Comparative Genomics of an Emerging Multidrug-Resistant blaNDM-Carrying ST182 Lineage in Enterobacter cloacae Complex
Abstract
1. Introduction
2. Results
2.1. Bacterial Strains, Whole-Genome Sequences and Phylogenetic Analysis of ECC Isolates
2.2. In Silico Identification of Plasmids, Antimicrobial Resistance and Virulence Genes of the ECC NDM-Harboring Isolates
2.3. Genetic Background of blaNDM and Plasmid Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates, Genome Sequences and Phylogenetic Analysis
4.2. Identification of MGEs, Antimicrobial Resistance Genes and Virulence Factors and Plasmid Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Davin-Regli, A.; Lavigne, J.P.; Pagès, J.M. Enterobacter spp.: Update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 2019, 32, e00002-19. [Google Scholar] [CrossRef] [PubMed]
- Paauw, A.; Caspers, M.P.M.; Schuren, F.H.J.; Hall, M.A.L.-V.; Delétoile, A.; Montijn, R.C.; Verhoef, J.; Fluit, A.C. Genomic diversity within the Enterobacter cloacae complex. PLoS ONE 2008, 3, e3018. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.C. Multidrug-resistant Enterobacter cloacae Complex emerging as a global, diversifying threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef] [PubMed]
- Izdebski, R.; Baraniak, A.; Herda, M.; Fiett, J.; Bonten, M.J.; Carmeli, Y.; Goossens, H.; Hryniewicz, W.; Brun-Buisson, C.; Gniadkowski, M. MOSAR WP2, WP3 and WP5 Study Groups. MLST reveals potentially high-risk international clones of Enterobacter Cloacae. J. Antimicrob. Chemother. 2015, 70, 48–56. [Google Scholar] [CrossRef]
- Peirano, G.; Matsumura, Y.; Adams, M.D.; Bradford, P.; Motyl, M.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D. Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008–2014. Emerg. Infect. Dis. 2018, 24, 1010–1019. [Google Scholar] [CrossRef]
- Bolourchi, N.; Giske, C.G.; Nematzadeh, S.; Mirzaie, A.; Abhari, S.S.; Solgi, H.; Badmasti, F. Comparative resistome and virulome analysis of clinical NDM-1–producing carbapenem-resistant Enterobacter cloacae complex. J. Glob. Antimicrob. Resist. 2022, 28, 254–263. [Google Scholar] [CrossRef]
- Sugawara, Y.; Akeda, Y.; Hagiya, H.; Sakamoto, N.; Takeuchi, D.; Shanmugakani, R.K.; Motooka, D.; Nishi, I.; Zin, K.N.; Aye, M.M.; et al. Spreading patterns of NDM-producing Enterobacteriaceae in clinical and environmental settings in Yangon, Myanmar. Antimicrob. Agents Chemother. 2019, 63, e01924-18. [Google Scholar] [CrossRef]
- Papagiannitsis, C.C.; Studentova, V.; Chudackova, E.; Bergerova, T.; Hrabak, J.; Radej, J.; Novak, I. Identification of a New Delhi metallo-β-lactamase-4 (NDM-4)-producing Enterobacter cloacae from a Czech patient previously hospitalized in Sri Lanka. Folia Microbiol. 2013, 58, 547–549. [Google Scholar] [CrossRef]
- Paskova, V.; Medvecky, M.; Skalova, A.; Chudejova, K.; Bitar, I.; Jakubu, V.; Bergerova, T.; Zemlickova, H.; Papagiannitsis, C.C.; Hrabak, J. Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech hospitals. Front. Microbiol. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed]
- Merhi, G.; Amayri, S.; Bitar, I.; Araj, G.F.; Tokajian, S. Whole genome-based characterization of multidrug-resistant Enterobacter and Klebsiella aerogenes isolates from Lebanon. Microbiol. Spectr. 2023, 1, e0291722. [Google Scholar] [CrossRef] [PubMed]
- Gartzonika, K.; Politi, L.; Mavroidi, A.; Tsantes, A.G.; Spanakis, N.; Priavali, E.; Vrioni, G.; Tsakris, A. High prevalence of clonally related ST182 NDM-1-producing Enterobacter cloacae complex clinical isolates in Greece. Int. J. Antimicrob. Agents 2023, 62, 106837. [Google Scholar] [CrossRef] [PubMed]
- Mavroidi, A.; Gartzonika, K.; Spanakis, N.; Froukala, E.; Kittas, C.; Vrioni, G.; Tsakris, A. Comprehensive analysis of virulence determinants and genomic islands of blaNDM-1-producing Enterobacter hormaechei clinical isolates from Greece. Antibiotics 2023, 12, 1549. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-Y.; Wong, C.-F.; Chung, K.M.-K.; Jiang, J.-W.; Leung, F.C.-C. Comparative genome analysis of Enterobacter cloacae. PLoS ONE 2013, 8, e74487. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Sang, Y.; Teng, Q.; Ni, J.; Yang, Y.; Tsui, S.K.; Yao, Y.F. Heat shock proteins IbpA and IbpB are required for NlpI-participated cell division in Escherichia coli. Front. Microbiol. 2015, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Turkovicova, L.; Smidak, R.; Jung, G.; Turna, J.; Lubec, G.; Aradska, J. Proteomic analysis of the TerC interactome: Novel links to tellurite resistance and pathogenicity. J. Proteom. 2016, 136, 167–173. [Google Scholar] [CrossRef]
- Sukupolvi, S.; O’Connor, C.D. TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiol. Rev. 1990, 54, 331–341. [Google Scholar] [CrossRef]
- Gual-de-Torrella, A.; Delgado-Valverde, M.; Pérez-Palacios, P.; Oteo-Iglesias, J.; Rojo-Molinero, E.; Macià, M.D.; Oliver, A.; Pascual, Á.; Fernández-Cuenca, F. Prevalence of the fimbrial operon mrkABCD, mrkA expression, biofilm formation and effect of biocides on biofilm formation in carbapenemase-producing Klebsiella pneumoniae isolates belonging or not belonging to high-risk clones. Int. J. Antimicrob. Agents 2022, 60, 106663. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.; Guan, C.; Li, J.; Yang, H.; Zhao, G.; Liu, C.; Ma, J.; Tang, B. Characterization of an Escherichia coli isolate coharboring the virulence gene astA and tigecycline resistance gene tet(X4) from a dead piglet. Pathogens 2023, 12, 903. [Google Scholar] [CrossRef] [PubMed]
- Zong, B.; Liu, W.; Zhang, Y.; Wang, X.; Chen, H.; Tan, C. Effect of kpsM on the virulence of porcine extraintestinal pathogenic Escherichia coli. FEMS Microbiol. Lett. 2016, 363, fnw232. [Google Scholar]
- Moss, J.E.; Cardozo, T.J.; Zychlinsky, A.; Groisman, E.A. The selC-associated SHI-2 pathogenicity island of Shigellaflexneri. Mol. Microbiol. 1999, 33, 74–83. [Google Scholar] [CrossRef]
- Ho, P.-L.; Li, Z.; Lo, W.-U.; Cheung, Y.-Y.; Lin, C.-H.; Sham, P.-C.; Cheng, V.C.-C.; Ng, T.-K.; Que, T.-L.; Chow, K.-H. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg. Microbes Infect. 2012, 1, e39. [Google Scholar] [CrossRef] [PubMed]
- Wailan, A.M.; Paterson, D.L.; Kennedy, K.; Ingram, P.R.; Bursle, E.; Sidjabat, H.E. Genomic characteristics of NDM-producing Enterobacteriaceae isolates in Australia and their blaNDM genetic contexts. Antimicrob. Agents Chemother. 2015, 60, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Akeda, Y.; Sakamoto, N.; Takeuchi, D.; Motooka, D.; Nakamura, S.; Hagiya, H.; Yamamoto, N.; Nishi, I.; Yoshida, H.; et al. Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS ONE 2017, 12, e0184720. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Shao, C.; Geng, X.; Bai, Y.; Jin, Y.; Lu, Z. Genotypic and phenotypic characterization of clinical Escherichia coli Sequence Type 405 carrying IncN2 plasmid harboring blaNDM-1. Front. Microbiol. 2019, 10, 788. [Google Scholar] [CrossRef] [PubMed]
- Krishnaraju, M.; Kamatchi, C.; Jha, A.K.; Devasena, N.; Vennila, R.; Sumathi, G.; Vaidyanathan, R. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian. J. Med. Microbiol. 2015, 33, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.W.; Wang, J.T.; Lauderdale, T.L.; Liao, T.L.; Lai, J.F.; Tan, M.C.; Lin, A.C.; Chen, Y.T.; Tsai, S.F.; Chang, S.C. Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan. Antimicrob. Agents Chemother. 2013, 57, 4072–4076. [Google Scholar] [CrossRef]
- Phan, H.T.T.; Stoesser, N.; Maciuca, I.E.; Toma, F.; Szekely, E.; Flonta, M.; Hubbard, A.T.M.; Pankhurst, L.; Do, T.; Peto, T.E.A.; et al. Illumina short-read and MinION long-read WGS to characterize the molecular epidemiology of an NDM-1 Serratia marcescens outbreak in Romania. J. Antimicrob. Chemother. 2018, 73, 672–679. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Poirel, L.; Carattoli, A.; Nordmann, P. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS ONE 2012, 7, e34752. [Google Scholar] [CrossRef] [PubMed]
- Pitart, C.; Solé, M.; Roca, I.; Román, A.; Moreno, A.; Vila, J.; Marco, F. Molecular characterization ofblaNDM-5 carried on an IncFII plasmid in an Escherichia coli isolate from a nontraveler patient in Spain. Antimicrob. Agents Chemother. 2015, 59, 659–662. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, T.W.; Chen, T.L.; Chen, Y.T.; Lauderdale, T.L.; Liao, T.L.; Lee, Y.T.; Chen, C.P.; Liu, Y.M.; Lin, A.C.; Chang, Y.H.; et al. Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS ONE 2013, 8, e62774. [Google Scholar] [CrossRef] [PubMed]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef]
- Adler, A.; Ghosh, H.; Gross, A.; Rechavi, A.; Lasnoy, M.; Assous, M.V.; Geffen, Y.; Darawshe, B.; Wiener-Well, Y.; Grundmann, H.; et al. Molecular features and transmission of NDM-producing Enterobacterales in Israeli hospitals. J. Antimicrob. Chemother. 2023, 78, 719–723. [Google Scholar] [CrossRef]
- Acman, M.; Wang, R.; van Dorp, L.; Shaw, L.P.; Wang, Q.; Luhmann, N.; Yin, Y.; Sun, S.; Chen, H.; Wang, H.; et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat. Commun. 2022, 13, 1131. [Google Scholar] [CrossRef]
- David, S.; Cohen, V.; Reuter, S.; Sheppard, A.E.; Giani, T.; Parkhill, J. European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group; ESCMID Study Group for Epidemiological Markers (ESGEM); Rossolini GM, Feil EJ, Grundmann H, Aanensen DM. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA 2020, 117, 25043–25054. [Google Scholar] [CrossRef]
- Argimón, S.; David, S.; Underwood, A.; Abrudan, M.; Wheeler, N.E.; Kekre, M.; Abudahab, K.; Yeats, C.A.; Goater, R.; Taylor, B.; et al. NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance. 2021. Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clin Infect Dis 2021, 73 (Suppl. S4), S325–S335. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128–129. [Google Scholar] [CrossRef]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole genome phylogenies from short sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; Abudahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.; Zankari, E.; Aarestrup, F.M.; Lund, O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J. Antimicrob. Chemother. 2016, 71, 2484–2488. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, C.T. GC-Profile: A web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006, 34, W686–W691. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, Y.; Liu, M.; Tai, C.; Sun, J.; Deng, Z.; Ou, H.-Y. oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018, 46, W229–W234. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
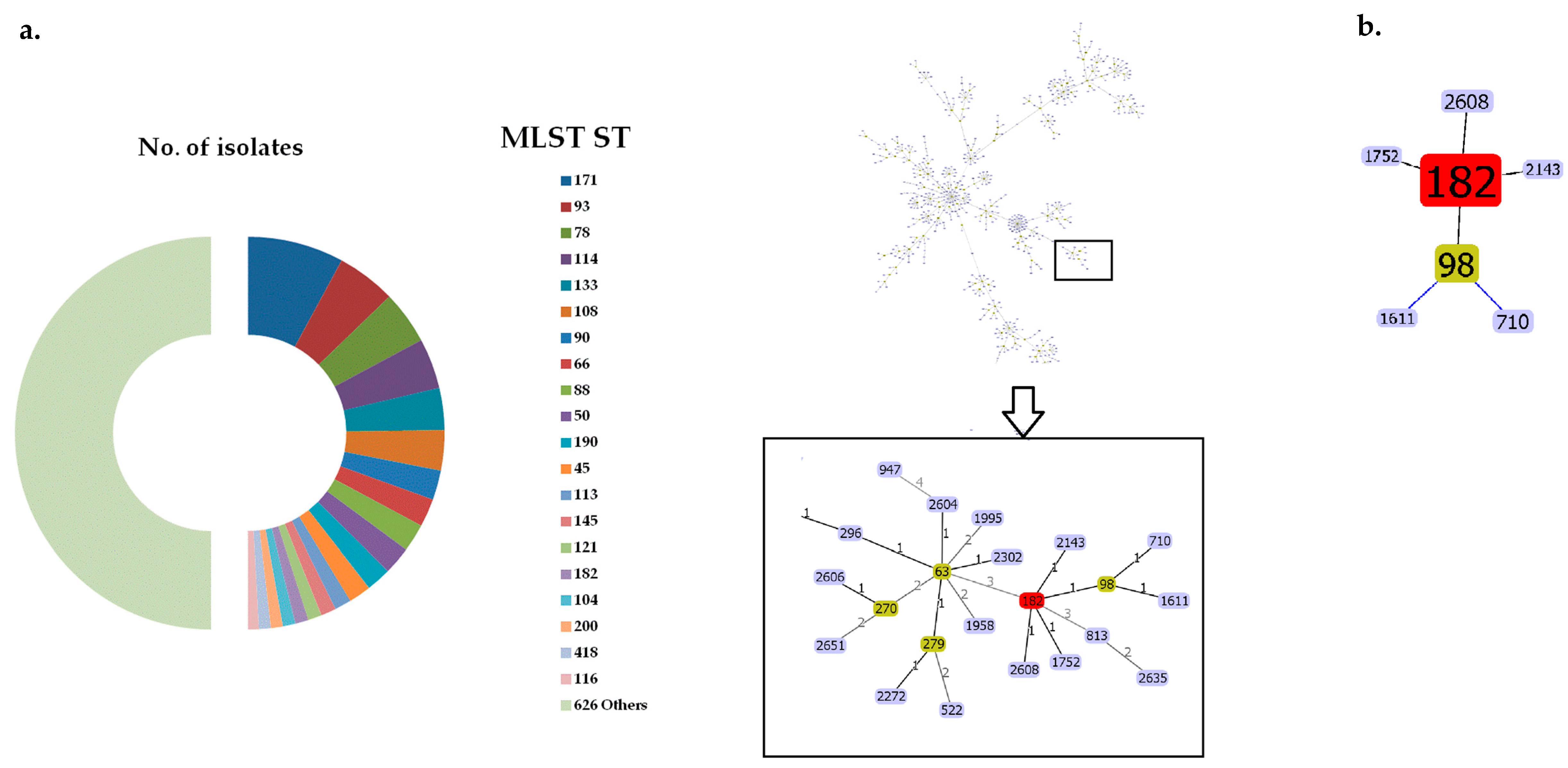
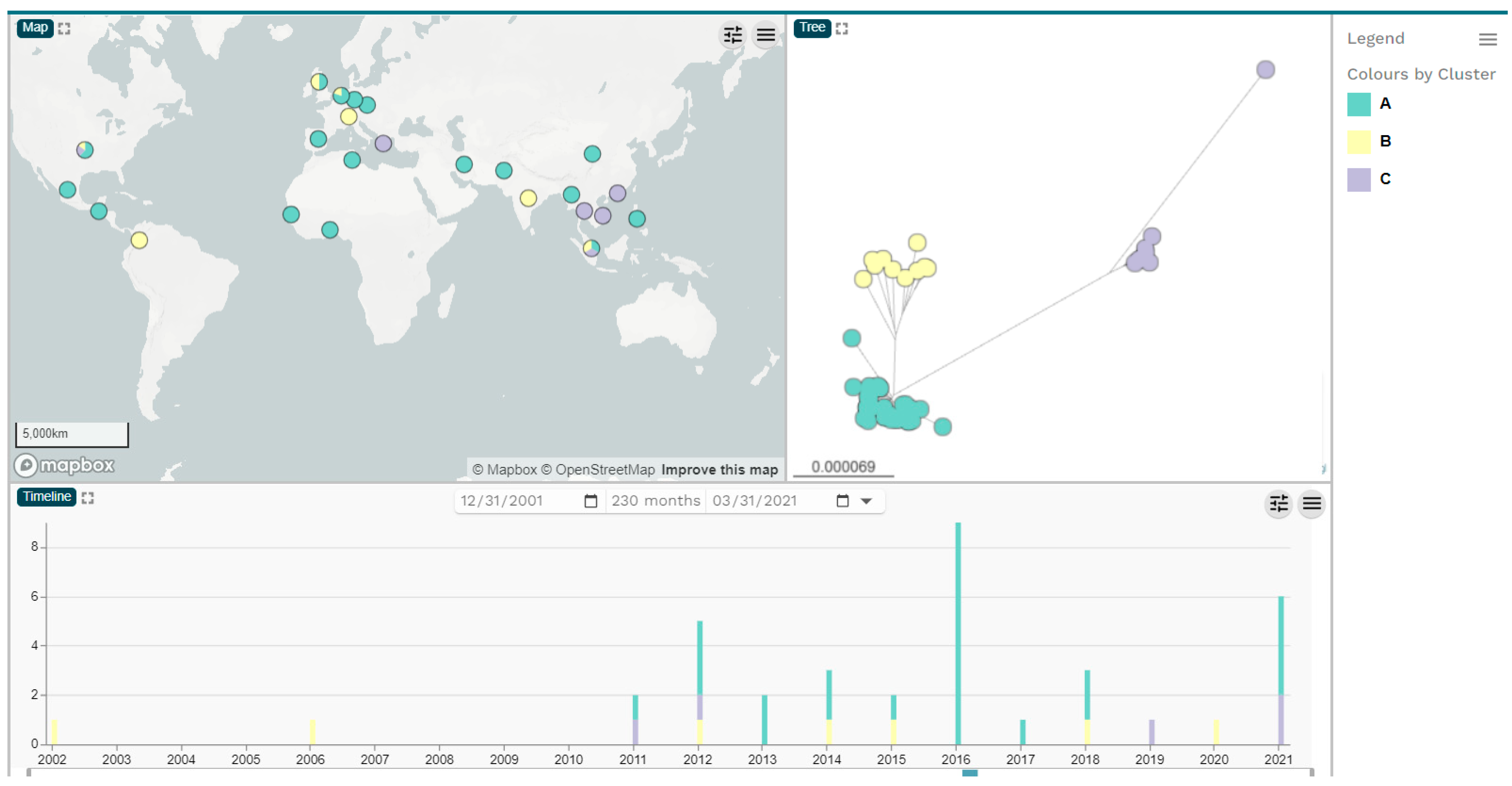

| Genomic Clusters | Continent/Country of Isolation | blaNDM Variants | Total | ||||
|---|---|---|---|---|---|---|---|
| blaNDM-1 | blaNDM-4 | blaNDM-5 | blaNDM-7 | None | |||
| Cluster A (No. of isolates) | 19 | 2 | 1 | 2 | 13 | 37 | |
| Africa | 1 | 3 | 4 | ||||
| Senegal | 2 | 2 | |||||
| Togo | 1 | 1 | |||||
| Tunisia | 1 | 1 | |||||
| Asia | 10 | 1 | 11 | ||||
| China | 2 | 1 | 3 | ||||
| Iran | 1 | 1 | |||||
| Myanmar | 3 | 3 | |||||
| Pakistan | 2 | 2 | |||||
| Philippines | 1 | 1 | |||||
| Singapore | 1 | 1 | |||||
| Europe | 4 | 2 | 1 | 5 | 12 | ||
| Czech Republic | 2 | 2 | |||||
| Germany | 1 | 1 | |||||
| Netherlands | 3 | 1 | 4 | ||||
| Spain | 3 | 3 | |||||
| United Kingdom | 1 | 1 | 2 | ||||
| North America | 4 | 1 | 4 | 9 | |||
| Guatemala | 1 | 1 | |||||
| USA | 3 | 1 | 4 | 8 | |||
| South America | 1 | 1 | |||||
| Mexico | 1 | 1 | |||||
| Cluster B (No. of isolates) | 1 | 9 | 10 | ||||
| Asia | 1 | 1 | 2 | ||||
| India | 1 | 1 | |||||
| Singapore | 1 | 1 | |||||
| Europe | 4 | 4 | |||||
| Netherlands | 1 | 1 | |||||
| Switzerland | 1 | 1 | |||||
| United Kingdom | 2 | 2 | |||||
| North America | 2 | 2 | |||||
| USA | 2 | 2 | |||||
| South America | 2 | 2 | |||||
| Colombia | 2 | 2 | |||||
| Cluster C (No. of isolates) | 4 | 1 | 3 | 8 | |||
| Asia | 3 | 1 | 4 | ||||
| China | 1 | 1 | |||||
| Singapore | 1 | 1 | |||||
| Thailand | 1 | 1 | |||||
| Viet Nam | 1 | 1 | |||||
| Europe | 1 | 1 | |||||
| Greece | 1 | 1 | |||||
| North America | 1 | 2 | 3 | ||||
| USA | 1 | 2 | 3 | ||||
| Total | 24 | 2 | 2 | 2 | 25 | 55 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mavroidi, A.; Froukala, E.; Tsakris, A. Comparative Genomics of an Emerging Multidrug-Resistant blaNDM-Carrying ST182 Lineage in Enterobacter cloacae Complex. Antibiotics 2024, 13, 535. https://doi.org/10.3390/antibiotics13060535
Mavroidi A, Froukala E, Tsakris A. Comparative Genomics of an Emerging Multidrug-Resistant blaNDM-Carrying ST182 Lineage in Enterobacter cloacae Complex. Antibiotics. 2024; 13(6):535. https://doi.org/10.3390/antibiotics13060535
Chicago/Turabian StyleMavroidi, Angeliki, Elisavet Froukala, and Athanasios Tsakris. 2024. "Comparative Genomics of an Emerging Multidrug-Resistant blaNDM-Carrying ST182 Lineage in Enterobacter cloacae Complex" Antibiotics 13, no. 6: 535. https://doi.org/10.3390/antibiotics13060535
APA StyleMavroidi, A., Froukala, E., & Tsakris, A. (2024). Comparative Genomics of an Emerging Multidrug-Resistant blaNDM-Carrying ST182 Lineage in Enterobacter cloacae Complex. Antibiotics, 13(6), 535. https://doi.org/10.3390/antibiotics13060535







