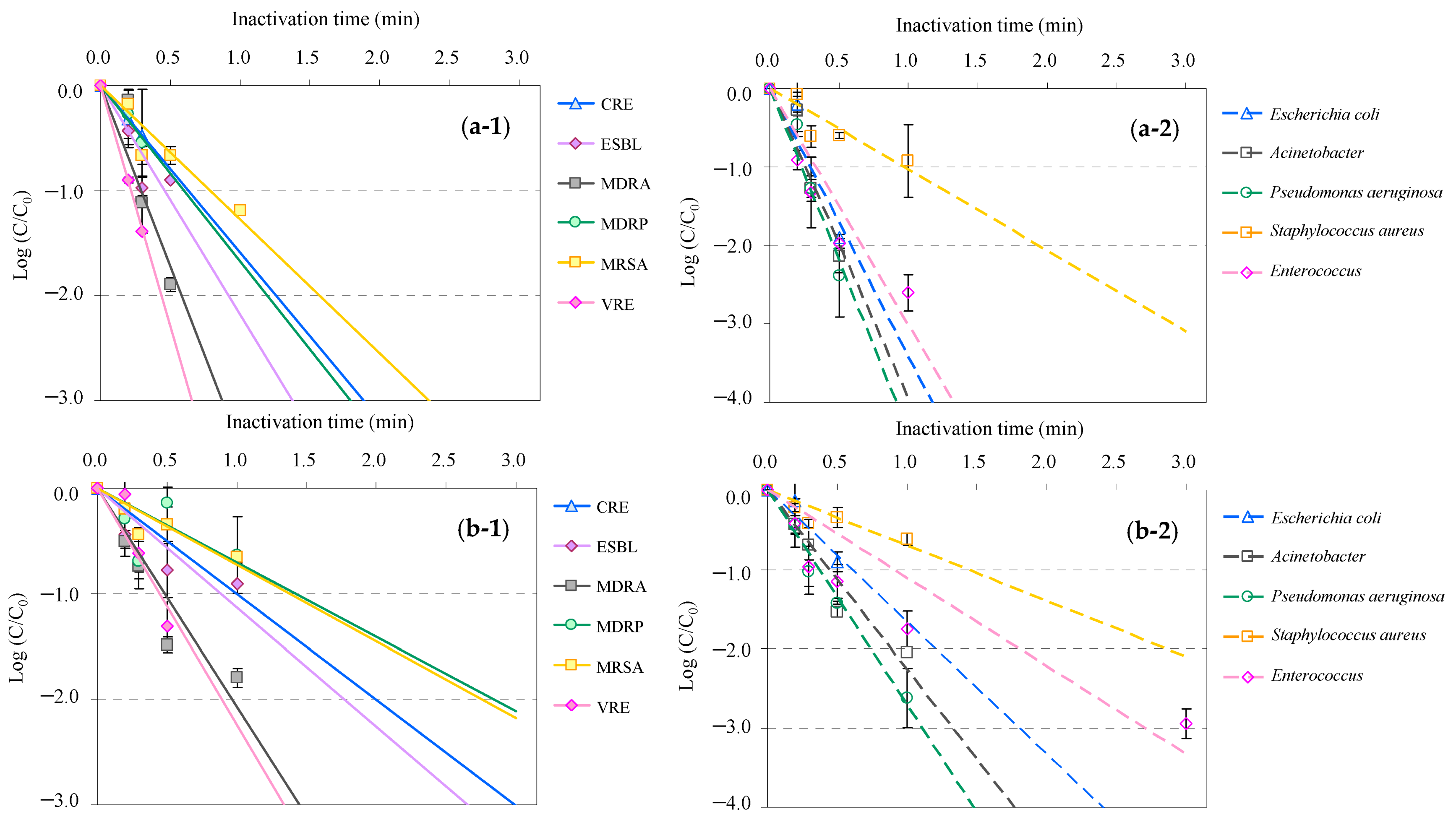

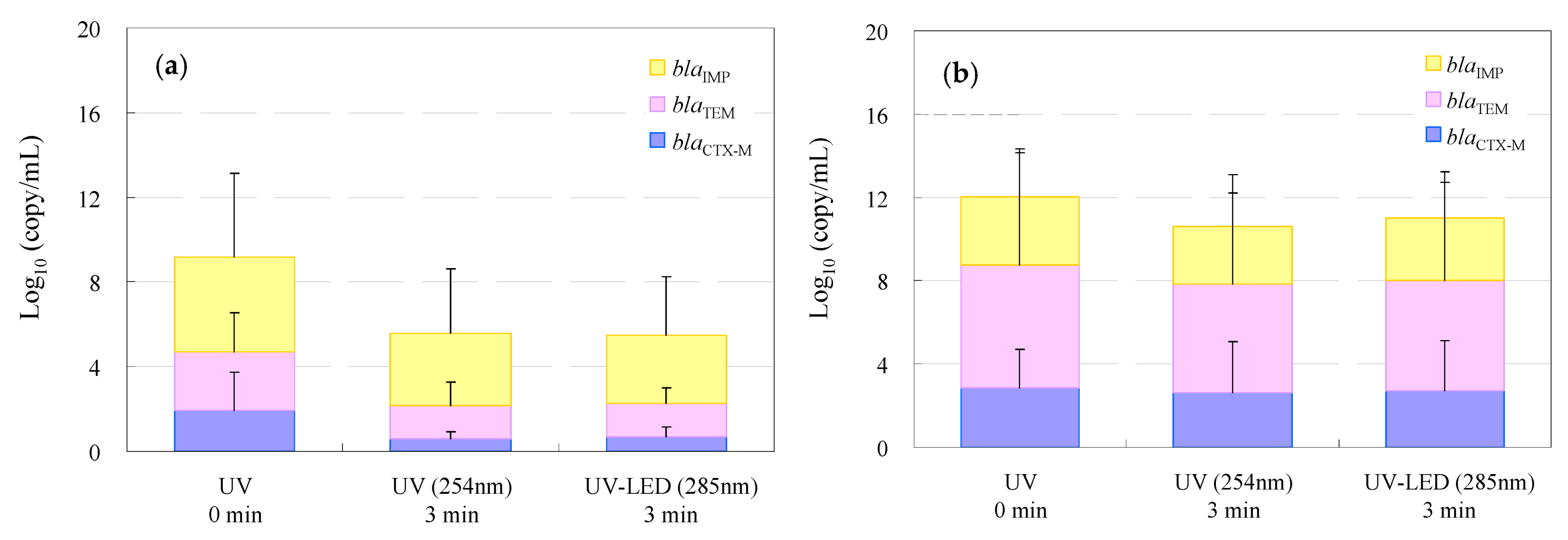
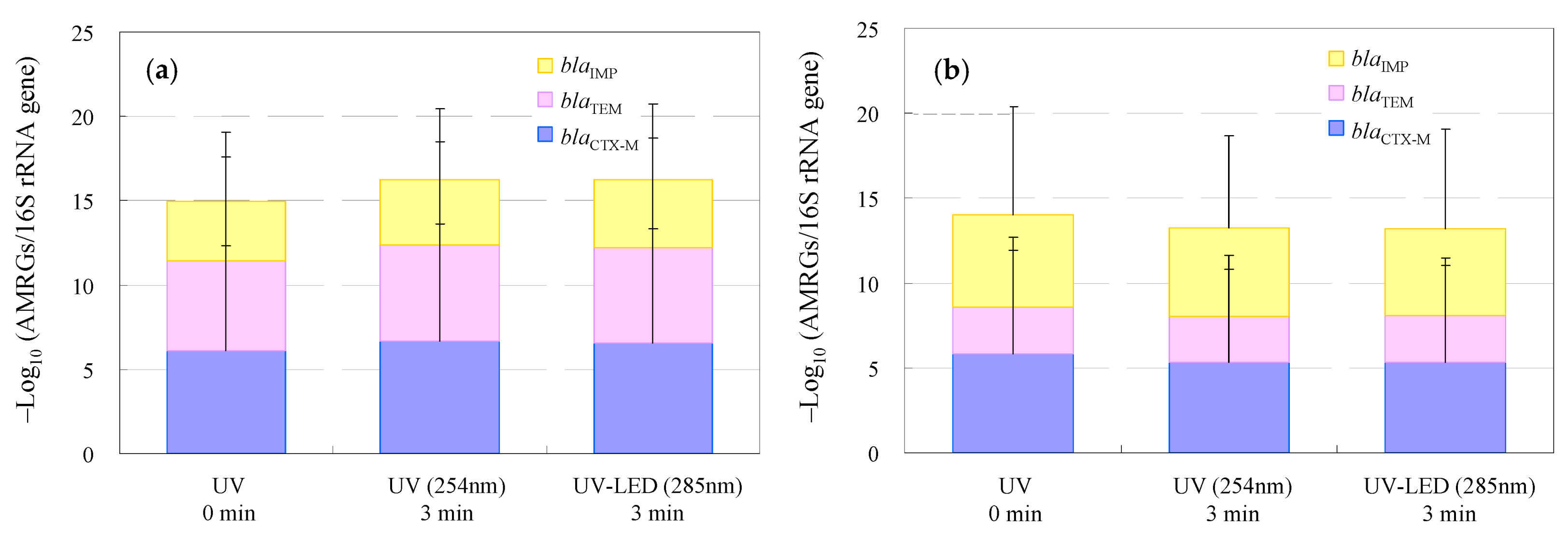
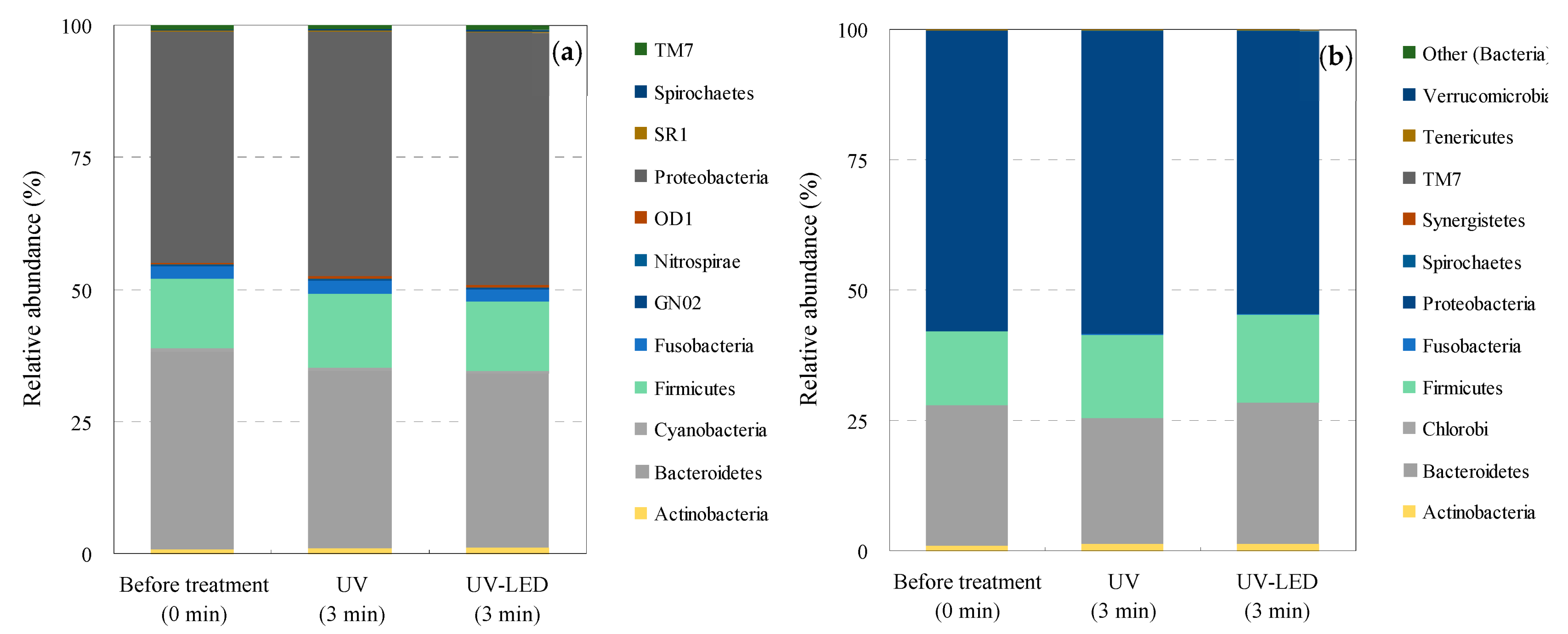
| Classification | Bacteria | Abbreviation |
|---|---|---|
| AMRB | Carbapenem-resistant Enterobacterales | CRE |
| Extended-spectrum β-lactamase (ESBL)-producing Enterobacterales | ESBL-E | |
| Multi-drug-resistant Acinetobacter | MDRA | |
| Multi-drug-resistant Pseudomonas aeruginosa | MDRP | |
| Methicillin-resistant Staphylococcus aureus | MRSA | |
| Vancomycin-resistant Enterococcus | VRE | |
| AMSB | Acinetobacter | Acinetobacter |
| Enterococcus | Enterococcus | |
| Escherichia col | E. coli | |
| Pseudomonas aeruginosa | P. aeruginosa | |
| Staphylococcus aureus | S. aureus |
| Bacteria | Bacteria Counts (CFU/mL) | ||||
|---|---|---|---|---|---|
| WWTP Influent | WWTP Secondary Effluent * | WWTP Effluent ** | WWTP Wastewater *** | Hospital Effluent | |
| CRE | 131 | 6 | 16 | 42 | 78 |
| ESBL-E | 963 | 10 | 9 | 73 | 131 |
| MDRA | 236 | 17 | 17 | 172 | 81 |
| MDRP | 43 | N.D. | 1 | 9 | 215 |
| MRSA | 47 | 7 | 6 | 21 | 31 |
| VRE | 35 | N.D. | N.D. | 27 | 151 |
| Acinetobacter | 257 | 34 | 15 | 91 | 307 |
| Enterococcus | 4267 | 664 | 140 | 1163 | 5736 |
| Escherichia coli | 20,000 | 145 | 5 | 3000 | 13,000 |
| Pseudomonas aeruginosa | 50 | N.D. | 1 | 21 | 1096 |
| Staphylococcus aureus | 126 | 8 | 17 | 24 | 63 |
| Bacteria | Inactivation Rate (min−1) | |||
|---|---|---|---|---|
| WWTP Wastewater | Hospital Effluent | |||
| UV (254 nm) | UV-LED (280 nm) | UV (254 nm) | UV-LED (280 nm) | |
| CRE | 5.794 | 2.351 | 6.932 | 11.992 |
| ESBL-E | 6.916 | 3.606 | 6.711 | 1.292 |
| MDRA | 8.440 | 4.763 | 1.497 | 1.511 |
| MDRP | 4.558 | 1.407 | 3.346 | 1.479 |
| MRSA | 3.215 | 1.663 | 1.928 | 0.804 |
| VRE | 10.649 | 5.417 | 2.985 | 3.554 |
| Acinetobacter | 8.920 | 5.126 | 2.524 | 1.465 |
| Enterococcus | 6.819 | 2.582 | 5.306 | 3.507 |
| Escherichia coli | 8.360 | 4.613 | 7.254 | 2.517 |
| Pseudomonas aeruginosa | 9.615 | 6.027 | 6.674 | 1.928 |
| Staphylococcus aureus | 2.501 | 1.601 | 2.047 | 1.506 |
| Bacteria | Fluences Required for Inactivation (mJ/cm2) (Mean (SD)) | |||
|---|---|---|---|---|
| WWTP Wastewater | Hospital Effluent | |||
| UV (254 nm) | UV-LED (280 nm) | UV (254 nm) | UV-LED (280 nm) | |
| CRE | 17.2 (3.2) | 34.3 (30.3) | 5.1 (0.8) | 9.6 (4.5) |
| ESBL-E | 15.0 (3.5) | 26.5 (11.4) | 5.4 (0.5) | 31.9 (11.1) |
| MDRA | 11.6 (2.4) | 18.7 (0.9) | 14.9 (11.7) | 32.8 (4) |
| MDRP | 14.6 (3.3) | 63.2 (35.3) | 10.3 (3.9) | 29.3 (3.9) |
| MRSA | 26.9 (1.7) | 65.2 (34.6) | 17.8 (6.8) | 70.6 (58) |
| VRE | 8.7 (9.0) | 16.4 (4.9) | 7.2 (4.1) | 10.4 (3.3) |
| Acinetobacter | 9.0 (0.3) | 17.2 (1.2) | 11.4 (1.3) | 40.8 (15.5) |
| Enterococcus | 11.8 (0.5) | 35 (2.9) | 4.2 (1.7) | 12.7 (0.5) |
| Escherichia coli | 9.5 (0.2) | 25.9 (8) | 6.0 (2.3) | 16.7 (5.9) |
| Pseudomonas aeruginosa | 8.2 (1.1) | 14.4 (1.5) | 4.7 (0.1) | 14.1 (3.2) |
| Staphylococcus aureus | 32.7 (0.0) | 55.7 (4.6) | 15.7 (3.0) | 28.3 (3.2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azuma, T.; Usui, M.; Hasei, T.; Hayashi, T. On-Site Inactivation for Disinfection of Antibiotic-Resistant Bacteria in Hospital Effluent by UV and UV-LED. Antibiotics 2024, 13, 711. https://doi.org/10.3390/antibiotics13080711
Azuma T, Usui M, Hasei T, Hayashi T. On-Site Inactivation for Disinfection of Antibiotic-Resistant Bacteria in Hospital Effluent by UV and UV-LED. Antibiotics. 2024; 13(8):711. https://doi.org/10.3390/antibiotics13080711
Chicago/Turabian StyleAzuma, Takashi, Masaru Usui, Tomohiro Hasei, and Tetsuya Hayashi. 2024. "On-Site Inactivation for Disinfection of Antibiotic-Resistant Bacteria in Hospital Effluent by UV and UV-LED" Antibiotics 13, no. 8: 711. https://doi.org/10.3390/antibiotics13080711
APA StyleAzuma, T., Usui, M., Hasei, T., & Hayashi, T. (2024). On-Site Inactivation for Disinfection of Antibiotic-Resistant Bacteria in Hospital Effluent by UV and UV-LED. Antibiotics, 13(8), 711. https://doi.org/10.3390/antibiotics13080711







