Prevalence, Characterization, and Epidemiological Relationships between ESBL and Carbapenemase-Producing Escherichia coli, Klebsiella pneumoniae, and Acinetobacter spp. Isolated from Humans and the Kitchen Environment of Two Greek Hospitals
Abstract
1. Introduction
2. Results
2.1. Prevalence and Seasonal and Regional Variations of Strains
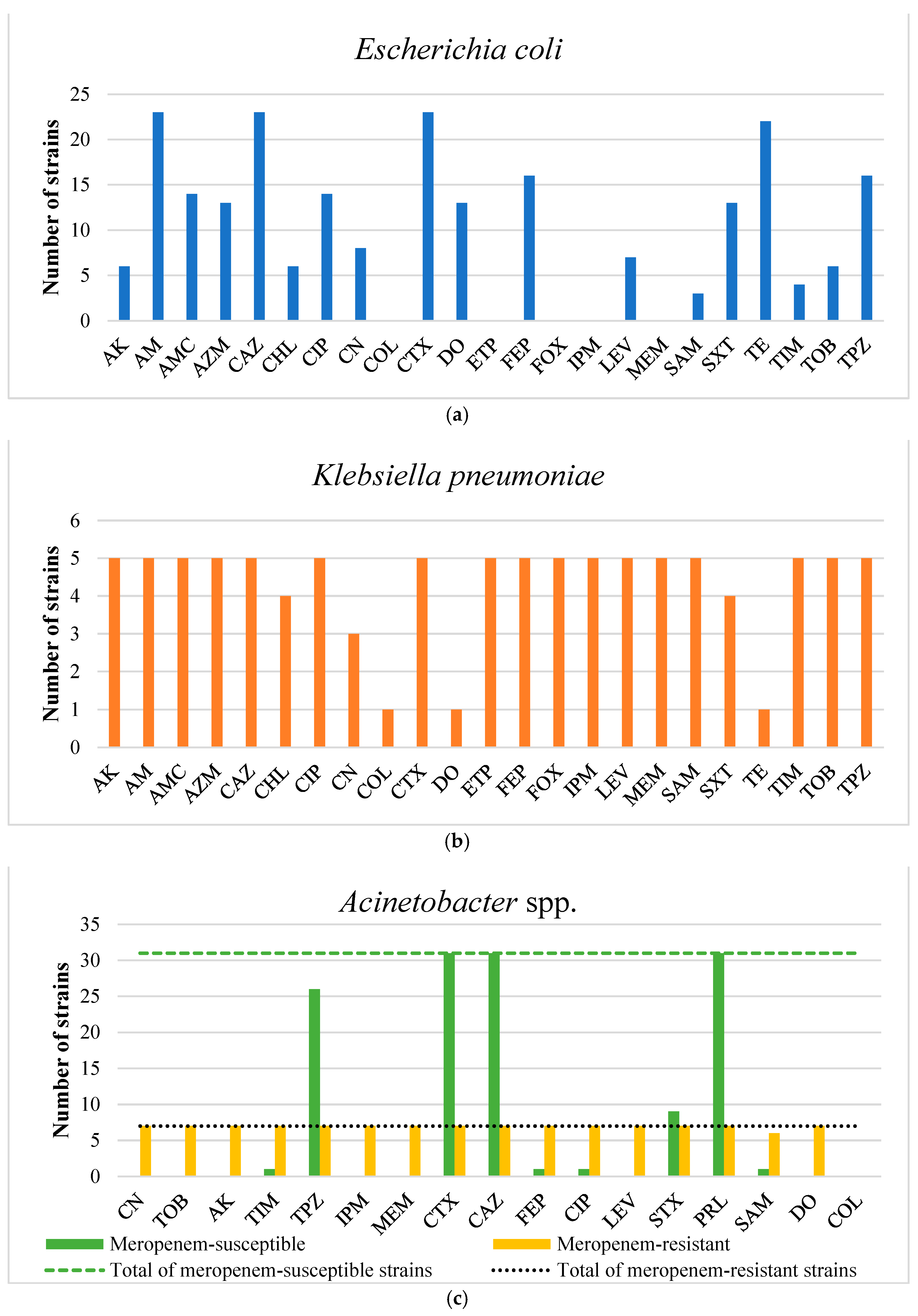
2.2. Antimicrobial Susceptibility Testing of the Isolated Strains
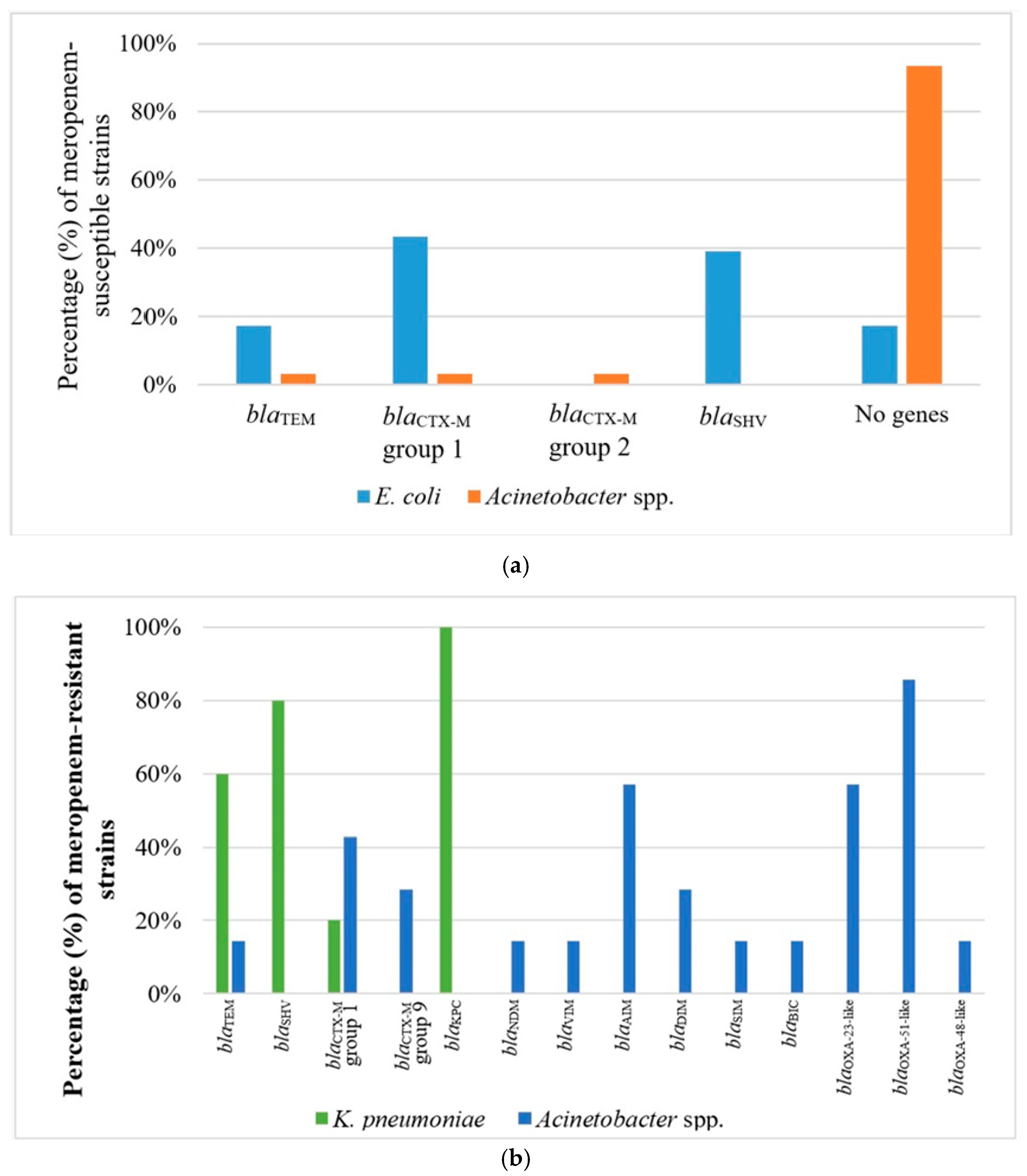
2.3. Phenotype of β-Lactam Resistance and Molecular Screening of β-Lactamase Genes
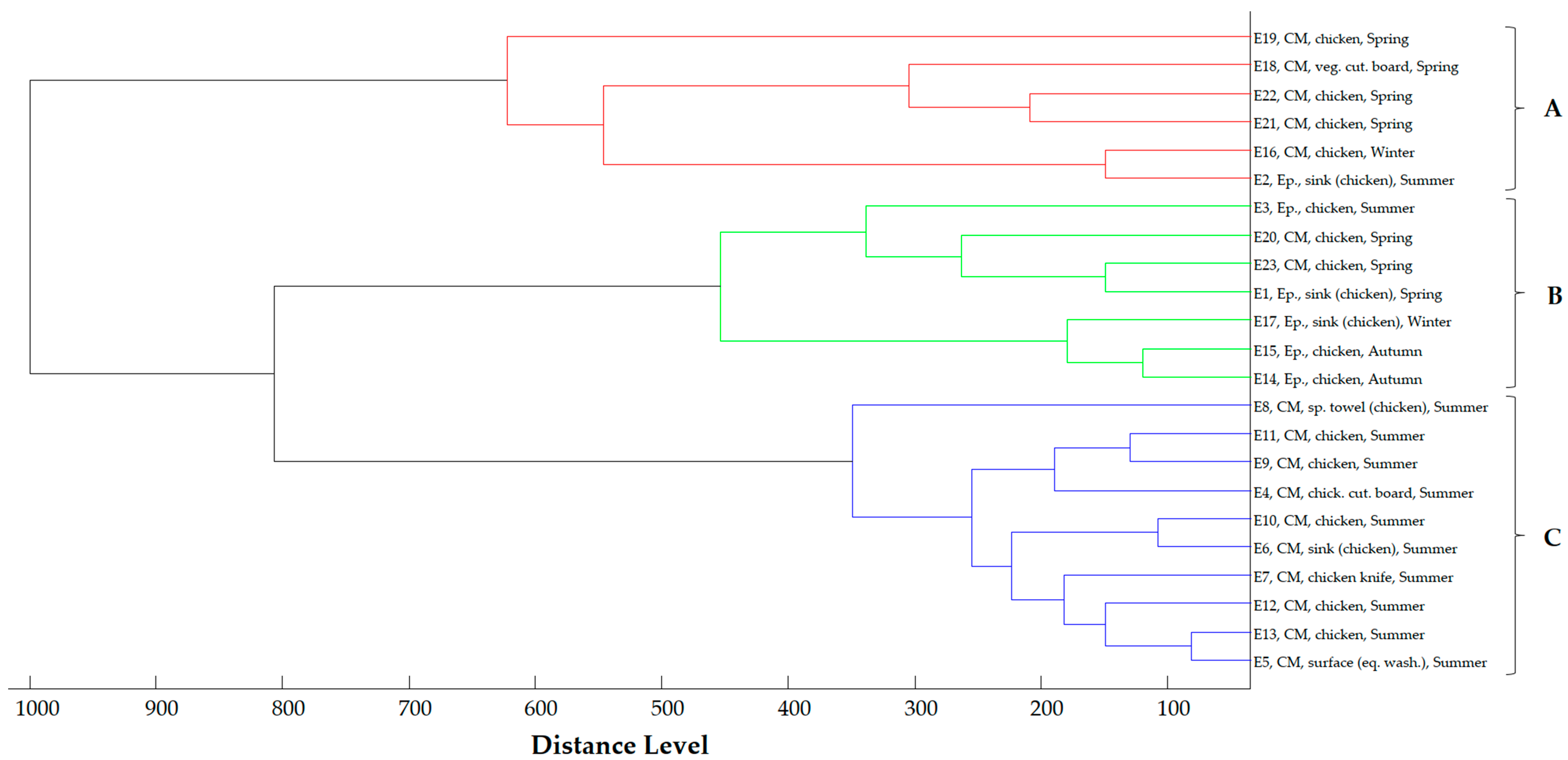
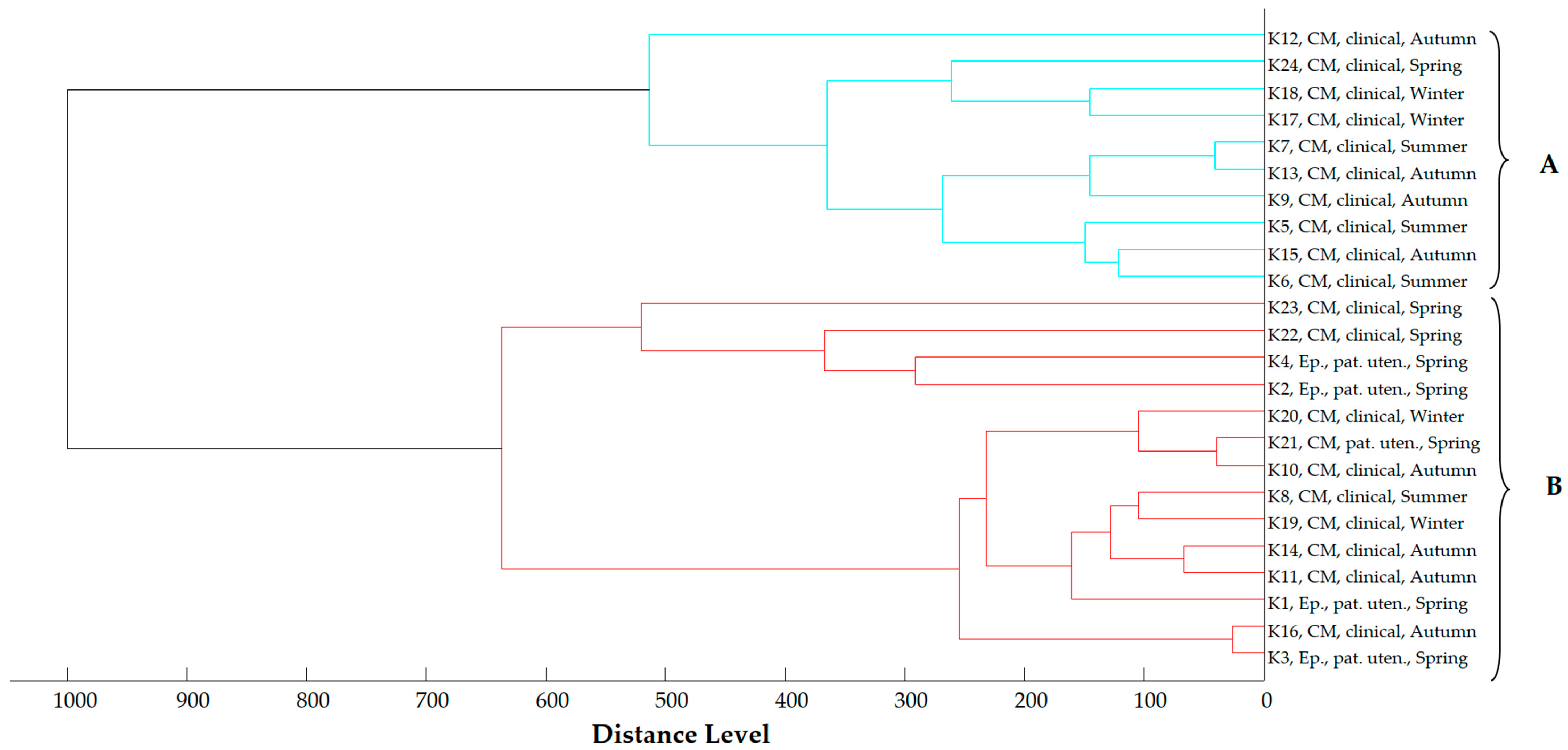
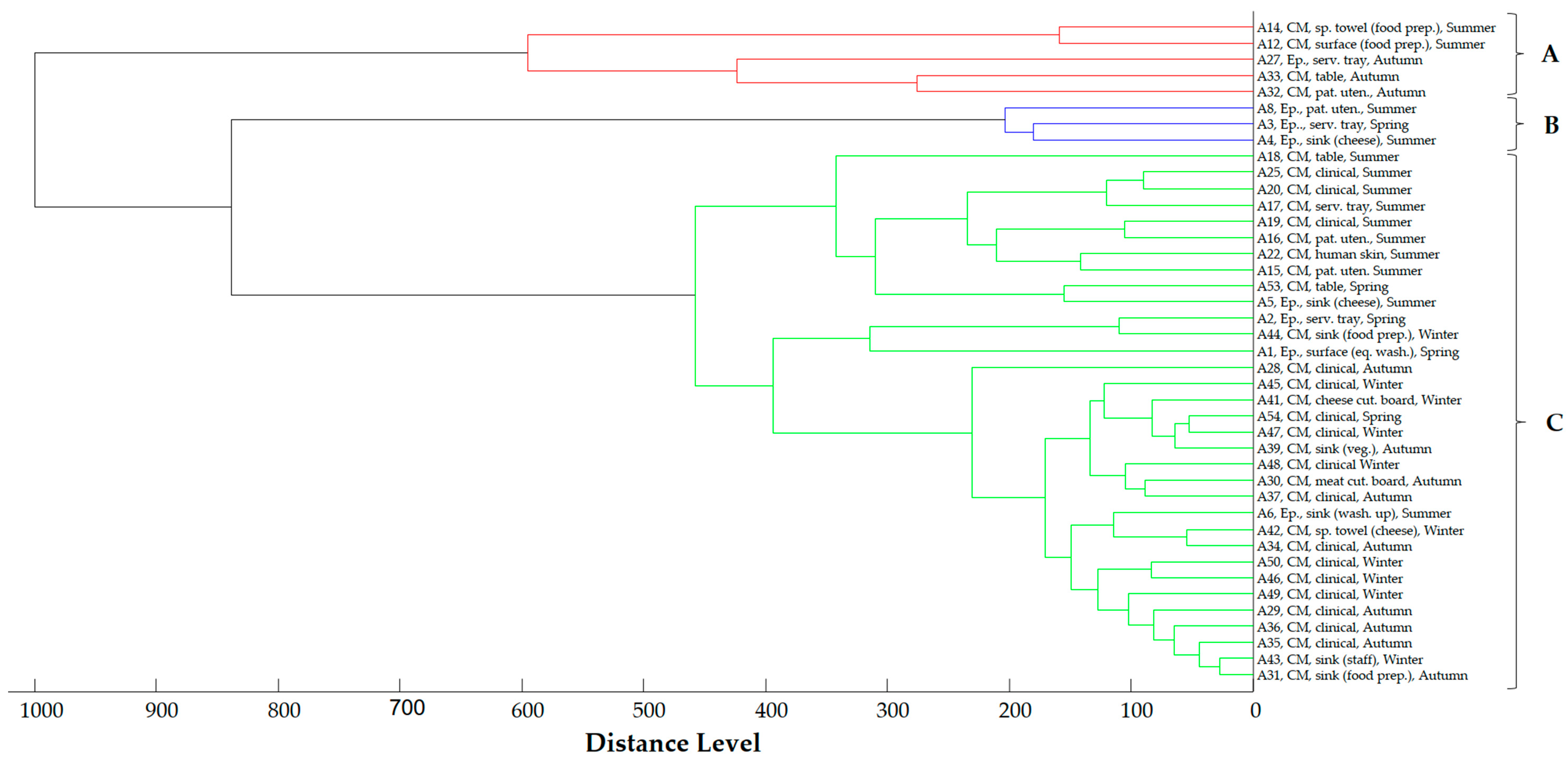
2.4. Proteomic Relationship of Strains
2.5. Phylogenetic Analysis of E. coli Strains
2.6. Medical History Results
3. Discussion
4. Materials and Methods
4.1. Sampling
4.2. Microbiological Examination, Identification, and Proteomic Relationship of the Isolated Strains
4.3. Antimicrobial Susceptibility Testing of the Isolated Strains
4.4. Confirmation of β-Lactamase Production
4.5. Molecular Screening of β-Lactamase Genes
4.6. Phylogenetic Analysis of E. coli Strains
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aljeldah, M.M. Antimicrobial Resistance and Its Spread Is a Global Threat. Antibiotics 2022, 11, 1082. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. The Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Chaw, P.S.; Höpner, J.; Mikolajczyk, R. The Knowledge, Attitude and Practice of Health Practitioners towards Antibiotic Prescribing and Resistance in Developing Countries—A Systematic Review. J. Clin. Pharm. Ther. 2018, 43, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.I.; Aqib, A.I.; Seleem, M.N.; Shabbir, M.A.; Hao, H.; Iqbal, Z.; Kulyar, M.F.-A.; Zaheer, T.; Li, K. Genetic Basis of Molecular Mechanisms in β-Lactam Resistant Gram-Negative Bacteria. Microb. Pathog. 2021, 158, 105040. [Google Scholar] [CrossRef]
- Cantón, R.; Akóva, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. Rapid Evolution and Spread of Carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-Spectrum β-Lactamases: An Update on Their Characteristics, Epidemiology and Detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Croxen, M.A.; Finlay, B.B. Molecular Mechanisms of Escherichia coli Pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef]
- Pitout, J.D.D. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Front. Microbiol. 2012, 3, 9. [Google Scholar] [CrossRef]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a Global Pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef]
- Hansen, G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021, 10, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Yang, X.; Chan, E.W.-C.; Zhang, R.; Chen, S. Klebsiella Species: Taxonomy, Hypervirulence and Multidrug Resistance. EBioMedicine 2022, 79, 103998. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, 1–33. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Gheorghe, I.; Czobor, I.B.; Chifiriuc, M.C. Antibiotic Resistance Profiles, Molecular Mechanisms and Innovative Treatment Strategies of Acinetobacter baumannii. Microorganisms 2020, 8, 935. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention Control & World Health Organization. Antimicrobial Resistance Surveillance in Europe 2023–2021 Data; European Centre for Disease Prevention and Control and World Health Organization: Stockholm, Sweden, 2023. [Google Scholar] [CrossRef]
- Overdevest, I.; Willemsen, I.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Heck, M.; Savelkoul, P.; Vandenbroucke-Grauls, C.; van der Zwaluw, K.; et al. Extended-Spectrum β-Lactamase Genes of Escherichia coli in Chicken Meat and Humans, the Netherlands. Emerg. Infect. Dis. 2011, 17, 1216–1222. [Google Scholar] [CrossRef]
- Carvalheira, A.; Casquete, R.; Silva, J.; Teixeira, P. Prevalence and Antimicrobial Susceptibility of Acinetobacter spp. Isolated from Meat. Int. J. Food Microbiol. 2017, 243, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Economou, V.; Delis, G.; Stavrou, D.; Gousia, P.; Tsitsos, A.; Mantzios, T.; Chouliara, E.; Kolovos, N.; Soultos, N. Characterization of Extended Spectrum Cephalosporin-Resistant Escherichia coli Strains Isolated from Raw Poultry Carcasses in Catering Services in Northern Greece. Vet. Sci. 2023, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Tschudin-Sutter, S.; Frei, R.; Stephan, R.; Hächler, H.; Nogarth, D.; Widmer, A.F. Extended-Spectrum β-Lactamase (ESBL)–Producing Enterobacteriaceae: A Threat from the Kitchen. Infect. Control Hosp. Epidemiol. 2014, 35, 581–584. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). Antimicrobial Consumption and Resistance in Bacteria from Humans and Food-producing Animals. EFSA J. 2024, 22, e8589. [Google Scholar] [CrossRef]
- Argudín, M.; Deplano, A.; Meghraoui, A.; Dodémont, M.; Heinrichs, A.; Denis, O.; Nonhoff, C.; Roisin, S. Bacteria from Animals as a Pool of Antimicrobial Resistance Genes. Antibiotics 2017, 6, 12. [Google Scholar] [CrossRef]
- Economou, V.; Gousia, P. Agriculture and Food Animals as a Source of Antimicrobial-Resistant Bacteria. Infect. Drug Resist. 2015, 49, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Borrusso, P.A.; Quinlan, J.J. Prevalence of Pathogens and Indicator Organisms in Home Kitchens and Correlation with Unsafe Food Handling Practices and Conditions. J. Food Prot. 2017, 80, 590–597. [Google Scholar] [CrossRef] [PubMed]
- El-Liethy, M.A.; Hemdan, B.A.; El-Taweel, G.E. Prevalence of E. coli, Salmonella, and Listeria spp. as Potential Pathogens: A Comparative Study for Biofilm of Sink Drain Environment. J. Food Saf. 2020, 40, e12816. [Google Scholar] [CrossRef]
- Stewardson, A.J.; Renzi, G.; Maury, N.; Vaudaux, C.; Brassier, C.; Fritsch, E.; Pittet, D.; Heck, M.; van der Zwaluw, K.; Reuland, E.A.; et al. Extended-Spectrum β-Lactamase–Producing Enterobacteriaceae in Hospital Food: A Risk Assessment. Infect. Control Hosp. Epidemiol. 2014, 35, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sanz, E.; Bagutti, C.; García-Martín, A.B.; Roth, J.A.; Alt Hug, M.; Maurer Pekerman, L.; Schindler, R.; Furger, R.; Eichenberger, L.; Steffen, I.; et al. Extended-Spectrum β-Lactamase-Producing Enterobacterales in Diverse Foodstuffs: A Prospective, Longitudinal Study in the City of Basel, Switzerland. Front. Microbiol. 2023, 14, 1295037. [Google Scholar] [CrossRef]
- Egervärn, M.; Börjesson, S.; Byfors, S.; Finn, M.; Kaipe, C.; Englund, S.; Lindblad, M. Escherichia coli with Extended-Spectrum Beta-Lactamases or Transferable AmpC Beta-Lactamases and Salmonella on Meat Imported into Sweden. Int. J. Food Microbiol. 2014, 171, 8–14. [Google Scholar] [CrossRef]
- Dramowski, A.; Aucamp, M.; Bekker, A.; Pillay, S.; Moloto, K.; Whitelaw, A.C.; Cotton, M.F.; Coffin, S. NeoCLEAN: A Multimodal Strategy to Enhance Environmental Cleaning in a Resource-Limited Neonatal Unit. Antimicrob. Resist. Infect. Control 2021, 10, 35. [Google Scholar] [CrossRef]
- Calbo, E.; Freixas, N.; Xercavins, M.; Riera, M.; Nicolas, C.; Monistrol, O.; Sole, M.d.m.; Sala, M.R.; Vila, J.; Garau, J. Foodborne Nosocomial Outbreak of SHV1 and CTX-M-15-Producing Klebsiella pneumoniae: Epidemiology and Control. Clin. Infect. Dis. 2011, 52, 743–749. [Google Scholar] [CrossRef]
- Cardinale, M.; Kaiser, D.; Lueders, T.; Schnell, S.; Egert, M. Microbiome Analysis and Confocal Microscopy of Used Kitchen Sponges Reveal Massive Colonization by Acinetobacter, Moraxella and Chryseobacterium Species. Sci. Rep. 2017, 7, 5791. [Google Scholar] [CrossRef]
- Araújo, B.C.; Moraes, M.S.; Costa, L.E.O.; Nascimento, J.S. Short Communication: Multidrug-Resistant Acinetobacter baumannii-Calcoaceticus Complex Isolated from Infant Milk Formula and Utensils in a Nursery in Rio de Janeiro, Brazil. J. Dairy. Sci. 2015, 98, 2303–2306. [Google Scholar] [CrossRef]
- Ranjbar, R.; Masoudimanesh, M.; Dehkordi, F.S.; Jonaidi-Jafari, N.; Rahimi, E. Shiga (Vero)-Toxin Producing Escherichia coli Isolated from the Hospital Foods; Virulence Factors, o-Serogroups and Antimicrobial Resistance Properties. Antimicrob. Resist. Infect. Control 2017, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Yousif, E.I.; Ashoush, I.S.; Donia, A.A.; Hala Goma, K.A. Critical Control Points for Preparing Chicken Meals in a Hospital Kitchen. Ann. Agric. Sci. 2013, 58, 203–211. [Google Scholar] [CrossRef][Green Version]
- Deeny, S.R.; van Kleef, E.; Bou-Antoun, S.; Hope, R.J.; Robotham, J.V. Seasonal Changes in the Incidence of Escherichia coli Bloodstream Infection: Variation with Region and Place of Onset. Clin. Microbiol. Infect. 2015, 21, 924–929. [Google Scholar] [CrossRef]
- Wielders, C.C.H.; Van Duijkeren, E.; Van Den Bunt, G.; Meijs, A.P.; Dierikx, C.M.; Bonten, M.J.M.; Van Pelt, W.; Franz, E.; De Greeff, S.C. Seasonality in Carriage of Extended-Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella Pneumoniae in the General Population: A Pooled Analysis of Nationwide Cross-Sectional Studies. Epidemiol. Infect. 2020, 148, e68. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Lavelle, K.; Huang, A.; Atwill, E.R.; Pitesky, M.; Li, X. Assessment of Prevalence and Diversity of Antimicrobial Resistant Escherichia coli from Retail Meats in Southern California. Antibiotics 2023, 12, 782. [Google Scholar] [CrossRef]
- Randall, L.P.; Lodge, M.P.; Elviss, N.C.; Lemma, F.L.; Hopkins, K.L.; Teale, C.J.; Woodford, N. Evaluation of Meat, Fruit and Vegetables from Retail Stores in Five United Kingdom Regions as Sources of Extended-Spectrum Beta-Lactamase (ESBL)-Producing and Carbapenem-Resistant Escherichia coli. Int. J. Food Microbiol. 2017, 241, 283–290. [Google Scholar] [CrossRef]
- Morgan, D.J.; Liang, S.Y.; Smith, C.L.; Johnson, J.K.; Harris, A.D.; Furuno, J.P.; Thorn, K.A.; Snyder, G.M.; Day, H.R.; Perencevich, E.N. Frequent Multidrug-Resistant Acinetobacter baumannii Contamination of Gloves, Gowns, and Hands of Healthcare Workers. Infect. Control Hosp. Epidemiol. 2010, 31, 716–721. [Google Scholar] [CrossRef]
- Bayuga, S.; Zeana, C.; Sahni, J.; Della-Latta, P.; El-Sadr, W.; Larson, E. Prevalence and Antimicrobial Patterns of Acinetobacter baumannii on Hands and Nares of Hospital Personnel and Patients: The Iceberg Phenomenon Again. Heart Lung 2002, 31, 382–390. [Google Scholar] [CrossRef]
- Giamarellou, H.; Antoniadou, A.; Kanellakopoulou, K. Acinetobacter baumannii: A Universal Threat to Public Health? Int. J. Antimicrob. Agents 2008, 32, 106–119. [Google Scholar] [CrossRef]
- Puig-Asensio, M.; Diekema, D.J.; Boyken, L.; Clore, G.S.; Salinas, J.L.; Perencevich, E.N. Contamination of Health-Care Workers’ Hands with Escherichia coli and Klebsiella Species after Routine Patient Care: A Prospective Observational Study. Clin. Microbiol. Infect. 2020, 26, 760–766. [Google Scholar] [CrossRef]
- Bitterman, R.; Geffen, Y.; Rabino, G.; Eluk, O.; Warman, S.; Greenblatt, A.S.; Neuberger, A.; Reisner, S.A.; Hussein, K.; Paul, M. Rate of Colonization of Health Care Workers by Carbapenem-Resistant Enterobacteriaceae in an Endemic Hospital: A Prospective Study. Am. J. Infect. Control 2016, 44, 1053–1054. [Google Scholar] [CrossRef] [PubMed]
- Zarras, C.; Karampatakis, T.; Pappa, S.; Iosifidis, E.; Vagdatli, E.; Roilides, E.; Papa, A. Genetic Characterization of Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates in a Tertiary Hospital in Greece, 2018–2022. Antibiotics 2023, 12, 976. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, M.; Deliolanis, I.; Polemis, M.; Vatopoulos, A.; Psichogiou, M.; Giakkoupi, P. Characteristics of the Genetic Spread of Carbapenem-Resistant Acinetobacter baumannii in a Tertiary Greek Hospital. Genes 2024, 15, 458. [Google Scholar] [CrossRef]
- Gong, Y.; Shen, X.; Huang, G.; Zhang, C.; Luo, X.; Yin, S.; Wang, J.; Hu, F.; Peng, Y.; Li, M. Epidemiology and Resistance Features of Acinetobacter baumannii Isolates from the Ward Environment and Patients in the Burn ICU of a Chinese Hospital. J. Microbiol. 2016, 54, 551–558. [Google Scholar] [CrossRef]
- Hu, H.; Lou, Y.; Feng, H.; Tao, J.; Shi, W.; Ni, S.; Pan, Q.; Ge, T.; Shen, P.; Zhong, Z.; et al. Molecular Characterization of Carbapenem-Resistant Acinetobacter baumannii Isolates Among Intensive Care Unit Patients and Environment. Infect. Drug Resist. 2022, 15, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, G.R.; Kim, E.-Y.; Jeong, J.; Kim, S.; Shin, J.H. Carbapenemase-Producing Eenterobacterales from Hospital Environment and Their Relation to Those from Patient Specimens. J. Infect. Public. Health 2022, 15, 241–244. [Google Scholar] [CrossRef]
- European Food Safety Authority. Scientific Opinion on Carbapenem Resistance in Food Animal Ecosystems. EFSA J. 2013, 11, 3501. [Google Scholar] [CrossRef]
- King, D.T.; Sobhanifar, S.; Strynadka, N.C.J. The Mechanisms of Resistance to β-Lactam Antibiotics. In Handbook of Antimicrobial Resistance; Springer: New York, NY, USA, 2014; pp. 1–22. [Google Scholar]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Woodford, N.; Ellington, M.; Coelho, J.; Turton, J.; Ward, M.; Brown, S.; Amyes, S.; Livermore, D. Multiplex PCR for Genes Encoding Prevalent OXA Carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 2006, 27, 351–353. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, R.A.; Jousset, A.B.; Emeraud, C.; Oueslati, S.; Dortet, L.; Naas, T. Genetic Diversity, Biochemical Properties, and Detection Methods of Minor Carbapenemases in Enterobacterales. Front. Med. 2021, 7, 616490. [Google Scholar] [CrossRef]
- Chen, L.; Tan, P.; Zeng, J.; Yu, X.; Cai, Y.; Liao, K.; Guo, P.; Chen, Y.; Wu, Z.; Qu, P.; et al. Impact of an Intervention to Control Imipenem-Resistant Acinetobacter baumannii and Its Resistance Mechanisms: An 8-Year Survey. Front. Microbiol. 2021, 11, 610109. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Costello, S.E.; Woosley, L.N.; Deshpande, L.M.; Davies, T.A.; Jones, R.N. Evaluation of Clonality and Carbapenem Resistance Mechanisms among Acinetobacter baumannii-Acinetobacter Calcoaceticus Complex and Enterobacteriaceae Isolates Collected in European and Mediterranean Countries and Detection of Two Novel β-Lactamases, GES-22 and VIM-35. Antimicrob. Agents Chemother. 2014, 58, 7358–7366. [Google Scholar] [CrossRef]
- Kızılyıldırım, S.; Kandemir, T.; Kendir, G.; Muhammed, M.T.; Köroğlu, A.; Köksal, F.; Ozogul, F. The Antibacterial Effect Mechanisms of Laurus Nobilis Extracts on Carbapenem-Resistant Acinetobacter Baumanii Isolates. Food Biosci. 2024, 59, 104011. [Google Scholar] [CrossRef]
- Starlander, G.; Yin, H.; Edquist, P.; Melhus, Å. Survival in the Environment Is a Possible Key Factor for the Expansion of Escherichia coli Strains Producing Extended-spectrum Β-lactamases. APMIS 2014, 122, 59–67. [Google Scholar] [CrossRef]
- Clarivet, B.; Grau, D.; Jumas-Bilak, E.; Jean-Pierre, H.; Pantel, A.; Parer, S.; Lotthé, A. Persisting Transmission of Carbapenemase-Producing Klebsiella Pneumoniae Due to an Environmental Reservoir in a University Hospital, France, 2012 to 2014. Eurosurveillance 2016, 21, 30213. [Google Scholar] [CrossRef]
- Benbow, A.; Clarke, M.; Yates, C.; Montgomery, R.; Staniforth, K.; Boswell, T.; Prescott, K.; Mahida, N. Hospital-Wide Healthcare-Associated Carbapenemase-Producing Enterobacterales Outbreak: Risks of Electric Floor Scrubbers in Catering Facilities and Kitchens. J. Hosp. Infect. 2024, 146, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Pais, S.; Costa, M.; Barata, A.R.; Rodrigues, L.; Afonso, I.M.; Almeida, G. Evaluation of Antimicrobial Resistance of Different Phylogroups of Escherichia coli Isolates from Feces of Breeding and Laying Hens. Antibiotics 2022, 12, 20. [Google Scholar] [CrossRef]
- Lagerstrom, K.M.; Hadly, E.A. Under-Appreciated Phylogroup Diversity of Escherichia coli within and between Animals at the Urban-Wildland Interface. Appl. Environ. Microbiol. 2023, 89, e00142-23. [Google Scholar] [CrossRef]
- European Food Safety Authority. Scientific Opinion on the Public Health Risks of Bacterial Strains Producing Extended-Spectrum β-Lactamases and/or AmpC β-Lactamases in Food and Food-Producing Animals. EFSA J. 2011, 9, 2322. [Google Scholar] [CrossRef]
- Carvalheira, A.; Ferreira, V.; Silva, J.; Teixeira, P. Enrichment of Acinetobacter spp. from Food Samples. Food Microbiol. 2016, 55, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Peratikos, P.; Tsitsos, A.; Damianos, A.; Kyritsi, M.A.; Hadjichristodoulou, C.; Soultos, N.; Economou, V. Listeria monocytogenes from Marine Fish and the Seafood Market Environment in Northern Greece: Prevalence, Molecular Characterization, and Antibiotic Resistance. Appl. Sci. 2024, 14, 2725. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 34th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2024. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2017; Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (accessed on 1 June 2024).
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important β-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli Phylo-typing Method Revisited: Improvement of Specificity and Detection of New Phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsitsos, A.; Damianos, A.; Boutel, M.; Gousia, P.; Soultos, N.; Papa, A.; Tirodimos, I.; Economou, V. Prevalence, Characterization, and Epidemiological Relationships between ESBL and Carbapenemase-Producing Escherichia coli, Klebsiella pneumoniae, and Acinetobacter spp. Isolated from Humans and the Kitchen Environment of Two Greek Hospitals. Antibiotics 2024, 13, 934. https://doi.org/10.3390/antibiotics13100934
Tsitsos A, Damianos A, Boutel M, Gousia P, Soultos N, Papa A, Tirodimos I, Economou V. Prevalence, Characterization, and Epidemiological Relationships between ESBL and Carbapenemase-Producing Escherichia coli, Klebsiella pneumoniae, and Acinetobacter spp. Isolated from Humans and the Kitchen Environment of Two Greek Hospitals. Antibiotics. 2024; 13(10):934. https://doi.org/10.3390/antibiotics13100934
Chicago/Turabian StyleTsitsos, Anestis, Alexandros Damianos, Maria Boutel, Panagiota Gousia, Nikolaos Soultos, Anna Papa, Ilias Tirodimos, and Vangelis Economou. 2024. "Prevalence, Characterization, and Epidemiological Relationships between ESBL and Carbapenemase-Producing Escherichia coli, Klebsiella pneumoniae, and Acinetobacter spp. Isolated from Humans and the Kitchen Environment of Two Greek Hospitals" Antibiotics 13, no. 10: 934. https://doi.org/10.3390/antibiotics13100934
APA StyleTsitsos, A., Damianos, A., Boutel, M., Gousia, P., Soultos, N., Papa, A., Tirodimos, I., & Economou, V. (2024). Prevalence, Characterization, and Epidemiological Relationships between ESBL and Carbapenemase-Producing Escherichia coli, Klebsiella pneumoniae, and Acinetobacter spp. Isolated from Humans and the Kitchen Environment of Two Greek Hospitals. Antibiotics, 13(10), 934. https://doi.org/10.3390/antibiotics13100934








