Four New Sequence Types and Molecular Characteristics of Multidrug-Resistant Escherichia coli Strains from Foods in Thailand
Abstract
1. Introduction
2. Results
2.1. Prevalence of Antibiotic-Resistant E. coli
2.2. New STs and Prevalence of Clermont Types A and B1 in Thailand
2.3. Clermont Type-Specific blaEC Genes and blaEC-13 Presence in ESBL-Producing Strains
2.4. Virulence Patterns in E. coli Isolates and Most Predicted Virulence Genes Found in SM47E1
2.5. IncFIB (AP001918) Is the Most Frequently Detected Plasmid Replicon among E. coli Isolates in This Study
2.6. Positive Correlation between Gene Cluster for Yersiniabactin Production and Colistin Resistance Phenotype in Clermont Type B1
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Cultivations
4.2. Antibiotic Resistance Susceptibility Testing
4.3. Genomic DNA Extraction and Whole Genome Sequencing
4.4. Genome Analysis
4.5. Correlation and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, F.; Zhang, W.; Niu, D. Editorial: Foodborne Enterobacteriaceae of Animal Origin. Front. Cell. Infect. Microbiol. 2021, 11, 772359. [Google Scholar] [CrossRef]
- Levine, M.M. Escherichia coli that cause diarrhea: Enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 1987, 155, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Bumyut, A.; Makkaew, P.; Yadee, K.; Hlamchoo, S.; Binyoosoh, I.; Precha, N. Assessment of food safety conditions at food service premises using Thai survey form and field fecal indicator testing in Pakpoon municipality of Nakhon Si Thammarat, Thailand. Food Sci. Technol. 2021, 42, e47521. [Google Scholar] [CrossRef]
- Ghafir, Y.; China, B.; Dierick, K.; De Zutter, L.; Daube, G. Hygiene indicator microorganisms for selected pathogens on beef, pork, and poultry meats in Belgium. J. Food Prot. 2008, 71, 35–45. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.E.; Shover, J.; Kallen, A.J.; Lonsway, D.R.; Watkins, S.; Miller, J.R. Investigation of First Identified mcr-1 Gene in an Isolate from a U.S. Patient—Pennsylvania, 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 977–978. [Google Scholar] [CrossRef]
- Bastidas-Caldes, C.; de Waard, J.H.; Salgado, M.S.; Villacis, M.J.; Coral-Almeida, M.; Yamamoto, Y.; Calvopina, M. Worldwide Prevalence of mcr-mediated Colistin-Resistance Escherichia coli in Isolates of Clinical Samples, Healthy Humans, and Livestock-A Systematic Review and Meta-Analysis. Pathogens 2022, 11, 659. [Google Scholar] [CrossRef]
- Li, W.; Yan, Y.; Chen, J.; Sun, R.; Wang, Y.; Wang, T.; Feng, Z.; Peng, K.; Wang, J.; Chen, S.; et al. Genomic characterization of conjugative plasmids carrying the mcr-1 gene in foodborne and clinical strains of Salmonella and Escherichia coli. Food Control. 2021, 125, 108032. [Google Scholar] [CrossRef]
- Asokan, G.V.; Ramadhan, T.; Ahmed, E.; Sanad, H. WHO Global Priority Pathogens List: A Bibliometric Analysis of Medline-PubMed for Knowledge Mobilization to Infection Prevention and Control Practices in Bahrain. Oman Med. J. 2019, 34, 184–193. [Google Scholar] [CrossRef]
- Pumart, P.; Phodha, T.; Thamlikitkul, V.; Riewpaiboon, A.; Prakongsai, P.; Limwattananon, S. Health and economic impacts of antimicrobial resistance in Thailand. J. Health Systems Res. 2012, 6, 352–360. [Google Scholar]
- Boonyasiri, A.; Tangkoskul, T.; Seenama, C.; Saiyarin, J.; Tiengrim, S.; Thamlikitkul, V. Prevalence of antibiotic resistant bacteria in healthy adults, foods, food animals, and the environment in selected areas in Thailand. Pathog. Glob. Health 2014, 108, 235–245. [Google Scholar] [CrossRef]
- NARST. Annual Report for Antibiogram and Global Antimicrobial Resistance Surveillance System in Thailand. Available online: http://narst.dmsc.moph.go.th/ (accessed on 2 October 2023).
- Tangcharoensathien, V.; Sommanustweechai, A.; Chanvatik, S.; Kosiyaporn, H.; Tisocki, K. Addressing the threat of antibiotic resistance in Thailand: Monitoring population knowledge and awareness. WHO South East Asia J. Public Health 2018, 7, 73–78. [Google Scholar] [CrossRef]
- Trongjit, S.; Angkittitrakul, S.; Chuanchuen, R. Occurrence and molecular characteristics of antimicrobial resistance of Escherichia coli from broilers, pigs and meat products in Thailand and Cambodia provinces. Microbiol. Immunol. 2016, 60, 575–585. [Google Scholar] [CrossRef]
- Ngamwongsatit, N.; Chaturongakul, S.; Aunpad, R. Development and Validation of an Efficient Multiplex PCR Assay for Simultaneous Detection of Six Common Foodborne Pathogens and Hygiene Indicators. Foodborne Pathog. Dis. 2023, 20, 222–229. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Ksiazczyk, M.; Dudek, B.; Kuczkowski, M.; O’Hara, R.; Korzekwa, K.; Wzorek, A.; Korzeniowska-Kowal, A.; Upton, M.; Junka, A.; Wieliczko, A.; et al. The Phylogenetic Structure of Reptile, Avian and Uropathogenic Escherichia coli with Particular Reference to Extraintestinal Pathotypes. Int. J. Mol. Sci. 2021, 22, 1192. [Google Scholar] [CrossRef]
- Abavisani, M.; Bostanghadiri, N.; Ghahramanpour, H.; Kodori, M.; Akrami, F.; Fathizadeh, H.; Hashemi, A.; Rastegari-Pouyani, M. Colistin resistance mechanisms in Gram-negative bacteria: A Focus on Escherichia coli. Lett. Appl. Microbiol. 2023, 76, ovad023. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Ewers, C.; Wieler, L.H. Extended-Spectrum Beta-Lactamases Producing E. coli in Wildlife, yet Another Form of Environmental Pollution? Front. Microbiol. 2011, 2, 246. [Google Scholar] [CrossRef]
- Yang, J.T.; Zhang, L.J.; Lu, Y.; Zhang, R.M.; Jiang, H.X. Genomic Insights into Global bla(CTX-M-55)-Positive Escherichia coli Epidemiology and Transmission Characteristics. Microbiol. Spectr. 2023, 11, e0108923. [Google Scholar] [CrossRef]
- Suay-Garcia, B.; Perez-Gracia, M.T. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics 2019, 8, 122. [Google Scholar] [CrossRef]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzym. Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Shaikh, S.; Fatima, J.; Shakil, S.; Rizvi, S.M.; Kamal, M.A. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J. Biol. Sci. 2015, 22, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, H.; Zhou, Z.; Miao, Y.; Li, R.; Yang, B.; Cao, C.; Xiao, S.; Wang, X.; Liu, H.; et al. Characterization of Extended-Spectrum beta-Lactamase-Producing Escherichia coli Isolates That Cause Diarrhea in Sheep in Northwest China. Microbiol. Spectr. 2022, 10, e0159522. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Yin, M.; Chen, J.; Li, X. Assembly and substrate recognition of curli biogenesis system. Nat. Commun. 2020, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.K.; Paul, K.; Blair, D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2010, 107, 9370–9375. [Google Scholar] [CrossRef]
- Peralta, D.R.; Adler, C.; Corbalan, N.S.; Paz Garcia, E.C.; Pomares, M.F.; Vincent, P.A. Enterobactin as Part of the Oxidative Stress Response Repertoire. PLoS ONE 2016, 11, e0157799. [Google Scholar] [CrossRef]
- Garnett, J.A.; Martinez-Santos, V.I.; Saldana, Z.; Pape, T.; Hawthorne, W.; Chan, J.; Simpson, P.J.; Cota, E.; Puente, J.L.; Giron, J.A.; et al. Structural insights into the biogenesis and biofilm formation by the Escherichia coli common pilus. Proc. Natl. Acad. Sci. USA 2012, 109, 3950–3955. [Google Scholar] [CrossRef]
- Schwan, W.R. Regulation of fim genes in uropathogenic Escherichia coli. World J. Clin. Infect. Dis. 2011, 1, 17–25. [Google Scholar] [CrossRef]
- Sandkvist, M. Biology of type II secretion. Mol. Microbiol. 2001, 40, 271–283. [Google Scholar] [CrossRef]
- Strozen, T.G.; Li, G.; Howard, S.P. YghG (GspSbeta) is a novel pilot protein required for localization of the GspSbeta type II secretion system secretin of enterotoxigenic Escherichia coli. Infect. Immun. 2012, 80, 2608–2622. [Google Scholar] [CrossRef] [PubMed]
- Addy, H.S.; Askora, A.; Kawasaki, T.; Fujie, M.; Yamada, T. Disruption of gspD and its Effects on Endoglucanase and Filamentous Phage Secretion in Ralstonia solanacearum. Procedia Environ. Sci. 2014, 20, 753–759. [Google Scholar] [CrossRef][Green Version]
- Mey, A.R.; Gomez-Garzon, C.; Payne, S.M. Iron Transport and Metabolism in Escherichia, Shigella, and Salmonella. EcoSal Plus 2021, 9, eESP00342020. [Google Scholar] [CrossRef]
- Chervy, M.; Barnich, N.; Denizot, J. Adherent-Invasive E. coli: Update on the Lifestyle of a Troublemaker in Crohn’s Disease. Int. J. Mol. Sci. 2020, 21, 3734. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Wenderoth, S.; Carlino, U.B.; Merrick, J.M.; Lesse, A.J. Identification, genomic organization, and analysis of the group III capsular polysaccharide genes kpsD, kpsM, kpsT, and kpsE from an extraintestinal isolate of Escherichia coli (CP9, O4/K54/H5). J. Bacteriol. 1998, 180, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.H.; Yang, Y.Y.; Hsieh, W.S.; Lee, C.H.; Leu, S.J.; Chen, M.R. OmpA is the critical component for Escherichia coli invasion-induced astrocyte activation. J. Neuropathol. Exp. Neurol. 2009, 68, 677–690. [Google Scholar] [CrossRef]
- Werneburg, G.T.; Thanassi, D.G. Pili Assembled by the Chaperone/Usher Pathway in Escherichia coli and Salmonella. EcoSal Plus 2018, 8, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Rakin, A.; Schneider, L.; Podladchikova, O. Hunger for iron: The alternative siderophore iron scavenging systems in highly virulent Yersinia. Front. Cell. Infect. Microbiol. 2012, 2, 151. [Google Scholar] [CrossRef] [PubMed]
- Dogan, O.; Vatansever, C.; Atac, N.; Albayrak, O.; Karahuseyinoglu, S.; Sahin, O.E.; Kilicoglu, B.K.; Demiray, A.; Ergonul, O.; Gonen, M.; et al. Virulence Determinants of Colistin-Resistant K. pneumoniae High-Risk Clones. Biology 2021, 10, 436. [Google Scholar] [CrossRef]
- Vignaroli, C.; Luna, G.M.; Rinaldi, C.; Di Cesare, A.; Danovaro, R.; Biavasco, F. New sequence types and multidrug resistance among pathogenic Escherichia coli isolates from coastal marine sediments. Appl. Environ. Microbiol. 2012, 78, 3916–3922. [Google Scholar] [CrossRef] [PubMed]
- Chotinantakul, K.; Woottisin, S.; Okada, S. The Emergence of CTX-M-55 in Extended-Sectrum beta-Lactamase-Producing Escherichia coli from Vegetables Sold in Local Markets of Northern Thailand. Jpn. J. Infect. Dis. 2022, 75, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Pungpian, C.; Lee, S.; Trongjit, S.; Sinwat, N.; Angkititrakul, S.; Prathan, R.; Srisanga, S.; Chuanchuen, R. Colistin resistance and plasmid-mediated mcr genes in Escherichia coli and Salmonella isolated from pigs, pig carcass and pork in Thailand, Lao PDR and Cambodia border provinces. J. Vet. Sci. 2021, 22, e68. [Google Scholar] [CrossRef] [PubMed]
- Aljohani, R.H.; ElFeky, D.S.; Alswaji, A.A.; Alrashidi, E.; Okdah, L.; Alalwan, B.; Aljohani, S.M.; Balkhy, H.H.; Redhwan, A.; Alghoribi, M.F. Genomic Characterization of Uropathogenic Escherichia coli Isolates from Tertiary Hospitals in Riyadh, Saudi Arabia. Int. J. Mol. Sci. 2023, 24, 7582. [Google Scholar] [CrossRef]
- Haley, B.J.; Kim, S.W.; Salaheen, S.; Hovingh, E.; Van Kessel, J.A.S. Genome-Wide Analysis of Escherichia coli Isolated from Dairy Animals Identifies Virulence Factors and Genes Enriched in Multidrug-Resistant Strains. Antibiotics 2023, 12, 1559. [Google Scholar] [CrossRef]
- Salaheen, S.; Kim, S.W.; Springer, H.R.; Hovingh, E.P.; Van Kessel, J.A.S.; Haley, B.J. Genomic diversity of antimicrobial-resistant and Shiga toxin gene-harboring non-O157 Escherichia coli from dairy calves. J. Glob. Antimicrob. Resist. 2023, 33, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Botelho, J.; Cazares, A.; Schulenburg, H. The ESKAPE mobilome contributes to the spread of antimicrobial resistance and CRISPR-mediated conflict between mobile genetic elements. Nucleic Acids Res. 2023, 51, 236–252. [Google Scholar] [CrossRef] [PubMed]
- Morales, G.; Abelson, B.; Reasoner, S.; Miller, J.; Earl, A.M.; Hadjifrangiskou, M.; Schmitz, J. The Role of Mobile Genetic Elements in Virulence Factor Carriage from Symptomatic and Asymptomatic Cases of Escherichia coli Bacteriuria. Microbiol. Spectr. 2023, 11, e0471022. [Google Scholar] [CrossRef]
- Feng, P.; Weagant, S.D.; Grant, M.A.; Burkhardt, W. Chapter 4: Enumeration of Escherichia coli and the coliform bacteria. In Bacteriological Analytical Manual (BAM); Food and Drug Administration: Silver Spring, MD, USA, 2017. [Google Scholar]
- CLSI. Performance Standard for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Benov, L. Effect of growth media on the MTT colorimetric assay in bacteria. PLoS ONE 2019, 14, e0219713. [Google Scholar] [CrossRef] [PubMed]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Thadtapong, N.; Chaturongakul, S.; Soodvilai, S.; Dubbs, P. Colistin and Carbapenem-Resistant Acinetobacter baumannii Aci46 in Thailand: Genome Analysis and Antibiotic Resistance Profiling. Antibiotics 2021, 10, 1054. [Google Scholar] [CrossRef]
- Andrews, S. FastQC A Quality Control tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 January 2019).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Molina-Mora, J.A.; Campos-Sanchez, R.; Rodriguez, C.; Shi, L.; Garcia, F. High quality 3C de novo assembly and annotation of a multidrug resistant ST-111 Pseudomonas aeruginosa genome: Benchmark of hybrid and non-hybrid assemblers. Sci. Rep. 2020, 10, 1392. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Huang, N.; Li, H. Compleasm: A faster and more accurate reimplementation of BUSCO. Bioinformatics 2023, 39, btad595. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Agama Study, G.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef] [PubMed]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder—An open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 2022, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]

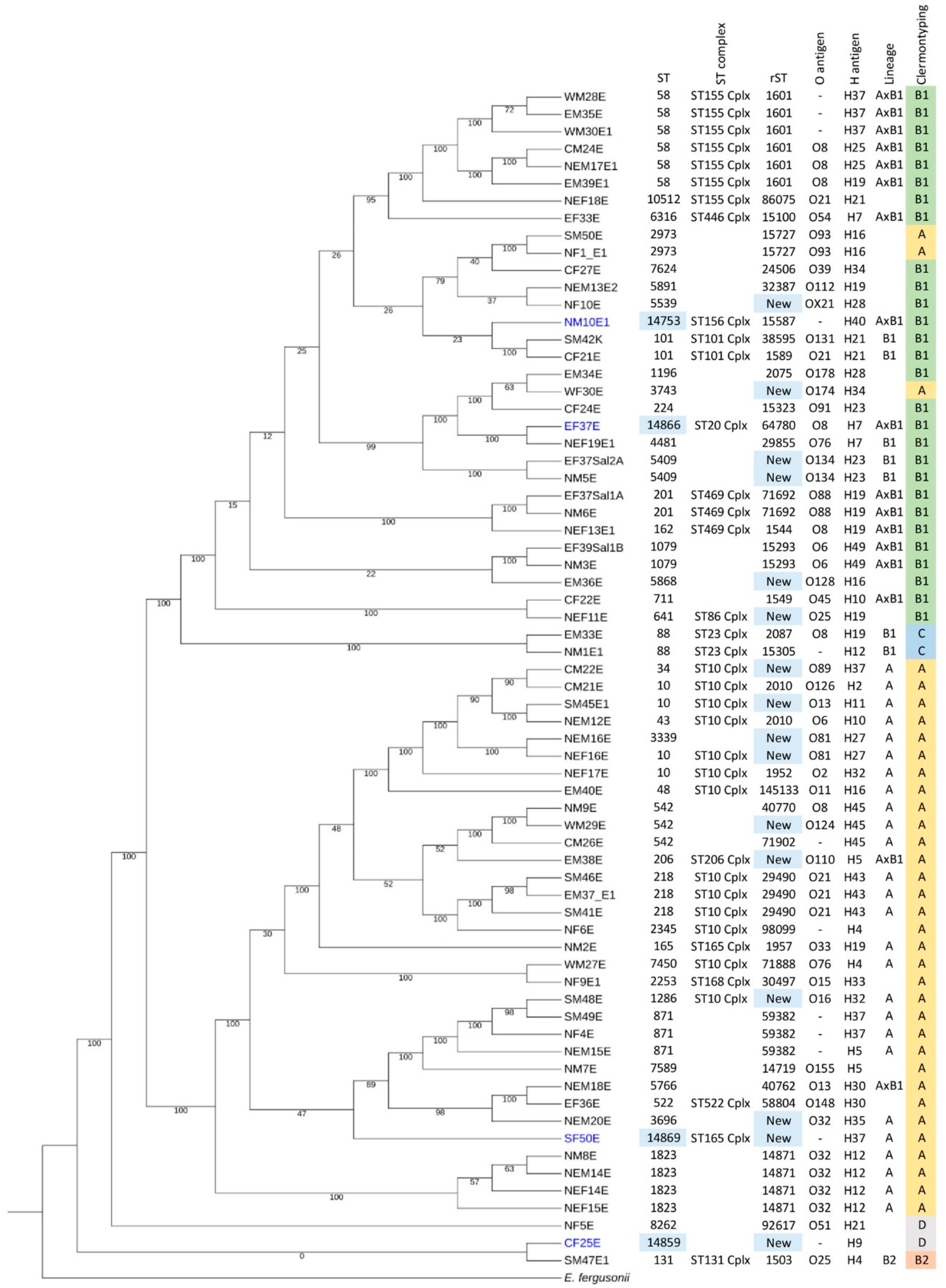
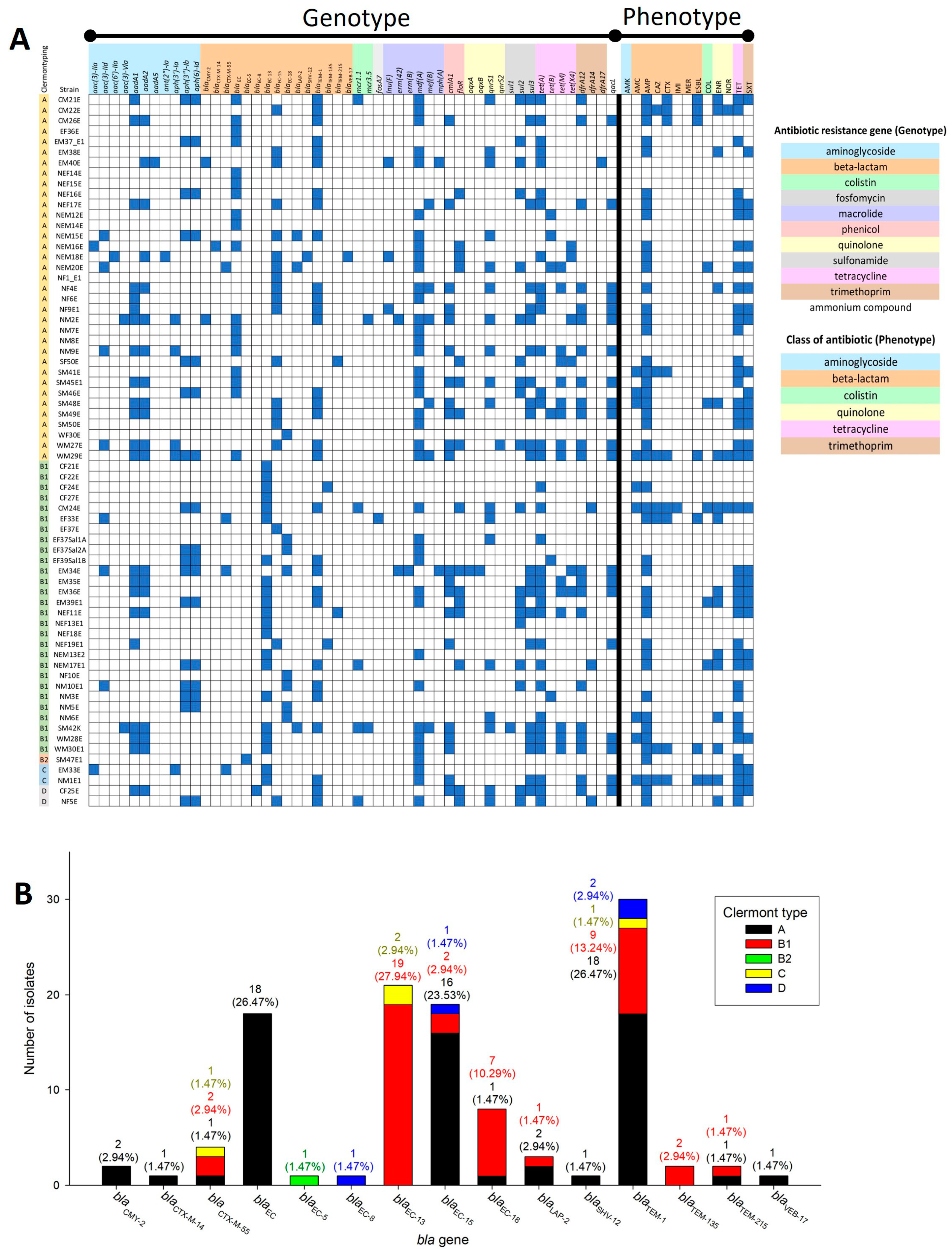
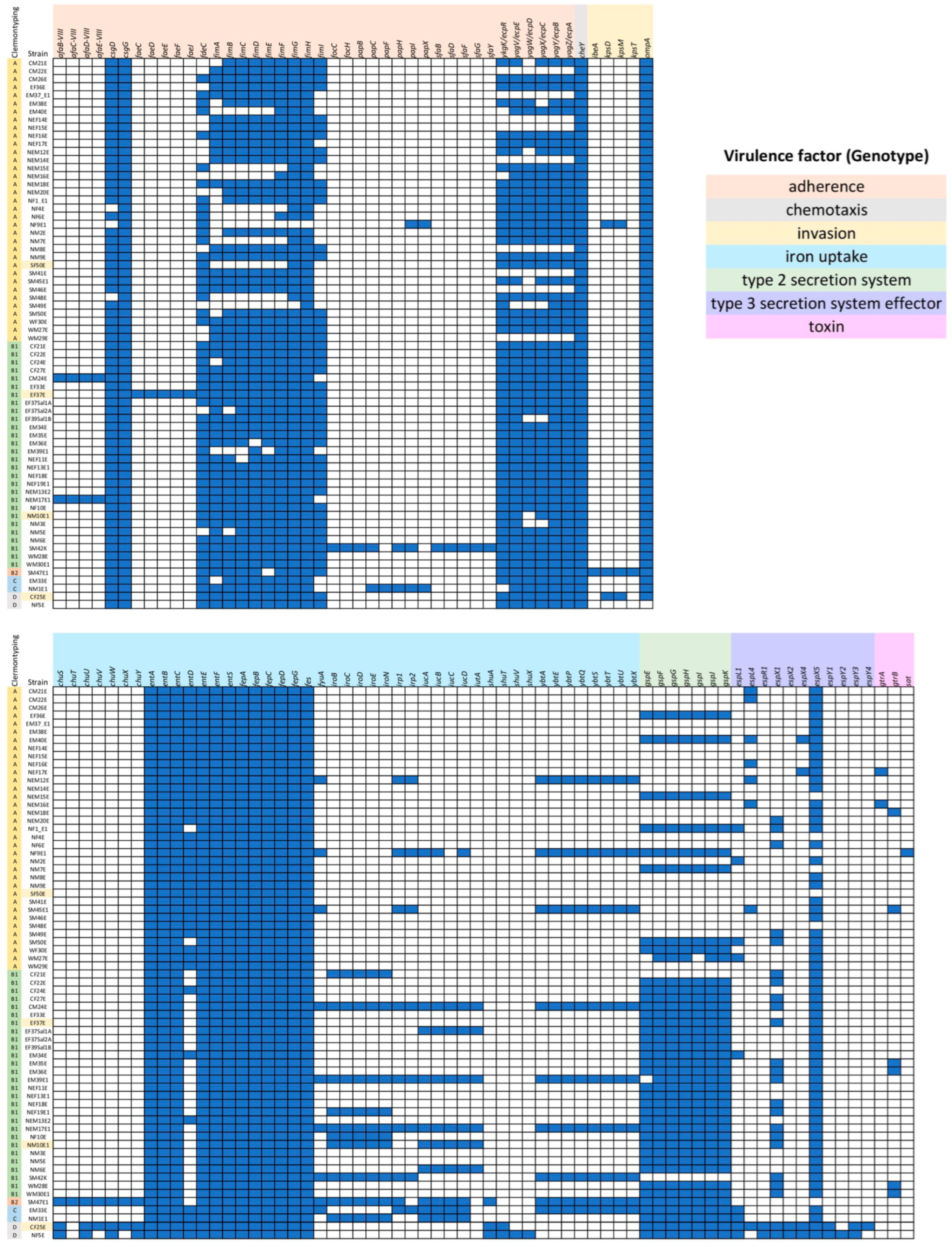

| AMR Pattern A | Number of Strains | % Population | MAR Index |
|---|---|---|---|
| AMC-AMP-CAZ-COL-CTX-ENR-IMI-NOR-TET-SXT | 1 | 1.47 | 0.83 |
| AMC-AMP-CAZ-COL-CTX-ENR-TET-SXT | 1 | 1.47 | 0.67 |
| AMC-AMP-CAZ-CTX-ENR-NOR-TET-SXT | 1 | 1.47 | 0.67 |
| AMC-AMP-CAZ-CTX-TET-SXT | 1 | 1.47 | 0.50 |
| AMC-AMP-COL-ENR-TET-SXT | 1 | 1.47 | 0.50 |
| AMP-CAZ-CTX-ENR-NOR-TET | 1 | 1.47 | 0.50 |
| AMP-COL-ENR-TET-SXT | 2 | 2.94 | 0.42 |
| AMP-CTX-ENR-TET-SXT | 1 | 1.47 | 0.42 |
| AMC-AMP-ENR-TET | 1 | 1.47 | 0.33 |
| AMC-AMP-TET-SXT | 1 | 1.47 | 0.33 |
| AMP-CAZ-CTX-ENR | 2 | 2.94 | 0.33 |
| AMP-COL-TET-SXT | 1 | 1.47 | 0.33 |
| AMP-ENR-TET-SXT | 3 | 4.41 | 0.33 |
| AMC-AMP-SXT | 1 | 1.47 | 0.25 |
| AMP-ENR-SXT | 1 | 1.47 | 0.25 |
| AMP-ENR-TET | 1 | 1.47 | 0.25 |
| AMP-TET-SXT | 11 | 16.18 | 0.25 |
| AMC-AMP | 1 | 1.47 | 0.17 |
| AMP-CTX | 1 | 1.47 | 0.17 |
| AMP-TET | 9 | 13.24 | 0.17 |
| TET-SXT | 2 | 2.94 | 0.17 |
| AMP | 2 | 2.94 | 0.08 |
| TET | 1 | 1.47 | 0.08 |
| ND (Not detected; sensitive for all selected antibiotics) | 21 | 30.88 | 0.00 |
| Comparison | Correlation Coefficient | p-Value | Interpretation |
|---|---|---|---|
| AMR genotype vs. AMR phenotype (Clermont type B1) | |||
| floR (phenicol resistance) vs. SXT | 0.759 | 2.87 × 10−6 | strong positive correlation |
| dfrA14 (trimethoprim resistance) vs. COL | 0.801 | 3.12 × 10−7 | strong positive correlation |
| Virulence genotype vs. AMR phenotype (Clermont type B1) | |||
| afaBCDE-VIII (adherence) vs. COL | 0.801 | 3.12 × 10−7 | strong positive correlation |
| fimI (adherence) vs. COL | −1.000 | 4.9 × 10−205 | very strong negative correlation |
| fyuA (iron uptake) vs. COL | 0.849 | 1.17 × 10−8 | strong positive correlation |
| irp12 (iron uptake) vs. COL | 0.849 | 1.17 × 10−8 | strong positive correlation |
| ybtAEPQSTUX (iron uptake) vs. COL | 0.849 | 1.17 × 10−8 | strong positive correlation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thadtapong, N.; Chaturongakul, S.; Tangphatsornruang, S.; Sonthirod, C.; Ngamwongsatit, N.; Aunpad, R. Four New Sequence Types and Molecular Characteristics of Multidrug-Resistant Escherichia coli Strains from Foods in Thailand. Antibiotics 2024, 13, 935. https://doi.org/10.3390/antibiotics13100935
Thadtapong N, Chaturongakul S, Tangphatsornruang S, Sonthirod C, Ngamwongsatit N, Aunpad R. Four New Sequence Types and Molecular Characteristics of Multidrug-Resistant Escherichia coli Strains from Foods in Thailand. Antibiotics. 2024; 13(10):935. https://doi.org/10.3390/antibiotics13100935
Chicago/Turabian StyleThadtapong, Nalumon, Soraya Chaturongakul, Sithichoke Tangphatsornruang, Chutima Sonthirod, Natharin Ngamwongsatit, and Ratchaneewan Aunpad. 2024. "Four New Sequence Types and Molecular Characteristics of Multidrug-Resistant Escherichia coli Strains from Foods in Thailand" Antibiotics 13, no. 10: 935. https://doi.org/10.3390/antibiotics13100935
APA StyleThadtapong, N., Chaturongakul, S., Tangphatsornruang, S., Sonthirod, C., Ngamwongsatit, N., & Aunpad, R. (2024). Four New Sequence Types and Molecular Characteristics of Multidrug-Resistant Escherichia coli Strains from Foods in Thailand. Antibiotics, 13(10), 935. https://doi.org/10.3390/antibiotics13100935






