Heterogeneous Antibiotic Resistance Gene Removal Impedes Evaluation of Constructed Wetlands for Effective Greywater Treatment
Abstract
1. Introduction
2. Results and Discussion
2.1. Physicochemical Analysis of Influent and Effluent
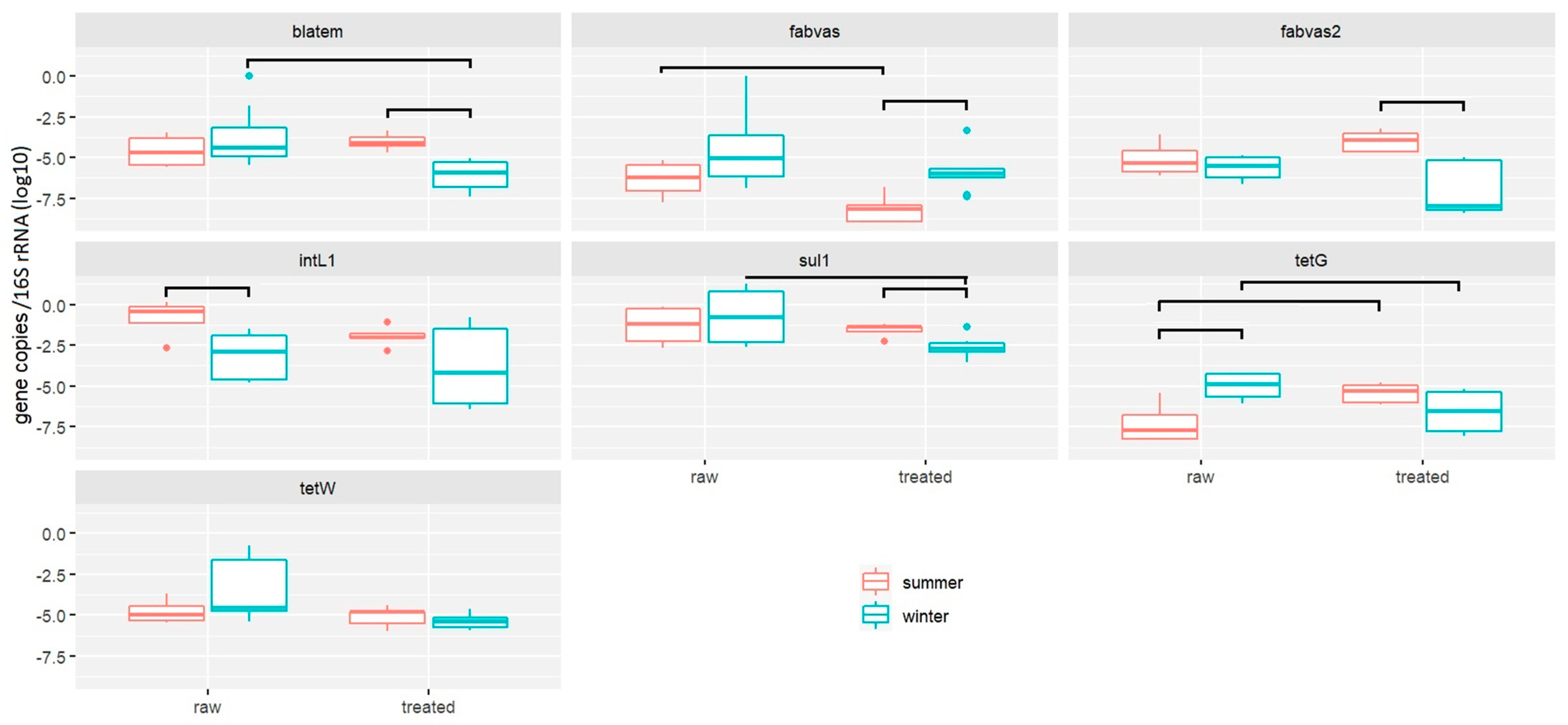
2.1.1. Performance of RVFCW for ARG Removal from Greywater
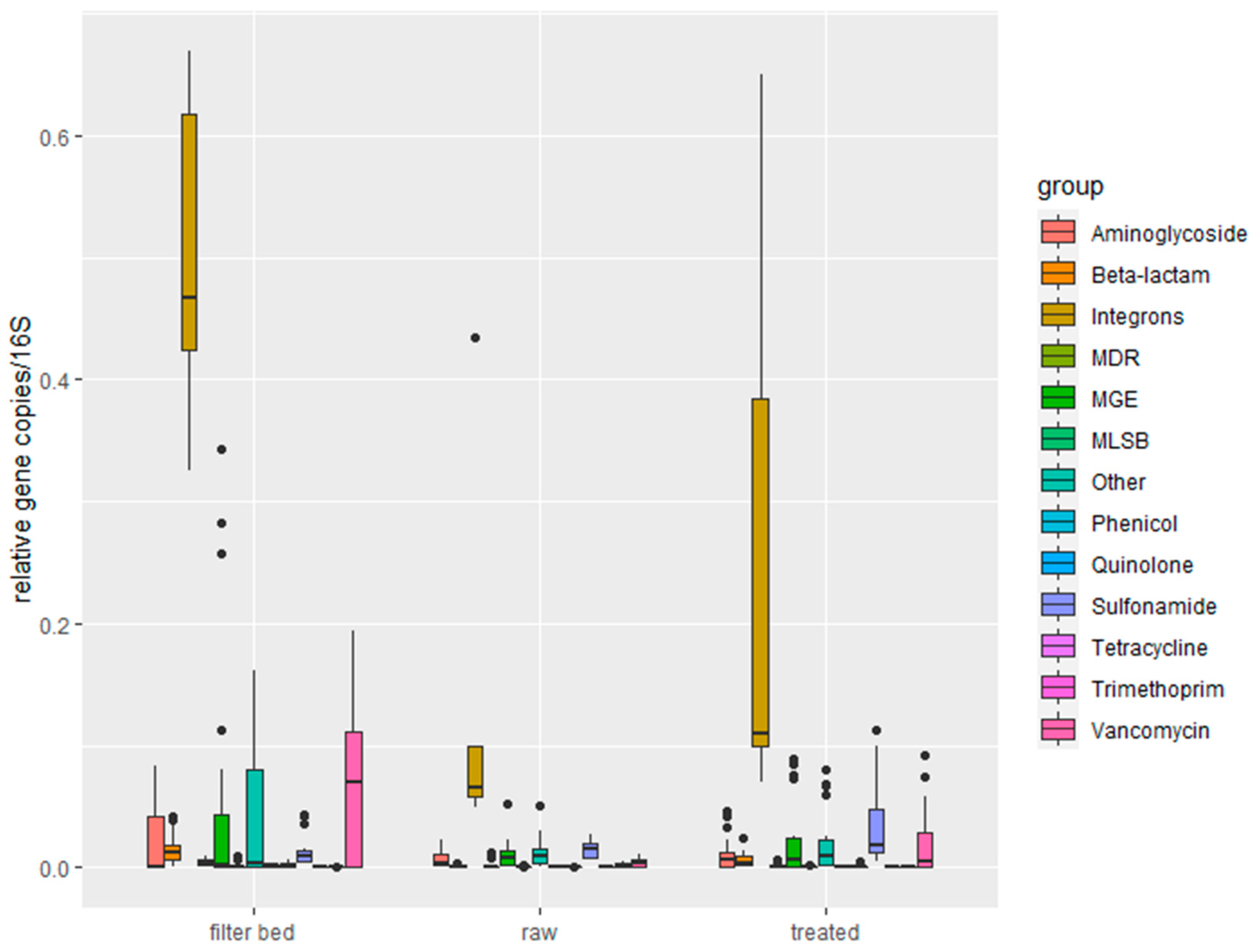
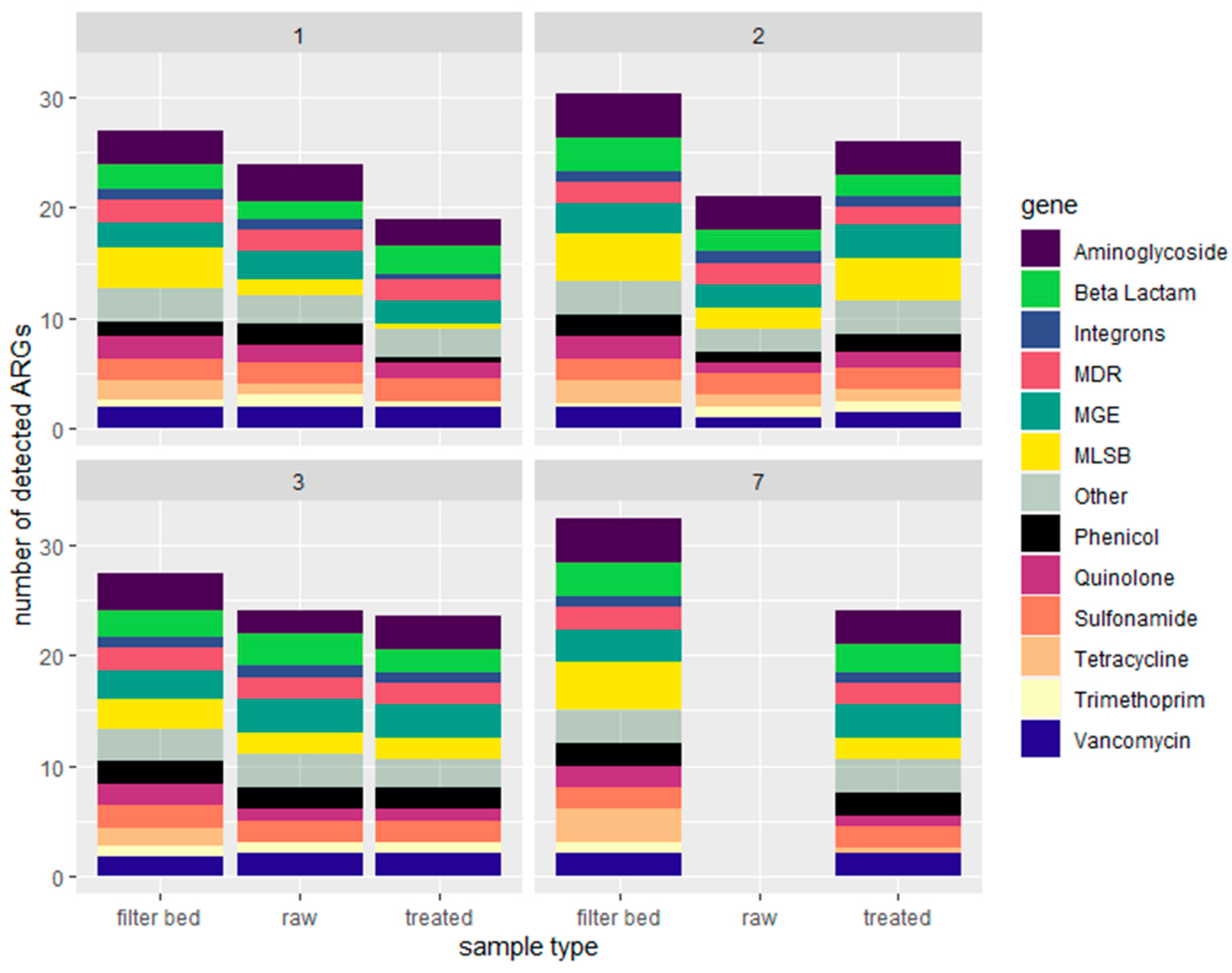
2.1.2. Quantification of Antimicrobial Resistance Genes by Resistomap
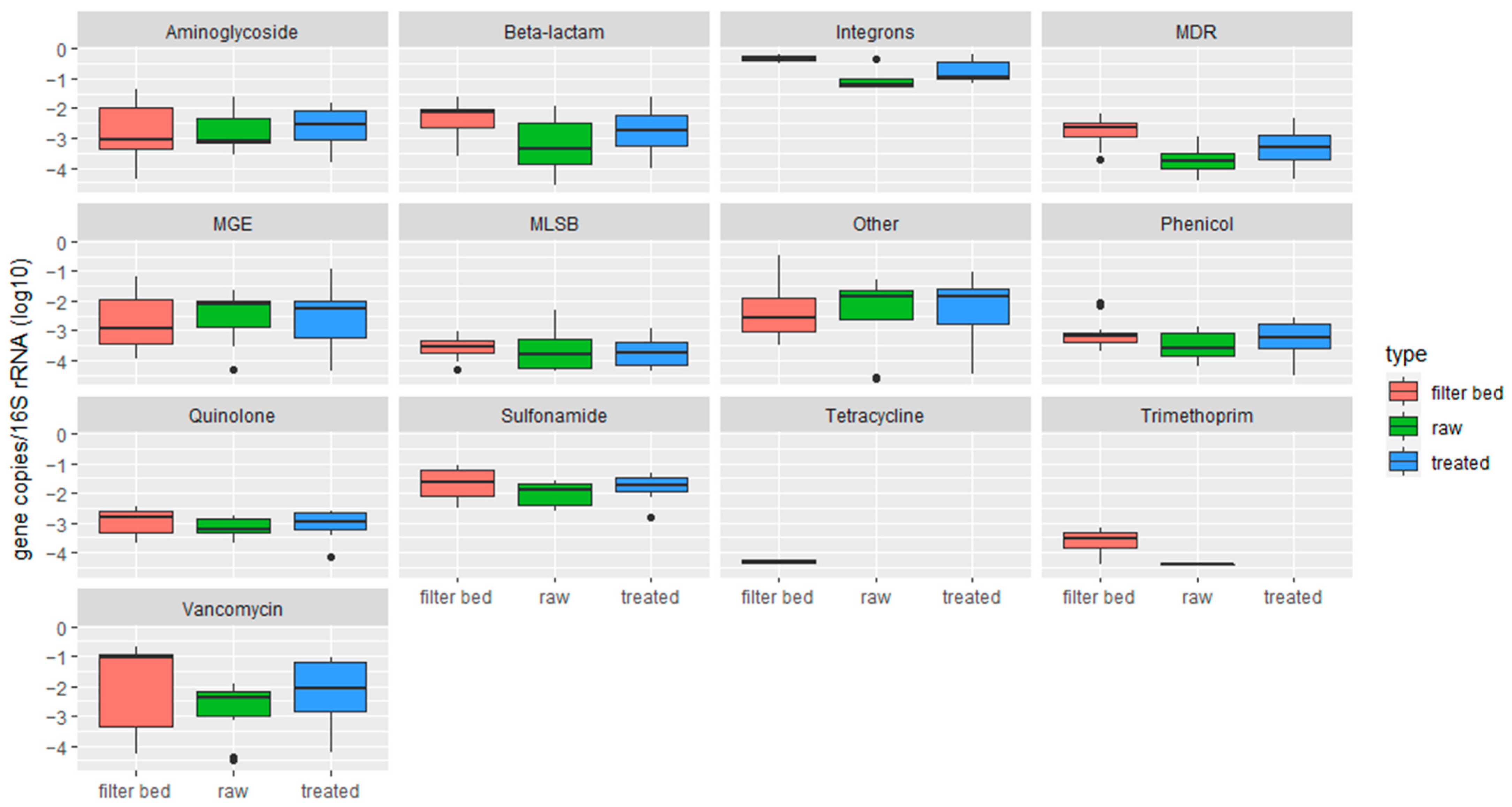
2.2. Characterization of Soil Samples and Quantification of Detected ARGs
2.2.1. Analysis and Quantification of Emerging Micropollutants in the Soil
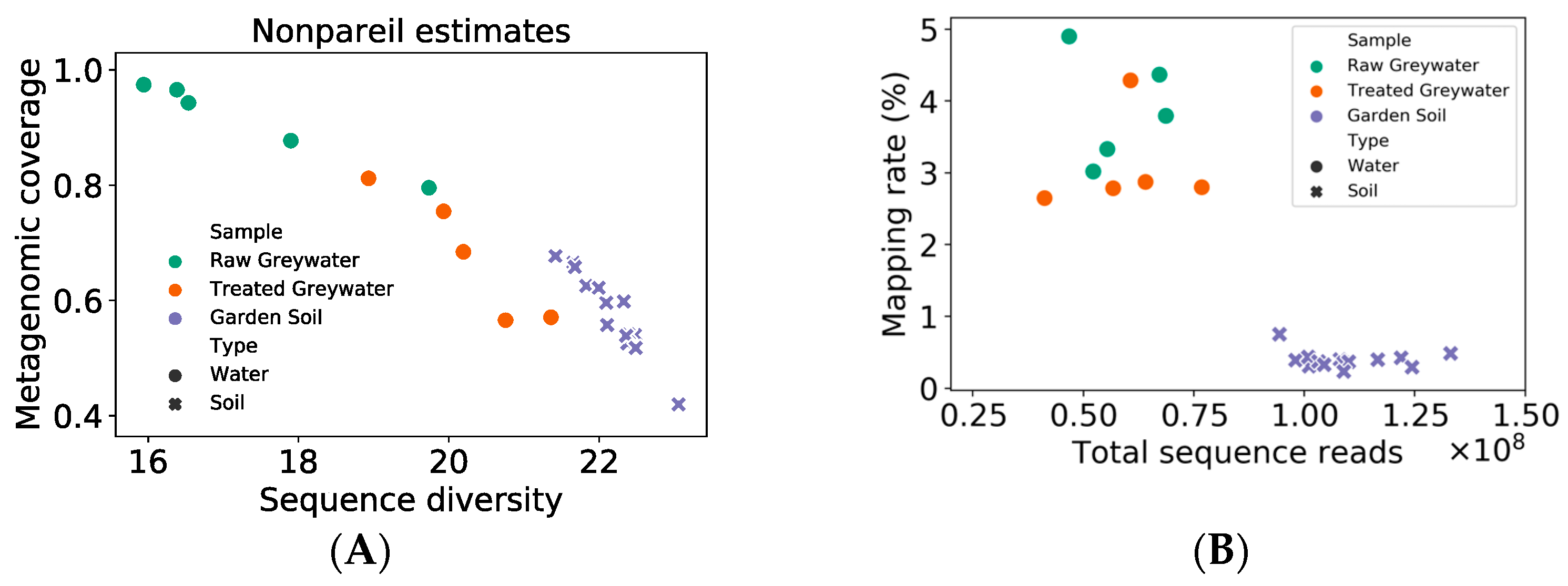
2.2.2. ARG Identification through Short Read-Based Metagenomic Analyses
3. Materials and Methods
3.1. DNA Extraction and qPCR
3.2. Detection of Micropollutants by LCMS
3.3. SmartChip™ Analysis
3.4. Metagenomics
3.5. Statistical Analysis
4. Conclusions
- (1)
- RVFCWs performance in relation to ARGs is highly variable: qPCR analysis presented that the total bacterial abundance was reduced in most households after treatment. In the winter season, most of the ARGs were significantly decreased after treatment, specifically for genes tetG, sul1, and blaTEM. Resistomap analysis analyzed 34 genes simultaneously; filter bed samples had the most microbial load compared to the raw and treated greywater samples. Genes conferring resistance to aminoglycoside, beta-lactam, vancomycin, and quinolone were increased in treated greywater;
- (2)
- Overall risks and impact on soil are low: The type of irrigation water (potable or treated greywater) had no specific influence on the soil’s total bacterial abundance (16S rRNA gene). No overlapping ARGs were found when tracing from treated greywater to soil irrigated with treated greywater;
- (3)
- More characterization of these systems will better reveal how they work, enabling more robust design and ensuring risks stay low: Future research should assess factors that modify the effect of wastewater irrigation on ARGs in soil. High throughput qPCR and metagenomics should be used to comprehensively evaluate ARGs in different soil types. Moreover, shotgun sequencing of the filter bed could give an insight into changes in the catabolic pathways and enzymes with the potential for biodegradation of micropollutants.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hazra, M.; Joshi, H.; Williams, J.B.; Watts, J.E.M. Antibiotics and antibiotic resistant bacteria/genes in urban wastewater: A comparison of their fate in conventional treatment systems and constructed wetlands. Chemosphere 2022, 303, 135148. [Google Scholar] [CrossRef]
- Bueno, I.; Verdugo, C.; Jimenez-Lopez, O.; Alvarez, P.P.; Gonzalez-Rocha, G.; Lima, C.A.; Travis, D.A.; Wass, B.; Zhang, Q.; Ishii, S.; et al. Role of wastewater treatment plants on environmental abundance of Antimicrobial Resistance Genes in Chilean rivers. Int. J. Hyg. Environ. Health 2020, 223, 56–64. [Google Scholar] [CrossRef]
- Lamba, M.; Ahammad, S.Z. Sewage treatment effluents in Delhi: A key contributor of Β-lactam resistant bacteria and genes to the environment. Chemosphere 2017, 188, 249–256. [Google Scholar] [CrossRef]
- Calderón-Franco, D.; Apoorva, S.; Medema, G.; van Loosdrecht, M.C.M.; Weissbrodt, D.G. Upgrading residues from wastewater and drinking water treatment plants as low-cost adsorbents to remove extracellular DNA and microorganisms carrying antibiotic resistance genes from treated effluents. Sci. Total Environ. 2021, 778, 146364. [Google Scholar] [CrossRef]
- Pant, A.; Shahadat, M.; Ali, S.W.; Ahammad, S.Z. Removal of antimicrobial resistance from secondary treated wastewater—A review. J. Hazard. Mater. Adv. 2022, 8, 100189. [Google Scholar] [CrossRef]
- Davies, J.; Davies, M. Origins and evolution of antibiotic resistance. Microbiología 2010, 12, 9–16. [Google Scholar] [CrossRef]
- Gatica, J.; Cytryn, E. Impact of treated wastewater irrigation on antibiotic resistance in the soil microbiome. Environ. Sci. Pollut. Res. 2013, 20, 3529–3538. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef]
- Giuliano, C.; Rybak, M. Efficacy of Triclosan as an Antimicrobial Hand Soap and Its Potential Impact on Antimicrobial Resistance: A Focused Review. Pharmacotherapy 2015, 35, 328–336. [Google Scholar] [CrossRef]
- Hartmann, E.M.; Hickey, R.; Hsu, T.; Román, C.M.B.; Chen, J.; Schwager, R.; Kline, J.; Brown, G.Z.; Halden, R.U.; Huttenhower, C.; et al. Antimicrobial Chemicals Are Associated with Elevated Antibiotic Resistance Genes in the Indoor Dust Microbiome. Environ. Sci. Technol. 2016, 50, 9807–9815. [Google Scholar] [CrossRef]
- Goudarzi, M.; Navidinia, M. Overview perspective of bacterial strategies of resistance to biocides and antibiotics. Arch. Clin. Infect. Dis. 2019, 14, e65744. [Google Scholar] [CrossRef]
- Romero, J.L.; Burgos, M.J.G.; Pérez-Pulido, R.; Gálvez, A.; Lucas, R. Resistance to Antibiotics, Biocides, Preservatives and Metals in Bacteria Isolated from Seafoods: Co-Selection of Strains Resistant or Tolerant to Different Classes of Compounds. Front. Microbiol. 2017, 8, 1650. [Google Scholar] [CrossRef]
- Chen, J.; Ying, G.-G.; Wei, X.-D.; Liu, Y.-S.; Liu, S.-S.; Hu, L.-X.; He, L.-Y.; Chen, Z.-F.; Chen, F.-R.; Yang, Y.-Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Effect of flow configuration and plant species. Sci. Total Environ. 2016, 571, 974–982. [Google Scholar] [CrossRef]
- Ávila, C.; Pelissari, C.; Sezerino, P.H.; Sgroi, M.; Roccaro, P.; García, J. Enhancement of total nitrogen removal through effluent recirculation and fate of PPCPs in a hybrid constructed wetland system treating urban wastewater. Sci. Total Environ. 2017, 584–585, 414–425. [Google Scholar] [CrossRef]
- Itzhari, D.; Ronen, Z. The Emergence of Antibiotics Resistance Genes, Bacteria, and Micropollutants in Grey Wastewater. Appl. Sci. 2023, 13, 2322. [Google Scholar] [CrossRef]
- Alfiya, Y.; Dubowski, Y.; Friedler, E. Diurnal patterns of micropollutants concentrations in domestic greywater. Urban Water J. 2018, 15, 399–406. [Google Scholar] [CrossRef]
- Leal, L.H.; Vieno, N.; Temmink, H.; Zeeman, G.; Buisman, C.J.N. Occurrence of xenobiotics in gray water and removal in three biological treatment systems. Environ. Sci. Technol. 2010, 44, 6835–6842. [Google Scholar] [CrossRef]
- Dordio, A.V.; Carvalho, A.J.P. Organic xenobiotics removal in constructed wetlands, with emphasis on the importance of the support matrix. J. Hazard. Mater. 2013, 252–253, 272–292. [Google Scholar] [CrossRef]
- Lv, M.; Zhang, D.; Niu, X.; Ma, J.; Lin, Z.; Fu, M. Insights into the fate of antibiotics in constructed wetland systems: Removal performance and mechanisms. J. Environ. Manag. 2022, 321, 116028. [Google Scholar] [CrossRef]
- Zhang, L.; Lv, T.; Zhang, Y.; Stein, O.R.; Arias, C.A.; Brix, H.; Carvalho, P.N. Effects of constructed wetland design on ibuprofen removal—A mesocosm scale study. Sci. Total Environ. 2017, 609, 38–45. [Google Scholar] [CrossRef]
- He, Y.; Sutton, N.B.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Pharmaceutical biodegradation under three anaerobic redox conditions evaluated by chemical and toxicological analyses. Sci. Total Environ. 2018, 618, 658–664. [Google Scholar] [CrossRef]
- Contreras, C.R.; López, D.; Leiva, A.M.; Domínguez, C.; Bayona, J.M.; Vidal, G. Removal of organic micropollutants in wastewater treated by activated sludge and constructed wetlands: A comparative study. Water 2019, 11, 2515. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, G.; Ng, W.J.; Tan, S.K. A review on removing pharmaceutical contaminants from wastewater by constructed wetlands: Design, performance and mechanism. Sci. Total Environ. 2014, 468–469, 908–932. [Google Scholar] [CrossRef]
- Carballa, M.; Omil, F.; Lema, J.M.; Llompart, M.; García-Jares, C.; Rodríguez, I.; Gómez, M.; Ternes, T. Behavior of pharmaceuticals, cosmetics and hormones in a sewage treatment plant. Water Res. 2004, 38, 2918–2926. [Google Scholar] [CrossRef]
- Alvarino, T.; Suarez, S.; Lema, J.M.; Omil, F. Understanding the removal mechanisms of PPCPs and the influence of main technological parameters in anaerobic UASB and aerobic CAS reactors. J. Hazard. Mater. 2014, 278, 506–513. [Google Scholar] [CrossRef]
- Verlicchi, P.; Zambello, E. How efficient are constructed wetlands in removing pharmaceuticals from untreated and treated urban wastewaters? A review. Sci. Total Environ. 2014, 470–471, 1281–1306. [Google Scholar] [CrossRef]
- Sklarz, M.Y.; Gross, A.; Yakirevich, A.; Soares, M.I.M. A recirculating vertical flow constructed wetland for the treatment of domestic wastewater. Desalination 2009, 246, 617–624. [Google Scholar] [CrossRef]
- Gross, A.; Shmueli, O.; Ronen, Z.; Raveh, E. Recycled vertical flow constructed wetland (RVFCW)-a novel method of recycling greywater for irrigation in small communities and households. Chemosphere 2007, 66, 916–923. [Google Scholar] [CrossRef]
- Gross, A.; Kaplan, D.; Baker, K. Removal of Microorganisms from Domestic Greywater Using a Recycling Vertical Flow Constructed Wetland (RVFCW). Proc. Water Environ. Fed. 2006, 6, 6133–6141. [Google Scholar] [CrossRef][Green Version]
- Troiano, E.; Beneduce, L.; Gross, A.; Ronen, Z. Antibiotic-resistant bacteria in greywater and greywater-irrigated soils. Front. Microbiol. 2018, 9, 2666. [Google Scholar] [CrossRef]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Monsalves, N.; Leiva, A.M.; Gómez, G.; Vidal, G. Antibiotic-Resistant Gene Behavior in Constructed Wetlands Treating Sewage: A Critical Review. Sustainability 2022, 14, 8524. [Google Scholar] [CrossRef]
- Chen, P.; Yu, X.; Zhang, J.; Wang, Y. New and traditional methods for antibiotic resistance genes removal: Constructed wetland technology and photocatalysis technology. Front. Microbiol. 2023, 13, 1110793. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Leiva, A.M.; Arismendi, W.; Vidal, G. Influence of design and operational parameters on the pathogens reduction in constructed wetland under the climate change scenario. Rev. Environ. Sci. Biotechnol. 2019, 18, 101–125. [Google Scholar] [CrossRef]
- Chandrasena, G.I.; Shirdashtzadeh, M.; Li, Y.L.; Deletic, A.; Hathaway, J.M.; McCarthy, D.T. Retention and survival of E. coli in stormwater biofilters: Role of vegetation, rhizosphere microorganisms and antimicrobial filter media. Ecol. Eng. 2017, 102, 166–177. [Google Scholar] [CrossRef]
- Shirdashtzadeh, M.; Chandrasena, G.I.; Henry, R.; McCarthy, D.T. Plants that can kill; improving E. coli removal in stormwater treatment systems using Australian plants with antibacterial activity. Ecol. Eng. 2017, 107, 120–125. [Google Scholar] [CrossRef]
- Du, L.; Zhao, Y.; Wang, C.; Zhang, H.; Chen, Q.; Zhang, X.; Zhang, L.; Wu, J.; Wu, Z.; Zhou, Q. Removal performance of antibiotics and antibiotic resistance genes in swine wastewater by integrated vertical-flow constructed wetlands with zeolite substrate. Sci. Total Environ. 2020, 721, 137765. [Google Scholar] [CrossRef] [PubMed]
- García, J.; García-Galán, M.J.; Day, J.W.; Boopathy, R.; White, J.R.; Wallace, S.; Hunter, R.G. A review of emerging organic contaminants (EOCs), antibiotic resistant bacteria (ARB), and antibiotic resistance genes (ARGs) in the environment: Increasing removal with wetlands and reducing environmental impacts. Bioresour. Technol. 2020, 307, 123228. [Google Scholar] [CrossRef]
- Leiva, A.M.; Piña, B.; Vidal, G. Antibiotic resistance dissemination in wastewater treatment plants: A challenge for the reuse of treated wastewater in agriculture. Rev. Environ. Sci. Biotechnol. 2021, 20, 1043–1072. [Google Scholar] [CrossRef]
- Liu, J.L.; Wong, M.H. Pharmaceuticals and personal care products (PPCPs): A review on environmental contamination in China. Environ. Int. 2013, 59, 208–224. [Google Scholar] [CrossRef]
- Dires, S.; Birhanu, T.; Ambelu, A.; Sahilu, G. Antibiotic resistant bacteria removal of subsurface flow constructed wetlands from hospital wastewater. J. Environ. Chem. Eng. 2018, 6, 4265–4272. [Google Scholar] [CrossRef]
- Aznar, R.; Sánchez-Brunete, C.; Albero, B.; Rodríguez, J.A.; Tadeo, J.L. Occurrence and analysis of selected pharmaceutical compounds in soil from Spanish agricultural fields. Environ. Sci. Pollut. Res. 2014, 21, 4772–4782. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Piernas, A.B.; Polo-López, M.I.; Fernández-Ibáñez, P.; Agüera, A. Validation and application of a multiresidue method based on liquid chromatography-tandem mass spectrometry for evaluating the plant uptake of 74 microcontaminants in crops irrigated with treated municipal wastewater. J. Chromatogr. A 2018, 1534, 10–21. [Google Scholar] [CrossRef]
- Christou, A.; Karaolia, P.; Hapeshi, E.; Michael, C.; Fatta-Kassinos, D. Long-term wastewater irrigation of vegetables in real agricultural systems: Concentration of pharmaceuticals in soil, uptake and bioaccumulation in tomato fruits and human health risk assessment. Water Res. 2017, 109, 24–34. [Google Scholar] [CrossRef]
- Martínez-Piernas, A.B.; Plaza-Bolaños, P.; García-Gómez, E.; Fernández-Ibáñez, P.; Agüera, A. Determination of organic microcontaminants in agricultural soils irrigated with reclaimed wastewater: Target and suspect approaches. Anal. Chim. Acta 2018, 1030, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Gielen, G.J.H.P.; Clinton, P.W.; Van den Heuvel, M.R.; Kimberley, M.O.; Greenfield, L.G. Influence of sewage and pharmaceuticals on soil microbial function. Environ. Toxicol. Chem. 2011, 30, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Henderson, M.; Ergas, S.J.; Ghebremichael, K.; Gross, A.; Ronen, Z. Occurrence of Antibiotic-Resistant Genes and Bacteria in Household Greywater Treated in Constructed Wetlands. Water 2022, 14, 758. [Google Scholar] [CrossRef]
- Hazra, M.; Durso, L.M. Performance Efficiency of Conventional Treatment Plants and Constructed Wetlands towards Reduction of Antibiotic Resistance. Antibiotics 2022, 11, 114. [Google Scholar] [CrossRef]
- Maiga, Y.; von Sperling, M.; Mihelcic, J.R. Constructed Wetlands. In Water and Sanitation for the 21st Century: Health and Microbiological Aspects of Excreta and Wastewater Management (Global Water Pathogen Project); Mihelcic, J.R., Verbyla, M.E., Eds.; Michigan State University: East Lansing, MI, USA, 2019. [Google Scholar] [CrossRef]
- Hastuti, Y.P.; Andina, Y.; Supriyono, E.; Fatma, Y.S.; Tridesianti, S. Identification and abundance of nitrifying-denitrifying bacteria in malang sand filter based culture environment for mud crabs Scylla serrata. IOP Conference Series: Earth and Environmental Science, Institute of Physics Publishing: Bristol, UK, 2019. [Google Scholar] [CrossRef]
- Laht, M.; Karkman, A.; Voolaid, V.; Ritz, C.; Tenson, T.; Virta, M.; Kisand, V. Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS ONE 2014, 9, e103705. [Google Scholar] [CrossRef]
- Shuai, W.; Itzhari, D.; Ronen, Z.; Hartmann, E.M. Mitigation of antimicrobial resistance genes in greywater treated at household level. Sci. Total Environ. 2023, 890, 164136. [Google Scholar] [CrossRef]
- Liu, X.; Guo, X.; Liu, Y.; Lu, S.; Xi, B.; Zhang, J.; Wang, Z.; Bi, B. A review on removing antibiotics and antibiotic resistance genes from wastewater by constructed wetlands: Performance and microbial response. Environ. Pollut. 2019, 254, 112996. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wilson, C.A.; Novak, J.T.; Riffat, R.; Aynur, S.; Murthy, S.; Pruden, A. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class i integrons. Environ. Sci. Technol. 2011, 45, 7855–7861. [Google Scholar] [CrossRef] [PubMed]
- Porob, S.; Craddock, H.A.; Motro, Y.; Sagi, O.; Gdalevich, M.; Ezery, Z.; Davidovitch, N.; Ronen, Z.; Moran-Gilad, J. Quantification and characterization of antimicrobial resistance in greywater discharged to the environment. Water 2020, 12, 1460. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.T.; Su, Z.X.; Lai, W.X.; Zhang, Y.B.; Liu, Y.W. Insights into the fate and removal of antibiotics and antibiotic resistance genes using biological wastewater treatment technology. Sci. Total Environ. 2021, 776, 145906. [Google Scholar] [CrossRef]
- Lucassen, R.; Rehberg, L.; Heyden, M.; Bockmuhl, D. Strong correlation of total phenotypic resistance of samples from household environments and the prevalence of class 1 integrons suggests for the use of the relative prevalence of intI1 as a screening tool for multi-resistance. PLoS ONE 2019, 14, e0218277. [Google Scholar] [CrossRef]
- Marshall, B.M.; Robleto, E.; Dumont, T.; Levy, S.B. The frequency of antibiotic-resistant bacteria in homes differing in their use of surface antibacterial agents. Curr. Microbiol. 2012, 65, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Quandt, E.M.; Gottell, N.; Gilbert, J.A. Examining the taxonomic distribution of tetracycline resistance in a wastewater plant. Sustain. Microbiol. 2024, 1, qvad003. [Google Scholar] [CrossRef] [PubMed]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef]
- Petrovich, M.; Chu, B.; Wright, D.; Griffin, J.; Elfeki, M.; Murphy, B.T.; Poretsky, R.; Wells, G. Antibiotic resistance genes show enhanced mobilization through suspended growth and biofilm-based wastewater treatment processes. FEMS Microbiol. Ecol. 2018, 94, fiy041. [Google Scholar] [CrossRef]
- Pu, C.; Liu, H.; Ding, G.; Sun, Y.; Yu, X.; Chen, J.; Ren, J.; Gong, X. Impact of direct application of biogas slurry and residue in fields: In situ analysis of antibiotic resistance genes from pig manure to fields. J. Hazard. Mater. 2018, 344, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, C.; Zheng, J.; Huang, X.; Wang, Z.; Liu, Y.; Zhu, G. Elimination of veterinary antibiotics and antibiotic resistance genes from swine wastewater in the vertical flow constructed wetlands. Chemosphere 2013, 91, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Nøhr-Meldgaard, K.; Struve, C.; Ingmer, H.; Agersø, Y. The Tetracycline Resistance Gene, tet(W) in Bifidobacterium animalis subsp. lactis Follows Phylogeny and Differs From tet(W) in Other Species. Front. Microbiol. 2021, 12, 658943. [Google Scholar] [CrossRef] [PubMed]
- Orlofsky, E.; Bernstein, N.; Sacks, M.; Vonshak, A.; Benami, M.; Kundu, A.; Maki, M.; Smith, W.; Wuertz, S.; Shapiro, K.; et al. Comparable levels of microbial contamination in soil and on tomato crops after drip irrigation with treated wastewater or potable water. Agric. Ecosyst. Environ. 2016, 215, 140–150. [Google Scholar] [CrossRef]
- Benami, M.; Gross, A.; Herzberg, M.; Orlofsky, E.; Vonshak, A.; Gillor, O. Assessment of pathogenic bacteria in treated graywater and irrigated soils. Sci. Total Environ. 2013, 458–460, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Ibekwe, A.M.; Gonzalez-Rubio, A.; Suarez, D.L. Impact of treated wastewater for irrigation on soil microbial communities. Sci. Total Environ. 2018, 622–623, 1603–1610. [Google Scholar] [CrossRef]
- Wang, F.H.; Qiao, M.; Su, J.Q.; Chen, Z.; Zhou, X.; Zhu, Y.G. High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ. Sci. Technol. 2014, 48, 9079–9085. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Zhu, L.; Wang, J.; Xing, B. Antibiotic resistance in agricultural soils: Source, fate, mechanism and attenuation strategy. Crit. Rev. Environ. Sci. Technol. 2022, 52, 847–889. [Google Scholar] [CrossRef]
- Slobodiuk, S.; Niven, C.; Arthur, G.; Thakur, S.; Ercumen, A. Does irrigation with treated and untreated wastewater increase antimicrobial resistance in soil and water: A systematic review. Int. J. Environ. Res. Public Health 2021, 18, 11046. [Google Scholar] [CrossRef]
- Cetinić, K.A.; Grgić, I.; Previšić, A.; Rožman, M. The curious case of methylparaben: Anthropogenic contaminant or natural origin? Chemosphere 2022, 294, 133781. [Google Scholar] [CrossRef]
- Chase, D.A.; Karnjanapiboonwong, A.; Fang, Y.; Cobb, G.P.; Morse, A.N.; Anderson, T.A. Occurrence of synthetic musk fragrances in effluent and non-effluent impacted environments. Sci. Total Environ. 2012, 416, 253–260. [Google Scholar] [CrossRef]
- Nguyen, M.-K.; Lin, C.; Nguyen, H.-L.; Hung, N.T.Q.; La, D.D.; Nguyen, X.H.; Chang, S.W.; Chung, W.J.; Nguyen, D.D. Occurrence, fate, and potential risk of pharmaceutical pollutants in agriculture: Challenges and environmentally friendly solutions. Sci. Total Environ. 2023, 899, 165323. [Google Scholar] [CrossRef]
- Wu, W.; Ma, M.; Hu, Y.; Yu, W.; Liu, H.; Bao, Z. The fate and impacts of pharmaceuticals and personal care products and microbes in agricultural soils with long term irrigation with reclaimed water. Agric. Water Manag. 2021, 251, 106862. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Nowak-Lange, M.; Niedziałkowska, K.; Bernat, P.; Lisowska, K. In vitro study of the ecotoxicological risk of methylisothiazolinone and chloroxylenol towards soil bacteria. Sci. Rep. 2022, 12, 19068. [Google Scholar] [CrossRef] [PubMed]
- Vodyanitskii, Y.N.; Yakovlev, A.S. Contamination of soils and groundwater with new organic micropollutants: A review. Eurasian Soil Sci. 2016, 49, 560–569. [Google Scholar] [CrossRef]
- Harrow, D.I.; Felker, J.M.; Baker, K.H. Impacts of Triclosan in Greywater on Soil Microorganisms. Appl. Environ. Soil Sci. 2011, 2011, 646750. [Google Scholar] [CrossRef]
- Frenk, S.; Hadar, Y.; Minz, D. Resilience of soil bacterial community to irrigation with water of different qualities under Mediterranean climate. Environ. Microbiol. 2014, 16, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; He, R.; Yuan, K.; Chen, E.; Lin, L.; Chen, X.; Sha, S.; Zhong, J.; Lin, L.; Yang, L.; et al. Polycyclic aromatic hydrocarbons (PAHs) enriching antibiotic resistance genes (ARGs) in the soils. Environ. Pollut. 2017, 220, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Hemkemeyer, M.; Schwalb, S.A.; Heinze, S.; Joergensen, R.G.; Wichern, F. Functions of elements in soil microorganisms. Microbiol. Res. 2021, 252, 126832. [Google Scholar] [CrossRef] [PubMed]
- Bastida, F.; Torres, I.; Abadía, J.; Romero-Trigueros, C.; Ruiz-Navarro, A.; Alarcón, J.; García, C.; Nicolás, E. Comparing the impacts of drip irrigation by freshwater and reclaimed wastewater on the soil microbial community of two citrus species. Agric. Water Manag. 2018, 203, 53–62. [Google Scholar] [CrossRef]
- Baird, R.; Eaton, A.D.; Rice, E.W.; Bridgewater, L. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Public Health Association: Washington, DC, USA, 2017. [Google Scholar]
- Rocha, M.C.; Rosa, H.S.; Grady, J.P.; Blakely, E.L.; He, L.; Romain, N.; Haller, R.G.; Newman, J.; McFarland, R.; Ng, Y.S.; et al. Pathological mechanisms underlying single large-scale mitochondrial DNA deletions. Ann. Neurol. 2018, 83, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Pszczolinska, K.; Michel, M. The QuEChERS approach for the determination of pesticide residues in soil samples: An overview. J. AOAC Int. 2016, 99, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Avraham, A.; Nwaobi, A.F.; Sela-Adler, M.; Gross, A.; Bernstein, R. Towards a generic solution to onsite wastewater treatment. J. Water Process Eng. 2023, 56, 104517. [Google Scholar] [CrossRef]
- Kasuga, I.; Nagasawa, K.; Suzuki, M.; Kurisu, F.; Furumai, H. High-Throughput Screening of Antimicrobial Resistance Genes and Their Association With Class 1 Integrons in Urban Rivers in Japan. Front. Environ. Sci. 2022, 10, 1235–1237. [Google Scholar] [CrossRef]
- Li, J.; Cao, J.; Zhu, Y.-G.; Chen, Q.-L.; Shen, F.; Wu, Y.; Xu, S.; Fan, H.; Da, G.; Huang, R.-J.; et al. Global Survey of Antibiotic Resistance Genes in Air. Environ. Sci. Technol. 2018, 52, 10975–10984. [Google Scholar] [CrossRef] [PubMed]
- Muziasari, W.I.; Pitkänen, L.K.; Sørum, H.; Stedtfeld, R.D.; Tiedje, J.M.; Virta, M. The resistome of farmed fish feces contributes to the enrichment of antibiotic resistance genes in sediments below baltic sea fish farms. Front. Microbiol. 2017, 7, 2137. [Google Scholar] [CrossRef] [PubMed]
- Muziasari, W.I.; Pärnänen, K.; Johnson, T.A.; Lyra, C.; Karkman, A.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol. Ecol. 2016, 92, fiw052. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef]
- McIver, L.J.; Abu-Ali, G.; A Franzosa, E.; Schwager, R.; Morgan, X.C.; Waldron, L.; Segata, N.; Huttenhower, C. bioBakery: A meta’omic analysis environment. Bioinformatics 2017, 34, 1235–1237. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Gunturu, S.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. Nonpareil 3: Fast Estimation of Metagenomic Coverage and Sequence Diversity. mSystems 2018, 3, e00039-18. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Fitzpatrick, D.; Walsh, F. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol. Ecol. 2016, 92, fiv168. [Google Scholar] [CrossRef]
- Ben Maamar, S.; Glawe, A.J.; Brown, T.K.; Hellgeth, N.; Hu, J.; Wang, J.-P.; Huttenhower, C.; Hartmann, E.M. Mobilizable antibiotic resistance genes are present in dust microbial communities. PLoS Pathog. 2020, 16, e1008211. [Google Scholar] [CrossRef]
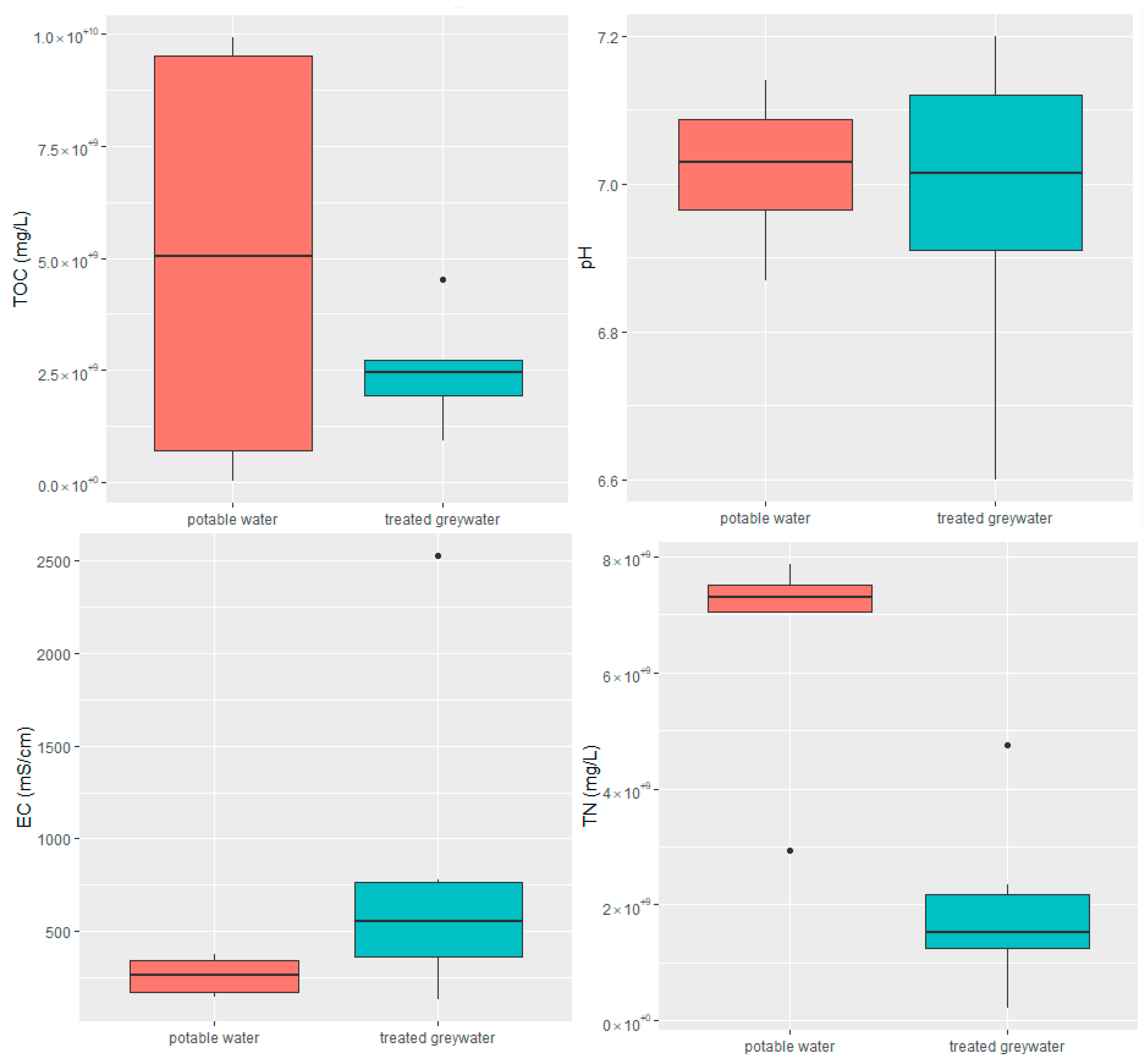
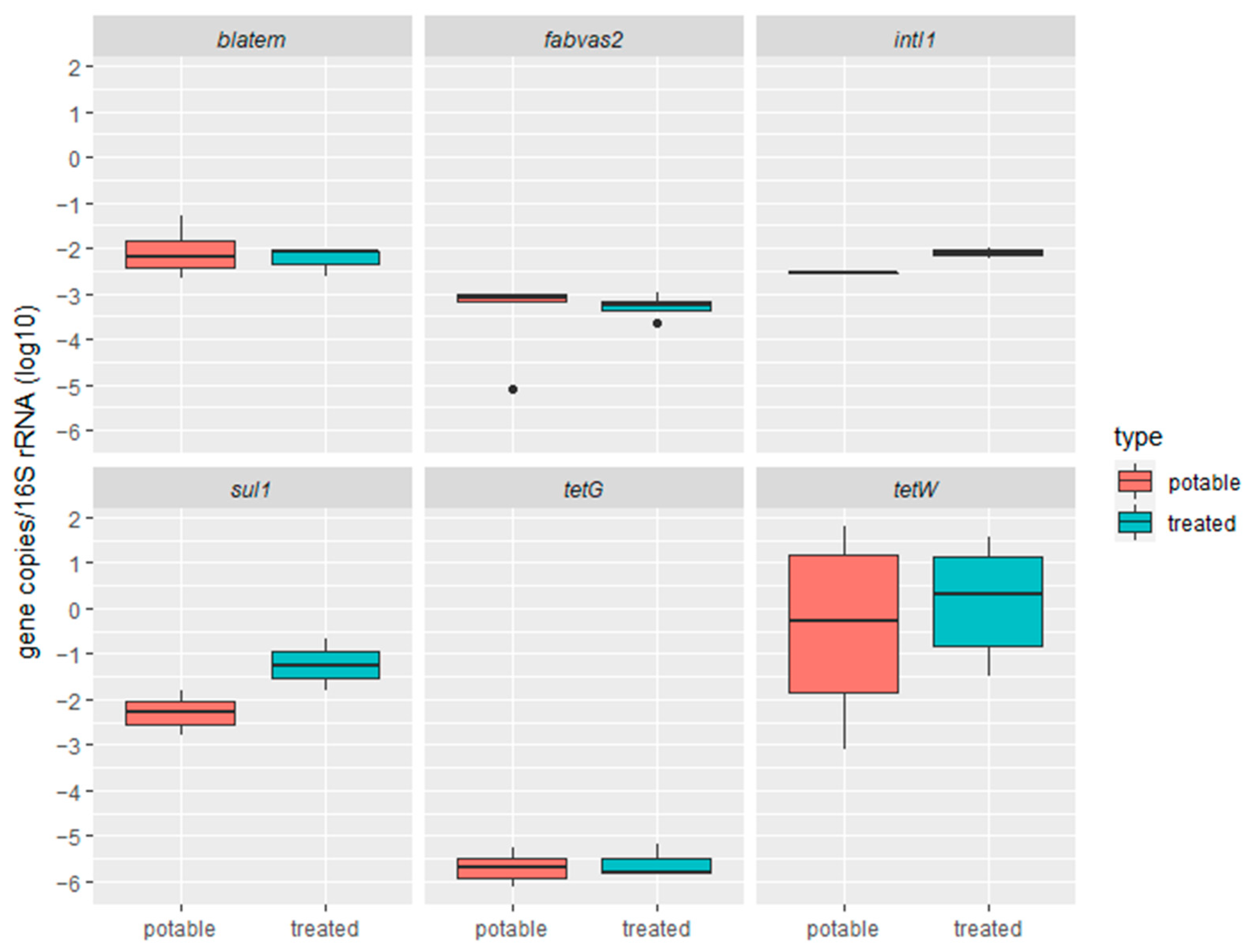
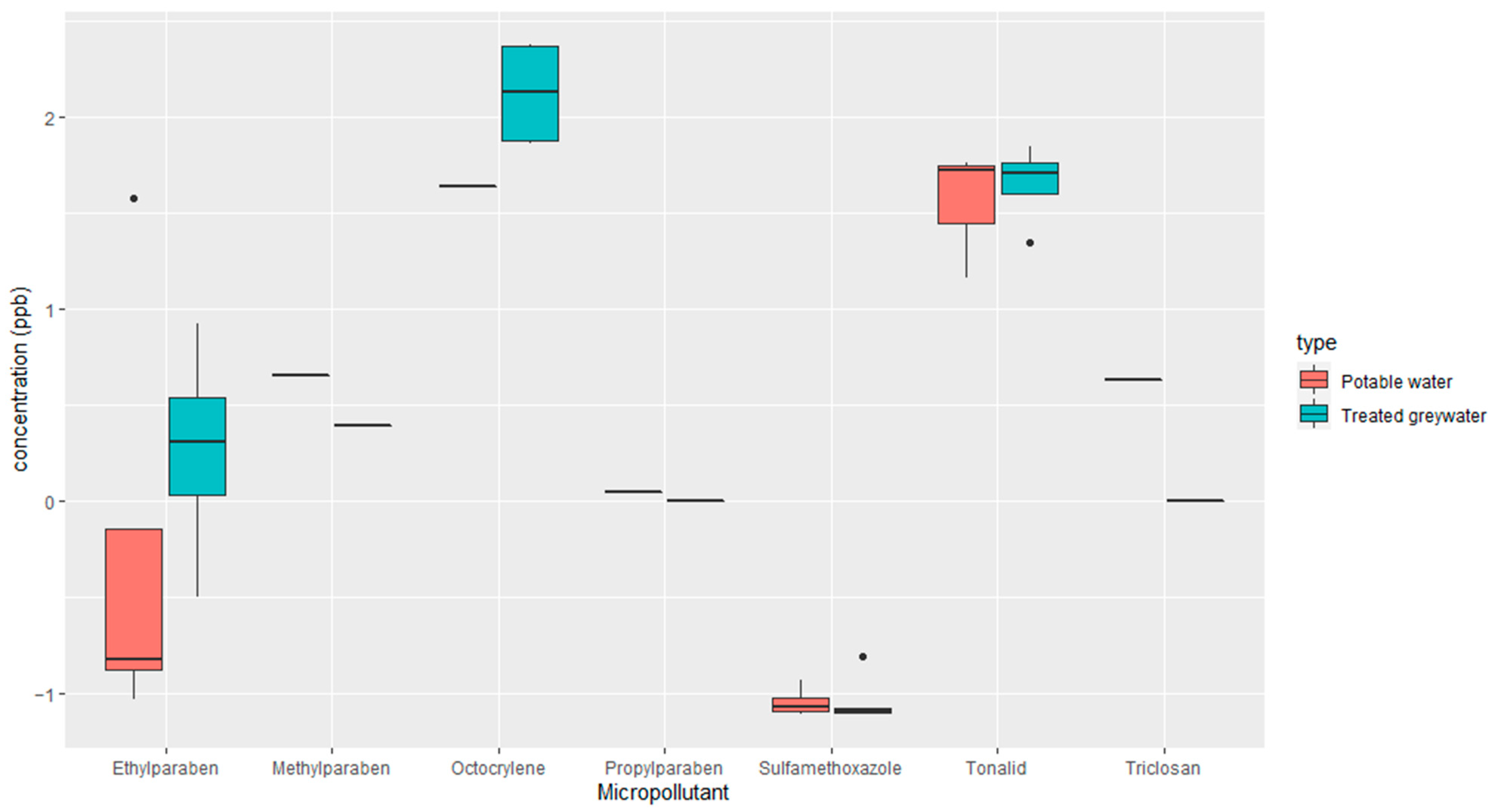
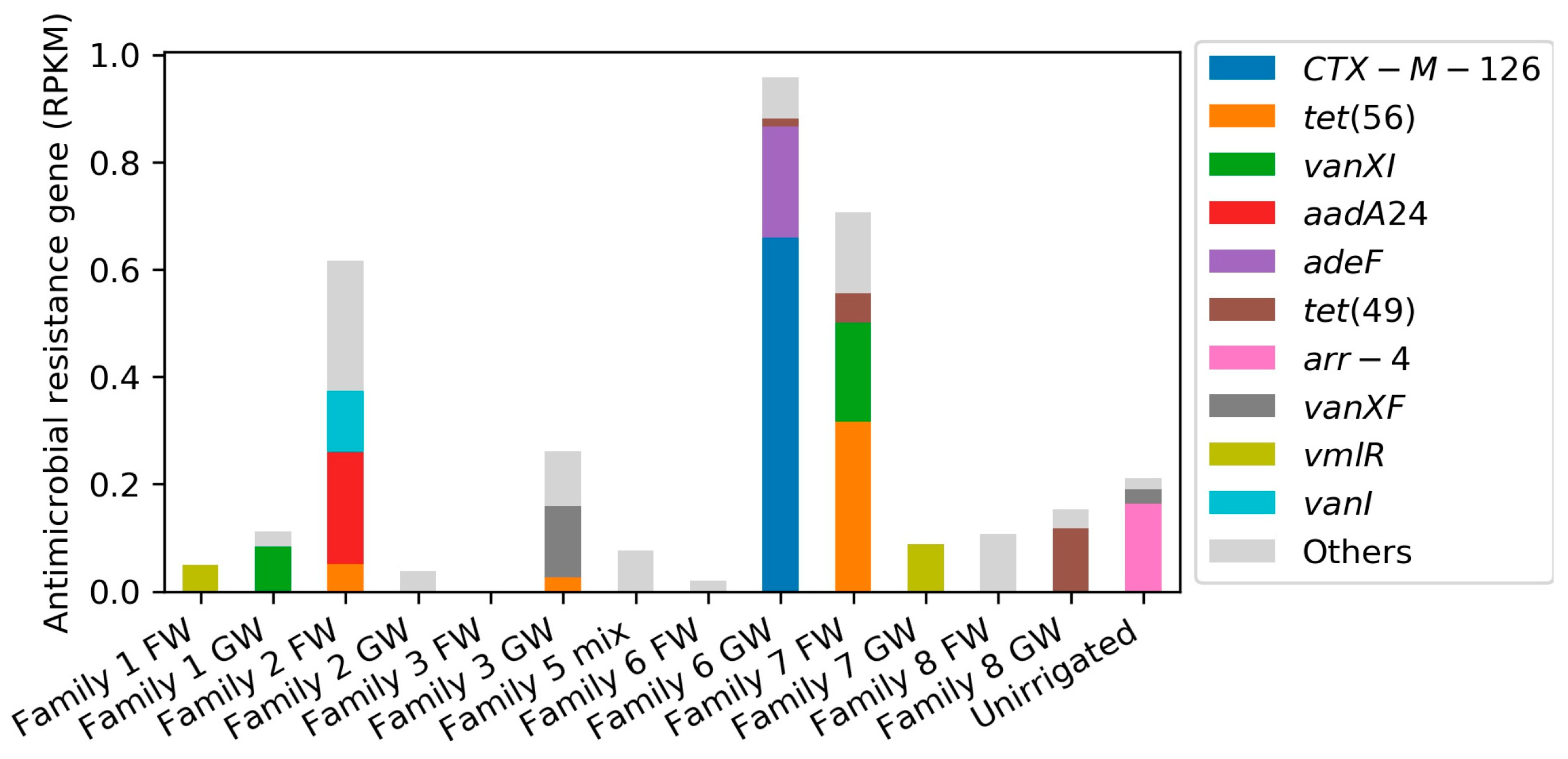
| Parameter | Raw Greywater | Treated Greywater |
|---|---|---|
| pH | 6.95–9.63 | 6.1–10.16 |
| EC [µs/cm] | 366–2150 | 321–995 |
| Turbidity [NTU] | 15.07–271.7 | 0–68 |
| BOD [mg/L] | 10.9–39.9 | 0.45–21.9 |
| TSS [g/L] | 12.43–26.92 | 0.001–47 |
| TOC [mg/L] | 5.47–43.78 | 1.68–39.1 |
| TN [mg/L] | 1.72–25.13 | 0.4–38.3 |
| Gene | Abundance | Sample Type | Gene | Abundance | Sample Type |
|---|---|---|---|---|---|
| intI1_1 | 8.73 × 106 | Filterbed | aac(6′)-II | 1.27 × 104 | Filterbed |
| vanA | 2.24 × 106 | Filterbed | aph6 | 2.22 × 104 | Raw |
| vanXD | 2.43 × 103 | Treated | blaCTX-M | 5.96 × 105 | Filterbed |
| ermD | 2.03 × 103 | Filterbed | blaCMY2 | 3.05 × 105 | Filterbed |
| mcr1_1 | 1.92 × 106 | Filterbed | blaOXA51 | 1.32 × 104 | Filterbed |
| qacE | 1.33 × 106 | Raw | adeA | 4.14 × 105 | Raw |
| fabK | 1.12 × 104 | Filterbed | pcoA | 5.27 × 104 | Filterbed |
| ISPps | 2.47 × 106 | Filterbed | qnrB | 7.45 × 104 | Filterbed |
| tnpA_1 | 6.00 × 105 | Filterbed | qnrD | 1.12 × 104 | Filterbed |
| Tn3 | 1.64 × 104 | Raw | cmlA_2 | 6.09 × 104 | Filterbed |
| sul2_1 | 3.50 × 106 | Treated | catB3 | 4.94 × 104 | Treated |
| sul1_2 | 9.19 × 105 | Treated | ermF | 9.56 × 104 | Filterbed |
| aph3-ib | 1.39 × 106 | Filterbed | erm34 | 2.06 × 104 | Filterbed |
| aadA_1 | 7.48 × 105 | Raw | ermX_1 | 4.90 × 103 | Filterbed |
| vgaA_1 | 5.94 × 103 | Filterbed | dfrA1 | 9.52 × 104 | Raw |
| tetA_1 | 4.09 × 103 | Filterbed | tetW | 1.51 × 104 | Treated |
| tetC_1 | 1.16 × 103 | Filterbed |
| Filter Bedding | Vegetation | Special Remarks | |
|---|---|---|---|
| House 1 | Small tuff stone | Iris pseudacorus | |
| House 2 | Big tuff stone | Nastrurtium officinale and Equisetum hyemale | |
| House 3 | Mainly big pebbles with some small tuff stones | No vegetation | The upper container is completely covered with a lid |
| House 4 | Small tuff stone | Iris pseudacorus | The system also receives water from the house swimming pool |
| House 5 | Plastic beads and some small tuff stones | Iris pseudacorus | |
| House 6 | Gravel stones | Solanum lycopersicum | |
| House 7 | Small tuff stone | Canna indica |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Itzhari, D.; Shuai, W.; Hartmann, E.M.; Ronen, Z. Heterogeneous Antibiotic Resistance Gene Removal Impedes Evaluation of Constructed Wetlands for Effective Greywater Treatment. Antibiotics 2024, 13, 315. https://doi.org/10.3390/antibiotics13040315
Itzhari D, Shuai W, Hartmann EM, Ronen Z. Heterogeneous Antibiotic Resistance Gene Removal Impedes Evaluation of Constructed Wetlands for Effective Greywater Treatment. Antibiotics. 2024; 13(4):315. https://doi.org/10.3390/antibiotics13040315
Chicago/Turabian StyleItzhari, Daniella, Weitao Shuai, Erica M. Hartmann, and Zeev Ronen. 2024. "Heterogeneous Antibiotic Resistance Gene Removal Impedes Evaluation of Constructed Wetlands for Effective Greywater Treatment" Antibiotics 13, no. 4: 315. https://doi.org/10.3390/antibiotics13040315
APA StyleItzhari, D., Shuai, W., Hartmann, E. M., & Ronen, Z. (2024). Heterogeneous Antibiotic Resistance Gene Removal Impedes Evaluation of Constructed Wetlands for Effective Greywater Treatment. Antibiotics, 13(4), 315. https://doi.org/10.3390/antibiotics13040315







