Abstract
Colistin is a last-resort antimicrobial for treating multidrug-resistant Gram-negative bacteria. Phenotypic colistin resistance is highly associated with plasmid-mediated mobile colistin resistance (mcr) genes. mcr-bearing Enterobacteriaceae have been detected in many countries, with the emergence of colistin-resistant pathogens a global concern. This study assessed the distribution of mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 genes with phenotypic colistin resistance in isolates from diarrheal infants and children in Bangladesh. Bacteria were identified using the API-20E biochemical panel and 16s rDNA gene sequencing. Polymerase chain reactions detected mcr gene variants in the isolates. Their susceptibilities to colistin were determined by agar dilution and E-test by minimal inhibitory concentration (MIC) measurements. Over 31.6% (71/225) of isolates showed colistin resistance according to agar dilution assessment (MIC > 2 μg/mL). Overall, 15.5% of isolates carried mcr genes (7, mcr-1; 17, mcr-2; 13, and mcr-3, with co-occurrence occurring in two isolates). Clinical breakout MIC values (≥4 μg/mL) were associated with 91.3% of mcr-positive isolates. The mcr-positive pathogens included twenty Escherichia spp., five Shigella flexneri, five Citrobacter spp., two Klebsiella pneumoniae, and three Pseudomonas parafulva. The mcr-genes appeared to be significantly associated with phenotypic colistin resistance phenomena (p = 0.000), with 100% colistin-resistant isolates showing MDR phenomena. The age and sex of patients showed no significant association with detected mcr variants. Overall, mcr-associated colistin-resistant bacteria have emerged in Bangladesh, which warrants further research to determine their spread and instigate activities to reduce resistance.
1. Introduction
Emerging antimicrobial resistance (AMR) is a significant public health concern [1]. In 2019, it was estimated there were 4.95 million deaths globally associated with bacterial AMR, including 1.27 million deaths directly attributable to bacterial AMR, with the highest mortality currently seen in South Asian and sub-Saharan African countries [2]. In addition, considerable morbidity and costs are associated with AMR [3,4,5,6], with AMR driven by the overuse and misuse of antibiotics [7,8,9,10]. While AMR is a universal phenomenon, the burden among low- and middle-income countries (LMICs) is appreciably higher due to economic, political, and environmental factors, including poor governance and infrastructures, as well as a limited number of national initiatives [11,12,13,14]. This is now changing with global initiatives, including the Global Action Plan (GAP) by the World Health Organization (WHO) to reduce AMR [1] and others from the World Bank and OECD [6,15]. The GAP has been translated into National Action Plan (NAP), which includes Bangladesh, with LMICs at different stages of their implementation due to resource and other issues [16,17,18,19].
Other important global initiatives include dividing antibiotics into three different categories based on their resistance potential, which includes the ‘Access’, ‘Watch’, and ‘Reserve’ categories [20,21]. Antibiotics in the ‘Reserve’ category, which include fifth-generation cephalosporins, some carbapenems, and linezolid, should only be prescribed in multidrug resistance cases, with the aim of curbing rising AMR rates [20,22,23,24].
Colistin is also classified as a ‘Reserve’ antibiotic following the global increase in the prevalence of carbapenem-resistant Enterobacteriaceae [21,25], with concerns with its overuse and resultant resistance development [26,27]. Concerns with its overuse in both animals and humans, with resultant resistance development via zoonotic gene transfers, coupled with its importance in treating resistant Gram-negative infections to reduce morbidity and mortality, have resulted in the WHO and others classifying colistin as an antibiotic of very high importance for use in humans, with its use reserved [20,28,29,30,31,32,33,34]. Alongside this, many countries now ban the use of colistin as a growth promoter in animal feeds and prophylactically to prevent bacterial infections [35,36,37], with such measures shown to be effective in reducing resistant strains [36,37,38]. This is important in Bangladesh, given extensive colistin-resistant Escherichia coli in broiler meat and chicken feces [39,40,41] exacerbating resistance among patients to colistin in Bangladesh [42,43,44,45]. Over-the-counter dispensing of antibiotics is also common in Bangladesh and is a concern, especially when this involves ‘Reserve’ antibiotics [46,47,48,49]
The use of colistin as an antibiotic of last resort is greatly threatened by its overuse and the associated rise of plasmid-borne mobile colistin resistance genes [50,51,52], spreading rapidly via horizontal gene transfer [53]. Resistance to colistin is generated by the chromosomally mediated modification of lipopolysaccharide (LPS) [54]. The acquisition of colistin resistance by a novel plasmid-mediated gene, mcr-1, was first described in Enterobacteriales from both farm-animal products and humans [55]. Earlier studies have shown the genotypic linkage of the mobile colistin resistance gene mcr-1 to phenotypic colistin resistance [56,57]. Since then, variants of mcr-carrying multiple species of Enterobacteriales have been detected in many countries from environments, animals, and humans [58,59,60,61]. Subsequently, more variants of the transferable colistin resistance mcr gene (mcr-1 to mcr-9) have been described in Enterobacteriaceae [62,63].
This is a concern. A recent outbreak involving colistin-resistant pathogens in China resulted in a very high case-fatality rate in humans [64]. Consequently, the identification of the root cause, transmission, and trajectories of colistin-resistant infections is an increasing priority globally. We are aware that mcr-gene variants can be detected in the environment, animals, human fecal samples, and food products [65]. However, only a limited number of studies have also shown the dissemination of plasmids carrying these variants in infants with acute diarrhea [66]. This is important in Bangladesh, with diarrhea being a major cause of childhood mortality in the country, combined with the increasing prevalence of resistant genes in children, exacerbated by the overuse of antibiotics [67,68,69].
Consequently, this study was designed to investigate different variants of the mcr gene and their association with colistin resistance among diarrheal pathogens in infants, children, and adults in Bangladesh, as well as the demographic factors associated with identified mcr variants. The findings can be used to suggest future policies and initiatives in Bangladesh and beyond where there are concerns.
2. Results
2.1. Study Patients
We collected a total of 179 diarrheal stool samples from infants, children, and adults in different locations in Bangladesh (Figure 1) throughout our study. We subsequently isolated 228 distinct bacteria from 168 culture-positive diarrheal patients. The study patients comprised 102 (57%) males and 77 (43%) females.
Figure 1.
Sampling Location. Different sites are marked, indicating the spatial distribution of diarrheal patients from whom samples were collected. The map displays sampling sites across various cities in Bangladesh.
In 11 stool samples, no bacterial growth appeared. The majority of the study patients were infants and children who needed hospitalization (admitted to Uttara Medical College, Dhaka), and the median and interquartile range (IQR) age was 1.17 (0.75–2.5) years. Additionally, 78.1% of patients were middle class, 21.1% were poor, and 0.8% were rich (Figure 2A). The duration of diarrhea among all patients ranged from 3 to 40 days, and the mean duration (standard deviation) was 7.2 ± 4.78 days.
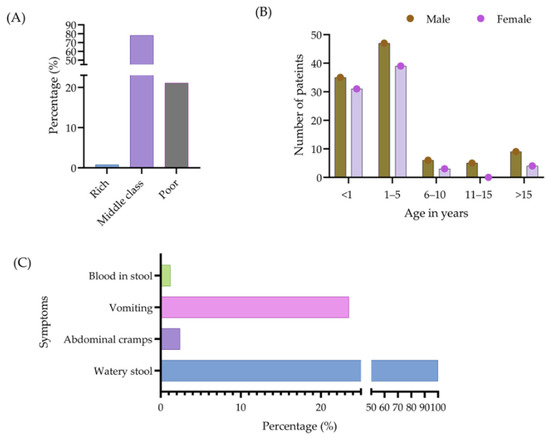
Figure 2.
Socioeconomic status of patients (A), as well as their age, sex (B), and clinical symptoms associated with diarrhea (C).
The ages of the patients were categorized into five groups, namely <1 year, 1–5 years, 6–10 years, 11–15 years, and >15 years, with disease occurrence highest in the 1–5 years age group (48%). Figure 2B provides further information regarding the incidence of diarrhea associated with each age group. Among all the patients, 179 (100%) appeared with watery stools, 4 (2.2%) presented with abdominal cramps, 39 (21.8%) with vomiting, and 2 (1.10%) patients with blood in their stools (Figure 2C).
2.2. Identification of Diarrheal Pathogens and Their Phenotypic Colistin Susceptibility
Of the 228 isolates, 140 were categorized as Escherichia spp. (61.40%), 20 as Citrobacter spp. (8.77%), 18 as Klebsiella spp. (7.89%), 13 as Shigella flexneri (5.70%), 10 as Enterobacter spp. (4.39%), and 7 as Stenotrophomonas maltophi (3.07%). Figure 3 contains details of all the pathogens identified. One Proteus sp. and two staphylococci were excluded from further study since colistin resistance is natural in Proteus and Staphylococcus species [70,71].
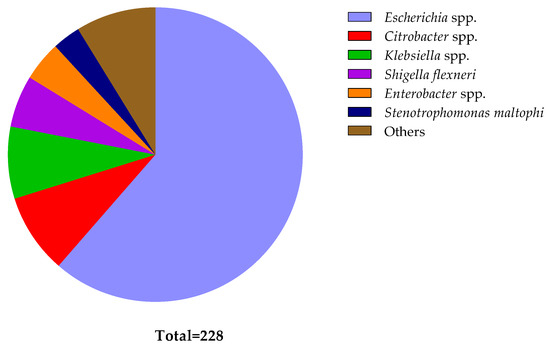
Figure 3.
Distribution of different bacteria identified from diarrheal patients. The pie chart illustrates the relative numbers of various bacteria identified among diarrheal patients. Each segment represents a specific bacterial species, with the size of each segment corresponding to the frequency of its occurrence in the sampled population. The ‘Other’ segment comprises bacteria found in singular instances (Bacillus cereus, Bacterium endosymbiont, Cronobacter sakazakii, Enterococcus faecium, Proteus mirabilis, Serratia marcescens, and Vibrio neocaledonicus), bacteria found in two (Acinetobacter spp. and Staphylococcus spp.), and bacteria found in three (Morganella morganii, Aeromonas caviae, and Pseudomonas parafulva).
The agar dilution method determined the test bacteria as susceptible (S) when there was no growth at ≤2 μg/mL colistin sulfate concentrations and resistant (R) when growth appeared at >2 μg/mL. In addition, the disk diffusion method was used to evaluate the antibacterial potency of colistin sulfate (25 µg) in vitro for the 225 isolates, and bacteria were considered colistin-resistant (R) if ≤10 mm diameter zone of inhibition was recorded (Materials and Methods Section (Section 4)). Among the 225 isolates, the agar dilution test revealed that 69 (30.7%) isolates were resistant to colistin sulfate. However, the colistin disk diffusion method showed that 180 (80.7%) isolates were resistant. Where possible, the results were validated by a minimum inhibitory concentration (MIC) assessment using the gold standard broth microdilution method. Overall, there was over 90% agreement in the results obtained between the agar dilution test and broth microdilution. In view of this, we considered the agar dilution test results as validated and subsequently disregarded the colistin disk diffusion results for subsequent analyses, similar to other authors [72].
2.3. Prevalence of mcr Genes in Diarrheal Isolates
All 225 isolates were subjected to polymerase chain reaction (PCR) to find mcr-1 to mcr-5 genes. Three types of mcr genes (mcr-1, mcr-2, and mcr-3) were detected in 35 isolates. These included 20 for Escherichia spp., five for Shigella flexneri, five for Citrobacter spp., two for Klebsiella pneumoniae and Enterobacter hormaechei, and one for Pseudomonas parafulva. Bacteria identified from 10 other genera did not appear with mcr genes (Table 1).

Table 1.
Identified diarrheal pathogens carrying mcr gene variants a.
Of the 35 mcr positive isolates, co-occurrence was identified in 2 isolates, one containing mcr-1 and mcr-2 and the other containing mcr-2, and mcr-3. The harborages of mcr-1, mcr-2, and mcr-3 were identified as 3.1% (7 isolates), 7.6% (17 isolates), and 5.8% (13 isolates), respectively. Combined, the presence of mcr variants was 15.56% (35/225).
2.4. Phenotypic–Genotypic Association
Of the 35 mcr-positive isolates, the agar dilution test identified 32 (91.4%) resistant isolates and 3 (8.6%) sensitive isolates, which revealed a very high statistical significance of mcr variant gene associations (p = 0.000). Further separate analyses showed very significantly high statistical associations of mcr-1, mcr-2, and mcr-3 with phenotypic colistin resistance (p = 0.000 for all three gene variants) (Table 2). All of the test isolates grew well on the control plate without colistin sulfate. As a susceptible control, the Escherichia coli ATCC 25922 strain with an MIC of 2 µg/mL was employed.

Table 2.
Phenotypic genotypic association of mcr genes and colistin resistance.
The MIC was determined by E-test and agar dilution test separately, using a range of ≤0.5 µg/mL to >256 µg/mL (Materials and Methods Section (Section 4)). The median and IQR MIC for mcr-positive isolates were 32 (8–128) µg/mL (Table 3). One isolate (E. coli) with co-carriage of mcr-1 and mcr-2 exhibited MIC values of 128 µg/mL. The other co-carrying bacterium (Escherichia coli) with mcr-2 and mcr-3 showed an MIC value of 8 µg/mL. The three mcr-positive bacteria that showed phenotypic susceptibilities to colistin sulfate were Citrobacter portucalensis, Citrobacter freundii, and Escherichia coli (Table 3). The median and IQR MIC for mcr-negative isolates was 1 (0.5–2) µg/mL, while 20% of the mcr-negative isolates (38/190) exhibited resistance to colistin sulfate in agar dilution, with the MIC range from 4 µg/mL to >128 µg/mL (Figure 4).

Table 3.
Diarrheal isolates with identified mcr-gene variants and associated minimum inhibitory concentration of colistin.
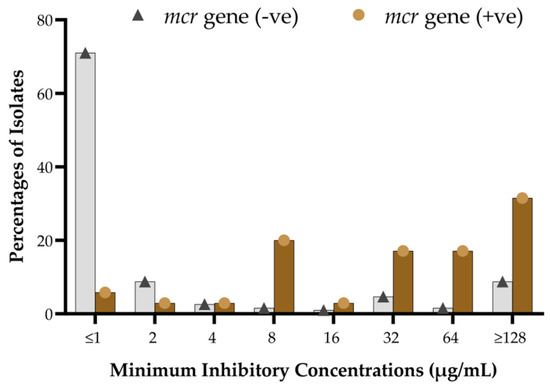
Figure 4.
Distribution of MIC levels of colistin sulfate among mcr-positive and mcr-negative isolates.
MIC analyses at each specific value between mcr-positive and mcr-negative isolates showed that most mcr-carrying isolates were identified with higher MIC levels of colistin sulfate, ranging from >8 µg/mL to >128 µg/mL (Figure 4). At the same time, the vast majority of the mcr-negative isolates exhibited lower MIC levels, ranging from ≤0.5 µg/mL to 2 µg/mL. A fraction of mcr-negative isolates demonstrated MIC values from 32 µg/mL to >128 µg/mL (Figure 4). The overall colistin resistance rate was 31.6% (71/225), with MIC50 and MIC90 at 1 µg/mL and 128 µg/mL, respectively. Conversely, MIC50/MIC90 for the mcr-positive and mcr-negative subpopulations were 32 µg/mL/128 µg/mL and 1.0 µg/mL/64 µg/mL, respectively.
2.5. Multidrug Resistance and mcr-Carriage
All mcr-positive isolates (carrying at least one mcr gene) were examined for the presence of MDR. The disk diffusion test was conducted to identify whether the 35 mcr-positive isolates exhibited susceptibility to the other 17 different antibiotics from 8 different groups or not. Remarkably, 100% of mcr-positive organisms exhibited MDR outcomes (Table 4), and mcr-negative isolates revealed a comparatively lower frequency of MDR outcomes.

Table 4.
Multidrug resistance (MDR) phenomena of mcr-positive isolates.
2.6. Demographic Factors Associated with mcr-Carriage
More than one type of bacterial isolate was isolated and analyzed from each stool sample. Consequently, 130 bacteria were evaluated from 95 male stool samples and the other 95 from 73 female stool samples. Bacteria isolated from female stool samples carried more proportionate mcr genes (16.8%, 16/95) in comparison to male-origin isolates (14.6%, 19/130). In the five different age groups of the study population, almost uniform gene distributions were reported. Overall, the results showed that the sex and age groups were not significantly associated with the presence of mcr-1, mcr-2, and mcr-3 (p = 0.445 to 0.781) (Table 5).

Table 5.
Demographic factors associated with mcr genes (n = 225).
3. Discussion
We believe this is one of the first studies in Bangladesh to investigate colistin-resistant genes in human diarrheal pathogens among infants and children, who comprised an appreciable percentage of participants. Our data clearly showed the association between phenotypic colistin resistance and the mcr genes (mcr-1 to mcr-5), similar to other published studies on colistin-resistant bacteria isolated from diarrhea [66,74,75]. This builds on earlier studies in Bangladesh, including among children and adults [60,76].
Isolates harboring mcr genes have been detected with high MIC values, showcasing disparities between the agar dilution test and the disk diffusion method [77]. This variation can be attributed to the slow diffusion of colistin disks on agar medium for the complex and large molecular structure of colistin sulfate. In parallel, some mcr-negative isolates exhibited resistance to colistin. Several potential reasons may account for this phenomenon. Firstly, mutations in the mgrB gene [78], responsible for binding polymyxin antibiotics in the Gram-negative cell wall, are particularly prevalent in Klebsiella pneumoniae [79,80]. Secondly, the absence of testing for other variants of mcr genes, including mcr-6, mcr-7, mcr-8, mcr-9, and mcr-10, which could also attribute phenotypic colistin resistance [81,82]. Thirdly, some resistant bacteria may develop capsules, polysaccharide coatings on the outer surface of the cell wall [83,84]. Fourthly, overexpression of efflux pump systems could contribute to resistance development [85,86]. Fifthly, modulation in the bacterial cell surface, including alterations in the structure of lipopolysaccharides (LPSs) of the cell membrane, might affect the binding of polymyxin antibiotics [87]. Additional investigations will be necessary to uncover potential molecular explanations for the observed differences between phenotypic and genotypic colistin resistance, and we will be looking at this more closely in the future.
On the other hand, three isolates appeared phenotypically susceptible to colistin (MIC values 0.5–2.0 µg/mL) in the presence of mcr-2/mcr-1 genes. The observation states that the carriage mcr gene variants are not sufficient to confer phenotypic colistin resistance. These findings contradict many earlier reports stating that mcr genes provide colistin resistance [34,38,41]. One research identified some genetic substitution mutations, and their reversions are associated with the acquisition and loss of colistin resistance, respectively [88]. The different susceptibilities exhibited could be explained by unstable mobile genetic elements, rare mutations, environmental modulations, and loss of cell membrane LPSs [89,90,91,92]. Comprehensive whole-genome sequencing (WGS) analysis of the bacterial population may decipher the factors that facilitate the evolution of colistin resistance.
In this study, male children had a higher incidence rate of diarrhea than female children, similar to previous studies [93,94]. However, the reason for this difference is unclear. We also found that 57.14% of mcr-positive isolates were resistant to amoxicillin/clavulanic acid, and 57 to 71% of mcr-carrying isolates were resistant to all generations of cephalosporins, although higher-generation cephalosporins were more susceptible. This is a concern, with clinicians now prescribing more carbapenems due to the decreased potency of cephalosporins, with meropenem showing more susceptibility among the 17 different antibiotics in the eight groups studied. However, recent studies showed carbapenem-resistant Enterobacteriaceae are now a global threat [95,96,97]. The MDR status of all mcr-positive isolates is positive, which is a threat to public health in Bangladesh and beyond, given the ensuing rise in untreatable infectious diseases [98,99,100]. High MDR associated with mcr gene acquisition may influence the pleiotropic resistance of the isolates against a broad group of antimicrobials [101].
Consequently, this study’s findings underscore the significance of establishing comprehensive surveillance systems for priority antibiotics, such as colistin, to help reduce rising AMR rates. Alongside this, we urge the government of Bangladesh to ban the use of colistin as a growth promoter in animal feeds and prophylactically to prevent bacterial infections, similar to other countries [35,36,37]. This is because such measures have been shown to be effective in reducing resistant strains [36,37,53]. Alongside this, instigate antimicrobial stewardship programs (ASPs) across care settings for colistin to other important antibiotics in Bangladesh to reduce resistant strains [102,103,104,105]. This includes ASPs among community pharmacists and drug sellers, building on ASPs in other sectors in Bangladesh [47,106].
We are aware that there are several limitations to this study. Firstly, it was challenging to accurately estimate the real-world scenario since adults experiencing diarrheal problems could readily seek treatment at a healthcare center, whereas children relied on their parents’ decisions and assistance to access medical care. Secondly, the small sample size posed significant obstacles to conducting fully powered statistical analyses, alongside the fact that we undertook convenient sampling. However, our research spanned 15 distinct districts across Bangladesh, and the outcomes are anticipated to be applicable if similar studies are conducted in other districts in the country. Thirdly, while this study examined mcr gene variants up to mcr-5, newer variants, such as mcr-6, mcr-7, mcr-8, and mcr-9, were not investigated. Fourthly, we are aware that a cross-sectional study could not ascertain patients’ health outcomes or treatment outcomes in linkage to mcr gene-associated colistin resistance. Having said this, efforts were made to maintain internal validity by conducting independent trials when necessary. Despite these limitations, we believe our findings are robust, providing guidance to all key stakeholders in Bangladesh to enhance future sensitivity to colistin.
4. Materials and Methods
4.1. Study Design and Sampling
A prospective cross-sectional study was conducted between January 2020 and December 2020 among children and young adults visiting the outpatient department of Uttara Adhunik Medical College Hospital, Dhaka, Bangladesh. A total of 179 children and adults with acute diarrhea participated in this study prior to treatment with any prescribed antibiotics.
Acute diarrhea was defined as three or more liquid, loose, mucous, or bloody stools within 24 h, lasting no longer than 14 days. Fever was defined as a temperature of ≥37.5 °C. Demographic data were obtained from each participant, and informed consent was obtained from the parents or guardians before sample collection. All relevant demographic, clinical, and laboratory data were recorded and transferred to the questionnaire prepared for this study.
The ages of the children and patients were categorized into five groups, namely <1 year, 1–5 years, 6–10 years, 11–15 years, and >15 years, based on previous studies [107]. The income of parents was classified as rich, middle-class, or poor; as we are aware, this can make a difference [108].
A sterilized cotton swab was dipped in the mucous, purulent, or bloody part of the freshly passed stool sample, placed immediately in CaryBlair Medium (Oxoid, Hampshire, UK), and transported to the laboratory for further analysis within six hours of collection.
4.2. Isolation and Identification of Bacteria
Collected samples were pre-enriched in buffered peptone water (Oxoid®, Hampshire, UK) at a dilution ratio of 1:10 and were incubated overnight at 37 °C. A loopful of each culture was streaked on MacConkey agar (Liofilchem Inc., Roseto degli Abruzzi TE, Italy) and cysteine-, lactose-, and electrolyte-deficient (CLED) agar (Liofilchem Inc., Roseto degli Abruzzi TE, Italy), and subsequently incubated simultaneously at 37 °C for 24 h in aerobic conditions. MacConkey agar supports Gram-negative diarrheal pathogens (Supplementary Figure S1A), while CLED agar aids in the growth of Gram-negative bacteria and Gram-positive cocci, if present in diarrheal samples. Colony counts of 103 or 105 CFU/mL were considered as a cutoff value for a probable diarrheal sample [109].
Gram’s staining and biochemical tests were initially performed to identify growth-positive bacteria. A rapid biochemical test kit API 20E (BioMe’rieux, Durham, NC, USA), consisting of carbohydrate batteries and enzymatic substrates in a set of chromogenic panels, was used to verify the isolated identity (Supplementary Figure S1B) [110]. Part of the bacterial identity was confirmed by the polymerase chain reaction (PCR) amplification and sequencing of the 16S rDNA gene [111]. In total, 228 different isolates were generated from all the diarrheal samples. Three bacteria (one Proteus and two staphylococci) were excluded from the study for the next level analyses since colistin resistance is a natural phenomenon of the excluded isolates. The remaining 225 isolates were subjected to an assessment of colistin susceptibility and mcr-1 to mcr-5 carriage. The isolates were preserved in 30% glycerol in Trypticase Soy Broth (TSB) at −80 °C until further use.
4.3. Phenotypic Colistin Susceptibility Testing
The phenotypic antibiogram profiles of diarrheal isolates against colistin were determined primarily using the Kirby–Bauer disk diffusion method, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Clinical and Laboratory Standards Institute (CLSI) guidelines (Supplementary Figure S1C) [112,113]. A 3-h bacterial suspension in Mueller–Hinton broth was prepared to a concentration of McFarland 0.5 equivalent and then streaked on Mueller–Hinton agar (MHA, Oxoid, Basingstoke, UK) plates using a cotton swab to ensure consistent growth. The susceptibility of the isolate to the following disks of antibiotics (Oxoid, Basingstoke, UK) was evaluated: colistin (25 µg), amoxycillin + clavulanic acid (30 µg), cefuroxime sodium (30 µg), cefixime (30 µg), cefepime (30 µg), imipenem (10 µg), meropenem (10 µg), nalidixic acid (10 µg), ciprofloxacin (5 µg), lomefloxacin (10 µg), levofloxacin (5 µg), gentamicin (30 µg), amikacin (30 µg), netilmicin (30 µg), tobramycin (10 µg), nitrofurantoin(300 µg), and trimethoprim-sulfamethoxazole (25 µg) by placing them on the bacterial lawns and incubating at 37 °C overnight. A clear zone was developed around the disk for sensitive bacteria, and the zone diameter was measured and evaluated to categorize bacteria as susceptible (S), intermediate (I), and resistant (R) from the CLSI guideline charts for the appropriate antibiotics tested [113].
The isolates’ phenotypic antimicrobial susceptibilities were further tested using the agar dilution method [114]. The lowest concentration of colistin adequate to inhibit the visible growth of bacterial test isolates was determined by minimal inhibitory concentration (MIC) measurement via the agar dilution method [114]. In order to determine the agar dilution MIC, different concentrations of colistin sulfate powder (Santa Cruz Biotechnology Inc., Dallas, TX, USA) from 0.50 µg/mL to 256 µg/mL in a two-fold dilution order were incorporated into the MHA medium accordingly. One pure culture colony was inoculated into Mueller–Hinton broth to prepare each test inoculum. Subsequently, it was incubated for three hours at 37 °C to develop a density of inoculum equivalent to 104 colony-forming units (CFU) per spot to drop on the MHA. The inoculum density was periodically compared to a 0.5 McFarland standard, equivalent to approximately 108 CFU/mL. The plates were incubated at 37 °C in air for 18–20 h. Agar dilution MICs were performed in duplicate. The experiments were repeated when some single colonies were seen or a thin haze growth was observed within the inoculated spot.
The epsilometer test (E-test) was performed partly—using a commercial strip containing a predefined gradient of colistin concentrations (Liofilchem Inc., Roseto degli Abruzzi TE, Italy) to validate colistin MIC determination using the agar dilution method [77,115]. Concordant results were found in independent MIC assessments and E-tests (Supplementary Figure S1D,E), as stated earlier [72]. The E. coli ATCC 25922 strain was used as the quality control strain for disk diffusion and MIC testing. In addition, a control plate without colistin sulfate was examined for the growth of both test and control isolates. Following EUCAST and CLSI guidelines, isolates were considered susceptible (S) when the MIC values exhibited ≤2 μg/mL and resistant (R) breakpoints when MICs appeared >2 μg/mL [112]. MIC50 and MIC90 values were calculated to report the concentration of colistin that can inhibit the growth of test pathogens by 50%/90%. Multidrug-resistant (MDR) isolates were described as those isolates that were found to be resistant to at least three different classes of antibiotics [116].
4.4. Detection of Colistin Resistance mcr Genes
All 225 isolates were subjected to a singleplex polymerase chain reaction (PCR) to detect the mcr-1 gene, yielding a 309 bp DNA band, using primers described elsewhere [55], and confirmed by sequencing. Amplicons were visualized under UV light after 1.2% agarose gel electrophoresis. The other four primer pairs to detect mcr-2, mcr-3, mcr-4, and mcr-5 gene amplicons were obtained from a recently published original study [117]. A multiplex polymerase chain reaction (PCR) was conducted to detect the mcr-1 to mcr-5 genes in the isolates. In brief, the modified protocol was as follows: prepared bacterial DNA (2 μL) was added to a 2× PCR premixture (15 μL, GeneON, Rhein, Germany), and five pmol of each primer (1 μL), and deionized water was added to obtain a final volume of 30 μL. Reactions went through an initial denaturation at 94 °C for 15 min followed by 25 cycles of amplification (Applied Biosystems 2720 Thermal Cycler, Singapore), consisting of denaturation for 30 s at 94 °C, annealing for 90 s at 55 °C, and extension for 1 min at 72 °C, and a final 10 min elongation at 72 °C. Expected amplicons for mcr-1 (309 bp), mcr-2 (715 bp), mcr-3 (929 bp), mcr-4 (1116 bp), and mcr-5 (1644 bp) underwent electrophoresis through 1.2% agarose gel, followed by staining with ethidium bromide, and were visualized under UV light (Supplementary Figure S1F). Lastly, the obtained results were validated by separate singleplex PCR analyses of the mcr genes.
4.5. Statistical Analysis
Using the IBM SPSS statistics data editor (version 21) and GraphPad prism software (version 9.5), verified data were entered and then examined. The bivariate analysis did not include missing data. The mcr gene variations carried by diarrheal pathogens and their phenotypic traits were described using both descriptive and inferential statistical methods. Any associations between categorical data were examined using Pearson’s chi-square test with the appropriate use of Yate’s continuity correction. Fisher’s exact test results of a 2 × 2 contingency table were presented in place of the chi-square test results if the predicted frequency of the test could not be assumed. Two-tailed p-values were computed, with a significance level of 0.05.
4.6. Ethics Statements
This study was authorized [No. UAMC/ERC/Recommend-62/2018, dated 9 July 2018] by the Ethics Review Committee of Uttara Adhunik Medical College. All research protocols complied with the Declaration of Helsinki regarding the use of human beings in research.
Each adult study participant provided written informed consent before the collection of their urine samples. For patients under the age of 18, parents or legal guardians were asked separately for written informed permission. The patients’ identities were anonymized.
5. Conclusions
Our findings indicate that multidrug-resistant pathogenic bacteria containing mcr genes are a major reservoir in the guts of young Bangladeshi children and adults. Overall, the mcr-1, mcr-2, and mcr-3 variants appear to predominate over other variants, such as mcr-4 and mcr-5, in the diarrheal bacteria of children and adults in Bangladesh. However, newer variants, such as mcr-6, mcr-7, mcr-8, and mcr-9, were not investigated in this study. Overall, we did not find any association between phenotypic colistin resistance and age or sex.
The advent of clinical MDR pathogens resistant to colistin, which is designated as an antibiotic of last resort, is a growing concern in Bangladesh and other countries, as it leads to infectious diseases that are subsequently difficult to treat. These findings require immediate monitoring and action from the government and other key stakeholders in Bangladesh in line with the goals of the NAP. Proposed activities include additional research activities to uncover potential molecular explanations for the observed differences between phenotypic and genotypic colistin resistance, as well as limiting the use of colistin as a growth promoter agent in animal husbandry in Bangladesh, similar to other countries. Concerted effort is especially important, as AMR is increasingly being transferred from animals to humans as a result of improper antibiotic use in animal feeding. Alongside this, there is a need to instigate the active monitoring of colistin resistance and the resistance patterns of other important antibiotics, which is also in line with the goals and activities of the Bangladesh NAP. There also needs to be increased patient education to address concerns with hygiene levels in the country, as well as instigating measures to improve the availability of safe drinking water. Other important activities in Bangladesh include instigating pertinent ASPs in ambulatory care to help physicians, pharmacists, and drug sellers reduce inappropriate prescribing and dispensing of colistin. This builds on successful ASPs in ambulatory care in other LMICs. We will continue to monitor the situation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13060534/s1. The supporting information on detailed procedures for bacterial isolation, antibiotic susceptibility testing, and mcr detection via Polymerase Chain Reaction (PCR), as outlined in Supplementary Figure S1.
Author Contributions
S.S. and R.M.N.: Sample collection, Methodology, Investigation, Formal analysis, and Manuscript drafting; M.B.H.: Data acquisition, Investigation, Data validation, Visualization; U.L.U. and M.A.A.: Methodology, Data curation, Writing—Reviewing, Validation, and Editing; A.S.M.M. and M.R.K.K.: Clinical and demographic data acquisition, Resources, Conceptualization, Visualization, Validation, and Manuscript reviewing; S.N.: Project administration and coordination, Resources, Methodology, Validation, and Visualization; B.G.: Conceptualization, Methodology, Rewriting and Editing, and Visualization; S.I.: Conceptualization, Supervision, Resources, Data curation and analysis, Writing–Reviewing, Editing, and Study coordination. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part from a research grant from Bangladesh Medical Research Council (BMRC, Award ID: HPNSDP 2019-20/605). The grant provided support in biospecimen collection, data collection, laboratory investigation, and a stipend for a co-author (Shafiuzzaman Sarker). The grant had no expenditure provision for manuscript processing and publication.
Institutional Review Board Statement
This study was authorized [No. UAMC/ERC/Recommend-62/2018, dated 9 July 2018] by the Ethics Review Committee of Uttara Adhunik Medical College, Dhaka, Bangladesh.
Informed Consent Statement
Informed consent was obtained from parents before the start of sample collection.
Data Availability Statement
Data are contained within the article and available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Global Action Plan on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- European Antimicrobial Resistance Collaborators. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: A cross-country systematic analysis. Lancet Public Health 2022, 7, e897–e913. [Google Scholar] [CrossRef]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef]
- Hofer, U. The cost of antimicrobial resistance. Nat. Rev. Microbiol. 2019, 17, 3. [Google Scholar] [CrossRef]
- World Bank Group. Pulling Together to Beat Superbugs Knowledge and Implementation Gaps in Addressing Antimicrobial Resistance; World Bank: Washington, DC, USA, 2019. [Google Scholar]
- Hossain, M.J.; Jabin, N.; Ahmmed, F.; Sultana, A.; Abdur Rahman, S.; Islam, M.R. Irrational use of antibiotics and factors associated with antibiotic resistance: Findings from a cross—sectional study in Bangladesh. Health Sci. Rep. 2023, 6, e1465. [Google Scholar] [CrossRef]
- Akram, F.; Imtiaz, M.; ul Haq, I. Emergent crisis of antibiotic resistance: A silent pandemic threat to 21st century. Microb. Pathog. 2023, 174, 105923. [Google Scholar] [CrossRef]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef]
- Van, T.T.H.; Yidana, Z.; Smooker, P.M.; Coloe, P.J. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J. Glob. Antimicrob. Resist. 2020, 20, 170–177. [Google Scholar] [CrossRef]
- Sulis, G.; Sayood, S.; Gandra, S. Antimicrobial resistance in low- and middle-income countries: Current status and future directions. Expert. Rev. Anti-Infect. Ther. 2022, 20, 147–160. [Google Scholar] [CrossRef]
- Iskandar, K.; Molinier, L.; Hallit, S.; Sartelli, M.; Hardcastle, T.C.; Haque, M.; Lugova, H.; Dhingra, S.; Sharma, P.; Islam, S.; et al. Surveillance of antimicrobial resistance in low- and middle-income countries: A scattered picture. Antimicrob. Resist. Infect. Control 2021, 10, 63. [Google Scholar] [CrossRef]
- Collignon, P.; Athukorala, P.C.; Senanayake, S.; Khan, F. Antimicrobial resistance: The major contribution of poor governance and corruption to this growing problem. PLoS ONE 2015, 10, e0116746. [Google Scholar] [CrossRef]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef]
- Fernandes, M.R.; Ignacio, A.; Rodrigues, V.A.; Groppo, F.C.; Cardoso, A.L.; Avila-Campos, M.J.; Nakano, V. Alterations of Intestinal Microbiome by Antibiotic Therapy in Hospitalized Children. Microb. Drug Resist. 2017, 23, 56–62. [Google Scholar] [CrossRef]
- Jouy, E.; Haenni, M.; Le Devendec, L.; Le Roux, A.; Chatre, P.; Madec, J.Y.; Kempf, I. Improvement in routine detection of colistin resistance in E. coli isolated in veterinary diagnostic laboratories. J. Microbiol. Methods 2017, 132, 125–127. [Google Scholar] [CrossRef]
- Chua, A.Q.; Verma, M.; Hsu, L.Y.; Legido-Quigley, H. An analysis of national action plans on antimicrobial resistance in Southeast Asia using a governance framework approach. Lancet Reg. Health West. Pac. 2021, 7, 100084. [Google Scholar] [CrossRef]
- Willemsen, A.; Reid, S.; Assefa, Y. A review of national action plans on antimicrobial resistance: Strengths and weaknesses. Antimicrob. Resist. Infect. Control 2022, 11, 90. [Google Scholar] [CrossRef]
- Godman, B.; Egwuenu, A.; Wesangula, E.; Schellack, N.; Kalungia, A.C.; Tiroyakgosi, C.; Kgatlwane, J.; Mwita, J.C.; Patrick, O.; Niba, L.L.; et al. Tackling antimicrobial resistance across sub-Saharan Africa: Current challenges and implications for the future. Expert. Opin. Drug Saf. 2022, 21, 1089–1111. [Google Scholar] [CrossRef]
- Sharland, M.; Pulcini, C.; Harbarth, S.; Zeng, M.; Gandra, S.; Mathur, S.; Magrini, N. Classifying antibiotics in the WHO Essential Medicines List for optimal use-be AWaRe. Lancet Infect. Dis. 2018, 18, 18–20. [Google Scholar] [CrossRef]
- Sharland, M.; Gandra, S.; Huttner, B.; Moja, L.; Pulcini, C.; Zeng, M.; Mendelson, M.; Cappello, B.; Cooke, G.; Magrini, N. Encouraging AWaRe-ness and discouraging inappropriate antibiotic use-the new 2019 Essential Medicines List becomes a global antibiotic stewardship tool. Lancet Infect. Dis. 2019, 19, 1278–1280. [Google Scholar] [CrossRef]
- Klein, E.Y.; Milkowska-Shibata, M.; Tseng, K.K.; Sharland, M.; Gandra, S.; Pulcini, C.; Laxminarayan, R. Assessment of WHO antibiotic consumption and access targets in 76 countries, 2000–2015: An analysis of pharmaceutical sales data. Lancet Infect. Dis. 2021, 21, 107–115. [Google Scholar] [CrossRef]
- Sharland, M.; Zanichelli, V.; Ombajo, L.A.; Bazira, J.; Cappello, B.; Chitatanga, R.; Chuki, P.; Gandra, S.; Getahun, H.; Harbarth, S.; et al. The WHO essential medicines list AWaRe book: From a list to a quality improvement system. Clin. Microbiol. Infect. 2022, 28, 1533–1535. [Google Scholar] [CrossRef]
- Hsia, Y.; Lee, B.R.; Versporten, A.; Yang, Y.; Bielicki, J.; Jackson, C.; Newland, J.; Goossens, H.; Magrini, N.; Sharland, M. Use of the WHO Access, Watch, and Reserve classification to define patterns of hospital antibiotic use (AWaRe): An analysis of paediatric survey data from 56 countries. Lancet Glob. Health 2019, 7, e861–e871. [Google Scholar] [CrossRef]
- Lu, Q.; Li, G.-H.; Qu, Q.; Zhu, H.-H.; Luo, Y.; Yan, H.; Yuan, H.-Y.; Qu, J. Clinical efficacy of polymyxin B in patients infected with carbapenem-resistant organisms. Infect. Drug Resist. 2021, 14, 1979–1988. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Zhang, S.; Abbas, M.; Rehman, M.U.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhu, D.; Zhao, X.; Gao, Q.; et al. Updates on the global dissemination of colistin-resistant Escherichia coli: An emerging threat to public health. Sci. Total Environ. 2021, 799, 149280. [Google Scholar] [CrossRef]
- WHO. Critically Important Antimicrobials for Human Medicine. 6th Revision 2018; Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance Due to Non-Human Use; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- EMA. Categorisation of Antibiotics Used in Animals Promotes Responsible Use to Protect Public and Animal Health; EMA: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Talat, A.; Miranda, C.; Poeta, P.; Khan, A.U. Farm to table: Colistin resistance hitchhiking through food. Arch. Microbiol. 2023, 205, 167. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm animals and aquaculture: Significant reservoirs of mobile colistin resistance genes. Environ. Microbiol. 2020, 22, 2469–2484. [Google Scholar] [CrossRef]
- Javed, H.; Saleem, S.; Zafar, A.; Ghafoor, A.; Shahzad, A.B.; Ejaz, H.; Junaid, K.; Jahan, S. Emergence of plasmid-mediated mcr genes from Gram-negative bacteria at the human-animal interface. Gut Pathog. 2020, 12, 54. [Google Scholar] [CrossRef]
- Yin, Y.; Qiu, L.; Wang, G.; Guo, Z.; Wang, Z.; Qiu, J.; Li, R. Emergence and Transmission of Plasmid-Mediated Mobile Colistin Resistance Gene mcr-10 in Humans and Companion Animals. Microbiol. Spectr. 2022, 10, e02097-22. [Google Scholar] [CrossRef]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef]
- Mendelson, M.; Brink, A.; Gouws, J.; Mbelle, N.; Naidoo, V.; Pople, T.; Schellack, N.; van Vuuren, M.; Rees, H. The One Health stewardship of colistin as an antibiotic of last resort for human health in South Africa. Lancet Infect. Dis. 2018, 18, e288–e294. [Google Scholar] [CrossRef]
- Ribeiro, S.; Mourão, J.; Novais, Â.; Campos, J.; Peixe, L.; Antunes, P. From farm to fork: Colistin voluntary withdrawal in Portuguese farms reflected in decreasing occurrence of mcr-1-carrying Enterobacteriaceae from chicken meat. Environ. Microbiol. 2021, 23, 7563–7577. [Google Scholar] [CrossRef]
- Usui, M.; Nozawa, Y.; Fukuda, A.; Sato, T.; Yamada, M.; Makita, K.; Tamura, Y. Decreased colistin resistance and mcr-1 prevalence in pig-derived Escherichia coli in Japan after banning colistin as a feed additive. J. Glob. Antimicrob. Resist. 2021, 24, 383–386. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef]
- Nath, C.; Das, T.; Islam, M.S.; Hasib, F.M.Y.; Singha, S.; Dutta, A.; Barua, H.; Islam, M.Z. Colistin Resistance in Multidrug-Resistant Escherichia coli Isolated from Retail Broiler Meat in Bangladesh. Microb. Drug Resist. 2023, 29, 523. [Google Scholar] [CrossRef]
- Uddin, M.B.; Alam, M.N.; Hasan, M.; Hossain, S.M.B.; Debnath, M.; Begum, R.; Samad, M.A.; Hoque, S.F.; Chowdhury, M.S.R.; Rahman, M.M.; et al. Molecular Detection of Colistin Resistance mcr-1 Gene in Multidrug-Resistant Escherichia coli Isolated from Chicken. Antibiotics 2022, 11, 97. [Google Scholar] [CrossRef]
- Islam, S.; Urmi, U.L.; Rana, M.; Sultana, F.; Jahan, N.; Hossain, B.; Iqbal, S.; Hossain, M.M.; Mosaddek, A.S.M.; Nahar, S. High abundance of the colistin resistance gene mcr-1 in chicken gut-bacteria in Bangladesh. Sci. Rep. 2020, 10, 17292. [Google Scholar] [CrossRef]
- Sonia, S.J.; Uddin, K.H.; Shamsuzzaman, S.M. Prevalence of Colistin Resistance in Klebsiella pneumoniae Isolated from a Tertiary Care Hospital in Bangladesh and Molecular Characterization of Colistin Resistance Genes among Them by Polymerase Chain Reaction and Sequencing. Mymensingh Med. J. 2022, 31, 733–740. [Google Scholar]
- Ara, B.; Urmi, U.L.; Haque, T.A.; Nahar, S.; Rumnaz, A.; Ali, T.; Alam, M.S.; Mosaddek, A.S.M.; Rahman, N.A.A.; Haque, M.; et al. Detection of mobile colistin-resistance gene variants (mcr-1 and mcr-2) in urinary tract pathogens in Bangladesh: The last resort of infectious disease management colistin efficacy is under threat. Expert Rev. Clin. Pharmacol. 2021, 14, 513–522. [Google Scholar] [CrossRef]
- Dutta, A.; Islam, M.Z.; Barua, H.; Rana, E.A.; Jalal, M.S.; Dhar, P.K.; Das, A.; Das, T.; Sarma, S.M.; Biswas, S.K.; et al. Acquisition of Plasmid-Mediated Colistin Resistance Gene mcr-1 in Escherichia coli of Livestock Origin in Bangladesh. Microb. Drug Resist. 2020, 26, 1058–1062. [Google Scholar] [CrossRef]
- Kawser, Z.; Shamsuzzaman, S.M. Association of Virulence with Antimicrobial Resistance among Klebsiella pneumoniae Isolated from Hospital Settings in Bangladesh. Int. J. Appl. Basic Med. Res. 2022, 12, 123–129. [Google Scholar] [CrossRef]
- Rousham, E.K.; Nahar, P.; Uddin, M.R.; Islam, M.A.; Nizame, F.A.; Khisa, N.; Akter, S.M.S.; Munim, M.S.; Rahman, M.; Unicomb, L. Gender and urban-rural influences on antibiotic purchasing and prescription use in retail drug shops: A one health study. BMC Public Health 2023, 23, 229. [Google Scholar] [CrossRef]
- Unicomb, L.E.; Nizame, F.A.; Uddin, M.R.; Nahar, P.; Lucas, P.J.; Khisa, N.; Akter, S.M.S.; Islam, M.A.; Rahman, M.; Rousham, E.K. Motivating antibiotic stewardship in Bangladesh: Identifying audiences and target behaviours using the behaviour change wheel. BMC Public Health 2021, 21, 968. [Google Scholar] [CrossRef]
- Orubu, E.S.F.; Samad, M.A.; Rahman, M.T.; Zaman, M.H.; Wirtz, V.J. Mapping the Antimicrobial Supply Chain in Bangladesh: A Scoping-Review-Based Ecological Assessment Approach. Glob. Health Sci. Pract. 2021, 9, 532–547. [Google Scholar] [CrossRef]
- Islam, M.A.; Akhtar, Z.; Hassan, M.Z.; Chowdhury, S.; Rashid, M.M.; Aleem, M.A.; Ghosh, P.K.; Mah-E-Muneer, S.; Parveen, S.; Ahmmed, M.K.; et al. Pattern of Antibiotic Dispensing at Pharmacies According to the WHO Access, Watch, Reserve (AWaRe) Classification in Bangladesh. Antibiotics 2022, 11, 247. [Google Scholar] [CrossRef]
- Liu, J.-H.; Liu, Y.-Y.; Shen, Y.-B.; Yang, J.; Walsh, T.R.; Wang, Y.; Shen, J. Plasmid-mediated colistin-resistance genes: mcr. Trends Microbiol. 2023, 32, 365–378. [Google Scholar] [CrossRef]
- Phuadraksa, T.; Wichit, S.; Songtawee, N.; Tantimavanich, S.; Isarankura-Na-Ayudhya, C.; Yainoy, S. Emergence of plasmid-mediated colistin resistance mcr-3.5 gene in Citrobacter amalonaticus and Citrobacter sedlakii isolated from healthy individual in Thailand. Front. Cell. Infect. Microbiol. 2023, 12, 1917. [Google Scholar] [CrossRef]
- Zelendova, M.; Papagiannitsis, C.C.; Sismova, P.; Medvecky, M.; Pomorska, K.; Palkovicova, J.; Nesporova, K.; Jakubu, V.; Jamborova, I.; Zemlickova, H. Plasmid-mediated colistin resistance among human clinical Enterobacterales isolates: National surveillance in the Czech Republic. Front. Microbiol. 2023, 14, 1147846. [Google Scholar] [CrossRef]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Ćwiek, K.; Woźniak-Biel, A.; Karwańska, M.; Siedlecka, M.; Lammens, C.; Rebelo, A.R.; Hendriksen, R.S.; Kuczkowski, M.; Chmielewska-Władyka, M.; Wieliczko, A. Phenotypic and genotypic characterization of mcr-1-positive multidrug-resistant Escherichia coli ST93, ST117, ST156, ST10, and ST744 isolated from poultry in Poland. Braz. J. Microbiol. 2021, 52, 1597–1609. [Google Scholar] [CrossRef]
- Elias, R.; Spadar, A.; Phelan, J.; Melo-Cristino, J.; Lito, L.; Pinto, M.; Gonçalves, L.; Campino, S.; Clark, T.G.; Duarte, A. A phylogenomic approach for the analysis of colistin resistance-associated genes in Klebsiella pneumoniae, its mutational diversity and implications for phenotypic resistance. Int. J. Antimicrob. Agents 2022, 59, 106581. [Google Scholar] [CrossRef]
- Karim, M.R.; Zakaria, Z.; Hassan, L.; Faiz, N.M.; Ahmad, N.I. The occurrence and molecular detection of mcr-1 and mcr-5 genes in Enterobacteriaceae isolated from poultry and poultry meats in Malaysia. Front. Microbiol. 2023, 14, 1208314. [Google Scholar] [CrossRef]
- Rebelo, A.; MH, C.L.; Bortolaia, V.; Kjeldgaard, J.; Hendriksen, R. PCR for Plasmid-Mediated Colistin Resistance Genes, mcr-1, mcr-2, mcr-3, mcr-4, mcr-5 and Variants (Multiplex); DTU National Food Institute: Kgs Lyngby, Denmark, 2018. [Google Scholar]
- Johura, F.-T.; Tasnim, J.; Barman, I.; Biswas, S.R.; Jubyda, F.T.; Sultana, M.; George, C.M.; Camilli, A.; Seed, K.D.; Ahmed, N. Colistin-resistant Escherichia coli carrying mcr-1 in food, water, hand rinse, and healthy human gut in Bangladesh. Gut Pathog. 2020, 12, 1–8. [Google Scholar] [CrossRef]
- Perdomo, A.; Webb, H.E.; Bugarel, M.; Friedman, C.R.; Francois Watkins, L.K.; Loneragan, G.H.; Calle, A. First Known Report of mcr-Harboring Enterobacteriaceae in the Dominican Republic. Int. J. Environ. Res. Public Health 2023, 20, 5123. [Google Scholar] [CrossRef]
- Ling, Z.; Yin, W.; Shen, Z.; Wang, Y.; Shen, J.; Walsh, T.R. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 2020, 75, 3087–3095. [Google Scholar] [CrossRef]
- Lemlem, M.; Aklilu, E.; Mohamed, M.; Kamaruzzaman, N.F.; Zakaria, Z.; Harun, A.; Devan, S.S.; Kamaruzaman, I.N.A.; Reduan, M.F.H.; Saravanan, M. Phenotypic and genotypic characterization of colistin-resistant Escherichia coli with mcr-4, mcr-5, mcr-6, and mcr-9 genes from broiler chicken and farm environment. BMC Microbiol. 2023, 23, 392. [Google Scholar] [CrossRef]
- Tian, G.-B.; Doi, Y.; Shen, J.; Walsh, T.R.; Wang, Y.; Zhang, R.; Huang, X. MCR-1-producing Klebsiella pneumoniae outbreak in China. Lancet Infect. Dis. 2017, 17, 577. [Google Scholar] [CrossRef]
- Schwarz, S.; Johnson, A.P. Transferable resistance to colistin: A new but old threat. J. Antimicrob. Chemother. 2016, 71, 2066–2070. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Chen, M.; Hu, J.; Zhang, H.; Xiang, Y.; Yang, H.; Qiu, S.; Song, H. Plasmid-borne colistin resistance gene mcr-1 in a multidrug resistant Salmonella enterica serovar Typhimurium isolate from an infant with acute diarrhea in China. Int. J. Infect. Dis. 2021, 103, 13–18. [Google Scholar] [CrossRef]
- Billah, S.M.; Raihana, S.; Ali, N.B.; Iqbal, A.; Rahman, M.M.; Khan, A.N.S.; Karim, F.; Karim, M.A.; Hassan, A.; Jackson, B.; et al. Bangladesh: A success case in combating childhood diarrhoea. J. Glob. Health 2019, 9, 020803. [Google Scholar] [CrossRef]
- Sharif, N.; Nobel, N.U.; Sakib, N.; Liza, S.M.; Khan, S.T.; Billah, B.; Parvez, A.K.; Haque, A.; Talukder, A.A.; Dey, S.K. Molecular and Epidemiologic Analysis of Diarrheal Pathogens in Children With Acute Gastroenteritis in Bangladesh During 2014–2019. Pediatr. Infect. Dis. J. 2020, 39, 580–585. [Google Scholar] [CrossRef]
- Islam, M.R.; Nuzhat, S.; Fahim, S.M.; Palit, P.; Flannery, R.L.; Kyle, D.J.; Mahfuz, M.; Islam, M.M.; Sarker, S.A.; Ahmed, T. Antibiotic exposure among young infants suffering from diarrhoea in Bangladesh. J. Paediatr. Child Health 2021, 57, 395–402. [Google Scholar] [CrossRef]
- Khedher, M.B.; Baron, S.A.; Riziki, T.; Ruimy, R.; Raoult, D.; Diene, S.M.; Rolain, J.-M. Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: Is there another role for these enzymes? Sci. Rep. 2020, 10, 5970. [Google Scholar] [CrossRef]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965. [Google Scholar] [CrossRef]
- Lo-Ten-Foe, J.R.; de Smet, A.M.G.; Diederen, B.M.; Kluytmans, J.A.; van Keulen, P.H. Comparative evaluation of the VITEK 2, disk diffusion, Etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob. Agents Chemother. 2007, 51, 3726–3730. [Google Scholar] [CrossRef]
- Cosentino, F.; Viale, P.; Giannella, M. MDR/XDR/PDR or DTR? Which definition best fits the resistance profile of Pseudomonas aeruginosa? Curr. Opin. Infect. Dis. 2023, 36, 564–571. [Google Scholar] [CrossRef]
- Gu, D.-x.; Huang, Y.-l.; Ma, J.-h.; Zhou, H.-w.; Fang, Y.; Cai, J.-c.; Hu, Y.-y.; Zhang, R. Detection of colistin resistance gene mcr-1 in hypervirulent Klebsiella pneumoniae and Escherichia coli isolates from an infant with diarrhea in China. Antimicrob. Agents Chemother. 2016, 60, 5099–5100. [Google Scholar] [CrossRef]
- Feng, J.; Zhuang, Y.; Luo, J.; Xiao, Q.; Wu, Y.; Chen, Y.; Chen, M.; Zhang, X. Prevalence of colistin-resistant mcr-1-positive Escherichia coli isolated from children patients with diarrhoea in Shanghai, 2016–2021. J. Glob. Antimicrob. Resist. 2023, 34, 166–175. [Google Scholar] [CrossRef]
- Monira, S.; Shabnam, S.A.; Ali, S.; Sadique, A.; Johura, F.-T.; Rahman, K.Z.; Alam, N.H.; Watanabe, H.; Alam, M. Multi-drug resistant pathogenic bacteria in the gut of young children in Bangladesh. Gut Pathog. 2017, 9, 19. [Google Scholar] [CrossRef]
- Maalej, S.M.; Meziou, M.R.; Rhimi, F.M.; Hammami, A. Comparison of disc diffusion, Etest and agar dilution for susceptibility testing of colistin against Enterobacteriaceae. Lett. Appl. Microbiol. 2011, 53, 546–551. [Google Scholar] [CrossRef]
- Sato, T.; Shiraishi, T.; Hiyama, Y.; Honda, H.; Shinagawa, M.; Usui, M.; Kuronuma, K.; Masumori, N.; Takahashi, S.; Tamura, Y. Contribution of novel amino acid alterations in pmrA or pmrB to Colistin resistance in mcr-negative Escherichia coli clinical isolates, including major multidrug-resistant lineages O25b: H4-ST131-H 30Rx and Non-x. Antimicrob. Agents Chemother. 2018, 62, e00864-18. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Bontron, S.; Villegas, M.-V.; Ozdamar, M.; Türkoglu, S.; Nordmann, P. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 75–80. [Google Scholar] [CrossRef]
- Cannatelli, A.; Giani, T.; D’Andrea, M.M.; Di Pilato, V.; Arena, F.; Conte, V.; Tryfinopoulou, K.; Vatopoulos, A.; Rossolini, G.M. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 2014, 58, 5696–5703. [Google Scholar] [CrossRef]
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front. Microbiol. 2020, 11, 80. [Google Scholar] [CrossRef]
- Hussein, N.H.; AL-Kadmy, I.; Taha, B.M.; Hussein, J.D. Mobilized colistin resistance (mcr) genes from 1 to 10: A comprehensive review. Mol. Biol. Rep. 2021, 48, 2897–2907. [Google Scholar] [CrossRef]
- Formosa, C.; Herold, M.; Vidaillac, C.; Duval, R.; Dague, E. Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J. Antimicrob. Chemother. 2015, 70, 2261–2270. [Google Scholar] [CrossRef]
- Campos, M.; Vargas, M.; Regueiro, V.; Llompart, C.; Albertí, S.; Bengoechea, J.A. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 2004, 72, 7107–7114. [Google Scholar] [CrossRef]
- Bengoechea, J.A.; Skurnik, M. Temperature—regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol. Microbiol. 2000, 37, 67–80. [Google Scholar] [CrossRef]
- Puja, H.; Bolard, A.; Noguès, A.; Plésiat, P.; Jeannot, K. The efflux pump MexXY/OprM contributes to the tolerance and acquired resistance of Pseudomonas aeruginosa to colistin. Antimicrob. Agents Chemother. 2020, 64, e02033-19. [Google Scholar] [CrossRef]
- NationRL, L. Colistin in the 21st century. Curr. Opin. Infect. Dis. 2009, 22, 535–543. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Park, Y.K.; Chung, E.S.; Na, I.Y.; Ko, K.S. Evolved resistance to colistin and its loss due to genetic reversion in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 25543. [Google Scholar] [CrossRef]
- Nicoloff, H.; Hjort, K.; Levin, B.R.; Andersson, D.I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 2019, 4, 504–514. [Google Scholar] [CrossRef]
- Hughes, D.; Andersson, D.I. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 374–391. [Google Scholar] [CrossRef]
- Carretero-Ledesma, M.; García-Quintanilla, M.; Martín-Peña, R.; Pulido, M.R.; Pachón, J.; McConnell, M.J. Phenotypic changes associated with Colistin resistance due to Lipopolysaccharide loss in Acinetobacter baumannii. Virulence 2018, 9, 930–942. [Google Scholar] [CrossRef]
- Jayol, A.; Nordmann, P.; Brink, A.; Poirel, L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 2015, 59, 2780–2784. [Google Scholar] [CrossRef]
- Jarman, A.F.; Long, S.E.; Robertson, S.E.; Nasrin, S.; Alam, N.H.; McGregor, A.J.; Levine, A.C. Sex and gender differences in acute pediatric diarrhea: A secondary analysis of the Dhaka study. J. Epidemiol. Glob. Health 2018, 8, 42. [Google Scholar] [CrossRef]
- Mero, W.; Jameel, A.Y.; Amidy, K.S.K. Microorganisms and viruses causing diarrhea in infants and primary school children and their relation with age and sex in Zakho city, Kurdistan Region, Iraq. Int. J. Res. Med. Sci. 2015, 3, 3266–3273. [Google Scholar] [CrossRef]
- Gupta, N.; Limbago, B.M.; Patel, J.B.; Kallen, A.J. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 2011, 53, 60–67. [Google Scholar] [CrossRef]
- Smith, H.Z.; Hollingshead, C.M.; Kendall, B. Carbapenem-Resistant Enterobacterales. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Hansen, G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021, 10, 75–92. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef]
- Sharma, J.; Sharma, D.; Singh, A.; Sunita, K. Colistin Resistance and Management of Drug Resistant Infections. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 4315030. [Google Scholar] [CrossRef]
- Davies, M.; Walsh, T.R. A colistin crisis in India. Lancet Infect. Dis. 2018, 18, 256–257. [Google Scholar] [CrossRef]
- Cantón, R.; Ruiz-Garbajosa, P. Co-resistance: An opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 2011, 11, 477–485. [Google Scholar] [CrossRef]
- Godman, B.; Egwuenu, A.; Haque, M.; Malande, O.O.; Schellack, N.; Kumar, S.; Saleem, Z.; Sneddon, J.; Hoxha, I.; Islam, S.; et al. Strategies to Improve Antimicrobial Utilization with a Special Focus on Developing Countries. Life 2021, 11, 528. [Google Scholar] [CrossRef]
- Haque, T.A.; Urmi, U.L.; Islam, A.B.M.M.K.; Ara, B.; Nahar, S.; Mosaddek, A.S.M.; Lugova, H.; Kumar, S.; Jahan, D.; Rahman, N.A.A.; et al. Detection of qnr genes and gyrA mutation to quinolone phenotypic resistance of UTI pathogens in Bangladesh and the implications. Appl. Pharm. Sci. 2022, 12, 185–198. [Google Scholar] [CrossRef]
- Shanta, A.S.; Islam, N.; Asad, M.A.; Akter, K.; Habib, M.B.; Hossain, M.J.; Nahar, S.; Godman, B.; Islam, S. Resistance and Co-resistance of Metallo-Beta-Lactamase Genes in Diarrheal and Urinary Tract Pathogens in Bangladesh. Preprints 2024. [Google Scholar] [CrossRef]
- Haque, M.; Godman, B. Potential strategies to improve antimicrobial utilisation in hospitals in Bangladesh building on experiences across developing countries. Bangladesh J. Med. Sci. 2021, 20, 469–477. [Google Scholar] [CrossRef]
- Harun, M.G.D.; Anwar, M.M.U.; Sumon, S.A.; Hassan, M.Z.; Mohona, T.M.; Rahman, A.; Abdullah, S.; Islam, M.S.; Kaydos-Daniels, S.C.; Styczynski, A.R. Rationale and guidance for strengthening infection prevention and control measures and antimicrobial stewardship programs in Bangladesh: A study protocol. BMC Health Serv. Res. 2022, 22, 1239. [Google Scholar] [CrossRef]
- Sears, C.L.; Islam, S.; Saha, A.; Arjumand, M.; Alam, N.H.; Faruque, A.; Salam, M.; Shin, J.; Hecht, D.; Weintraub, A. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin. Infect. Dis. 2008, 47, 797–803. [Google Scholar] [CrossRef]
- Banerjee, A.V.; Duflo, E. What is middle class about the middle classes around the world? J. Econ. Perspect. 2008, 22, 3–28. [Google Scholar] [CrossRef]
- Okuma, T.; Nakamura, M.; Totake, H.; Fukunaga, Y. Microbial contamination of enteral feeding formulas and diarrhea. Nutrition 2000, 16, 719–722. [Google Scholar] [CrossRef]
- Djim-Adjim-Ngana, K.; Oumar, L.A.; Mbiakop, B.W.; Njifon, H.L.M.; Crucitti, T.; Nchiwan, E.N.; Yanou, N.N.; Deweerdt, L. Prevalence of extended-spectrum beta-lactamase-producing enterobacterial urinary infections and associated risk factors in small children of Garoua, Northern Cameroon. Pan Afr. Med. J. 2020, 36, 157. [Google Scholar] [CrossRef]
- Van Der Zee, A.; Roorda, L.; Bosman, G.; Ossewaarde, J.M. Molecular diagnosis of urinary tract infections by semi-quantitative detection of uropathogens in a routine clinical hospital setting. PLoS ONE 2016, 11, e0150755. [Google Scholar] [CrossRef]
- Satlin, M.J.; Lewis, J.S.; Weinstein, M.P.; Patel, J.; Humphries, R.M.; Kahlmeter, G.; Giske, C.G.; Turnidge, J. Clinical and Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing Position Statements on Polymyxin B and Colistin Clinical Breakpoints. Clin. Infect. Dis. 2020, 71, e523–e529. [Google Scholar] [CrossRef]
- CLSI M100; Performance Standards for Antimicrobial Susceptibility Testing. The Clinical and Laboratory Standards Institute (CLSI): Malvern, PA, USA, 2018; Volume 28.
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Behera, B.; Mathur, P.; Das, A.; Kapil, A.; Gupta, B.; Bhoi, S.; Farooque, K.; Sharma, V.; Misra, M.C. Evaluation of susceptibility testing methods for polymyxin. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2010, 14, e596–e601. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018, 23, 17-00672. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).