Antibacterial and Antiviral Properties of Chenopodin-Derived Synthetic Peptides
Abstract
1. Introduction
2. Results
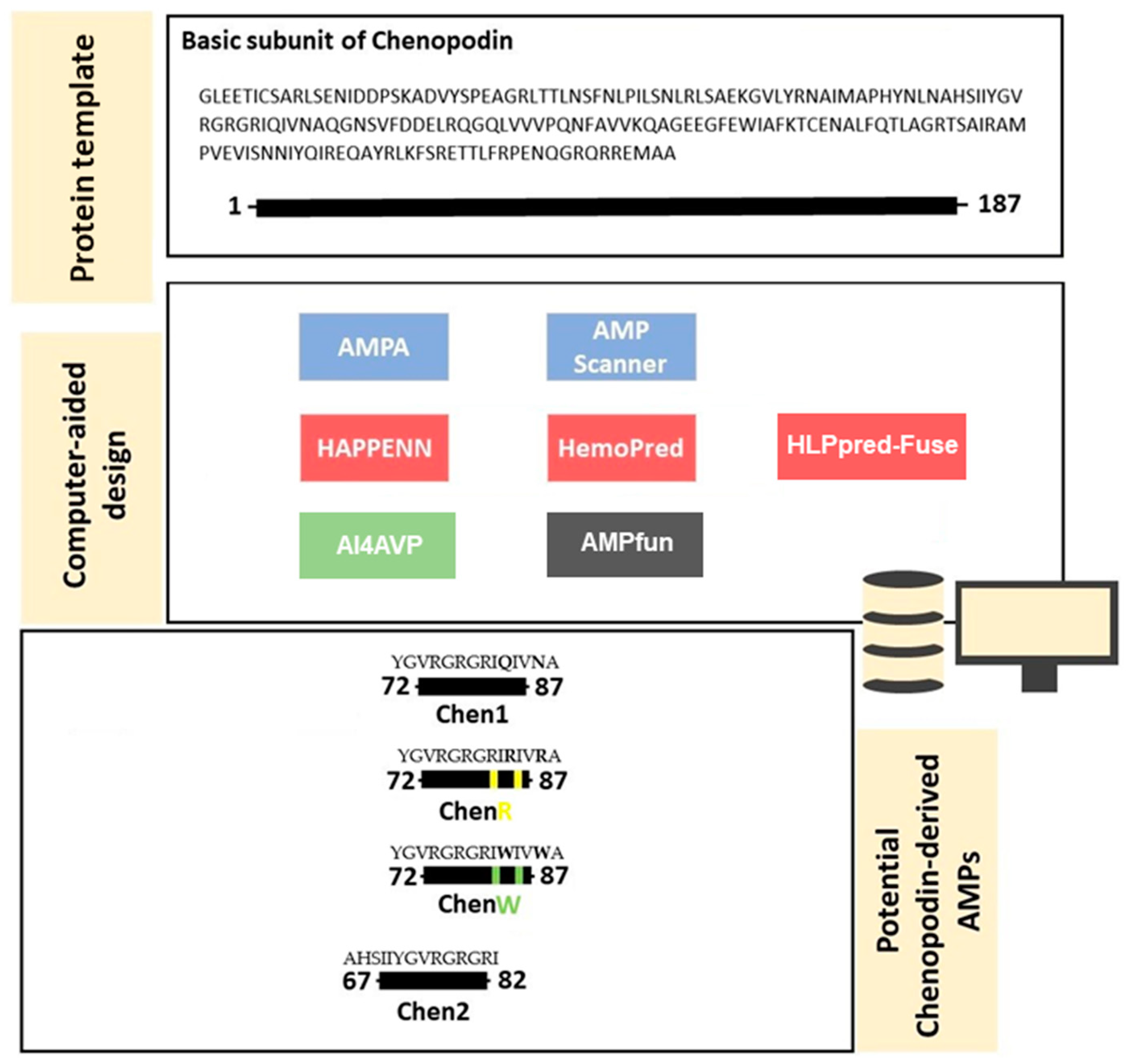
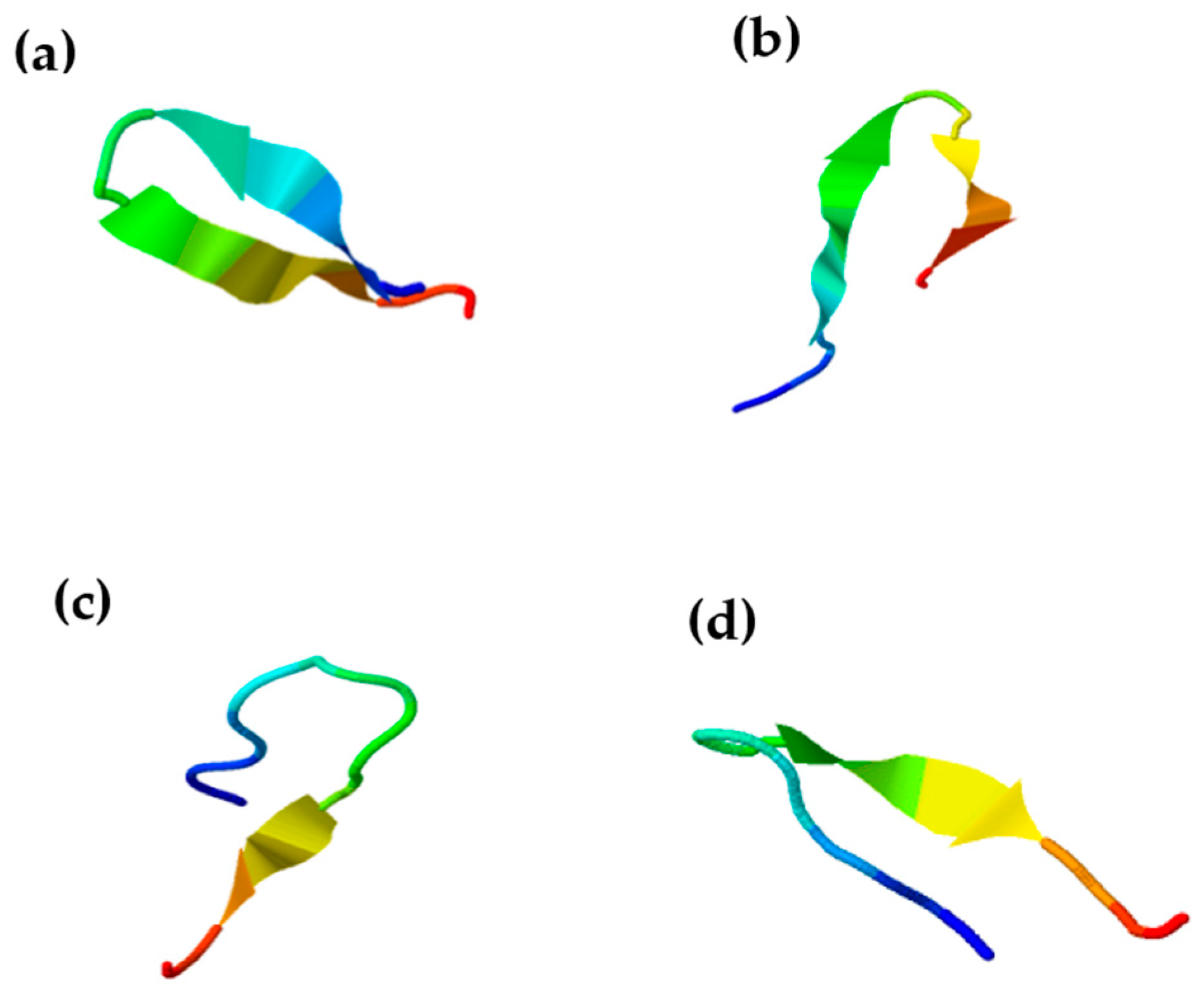
2.1. In Silico Design and Characterisation of Chenopodin-Derived Peptides
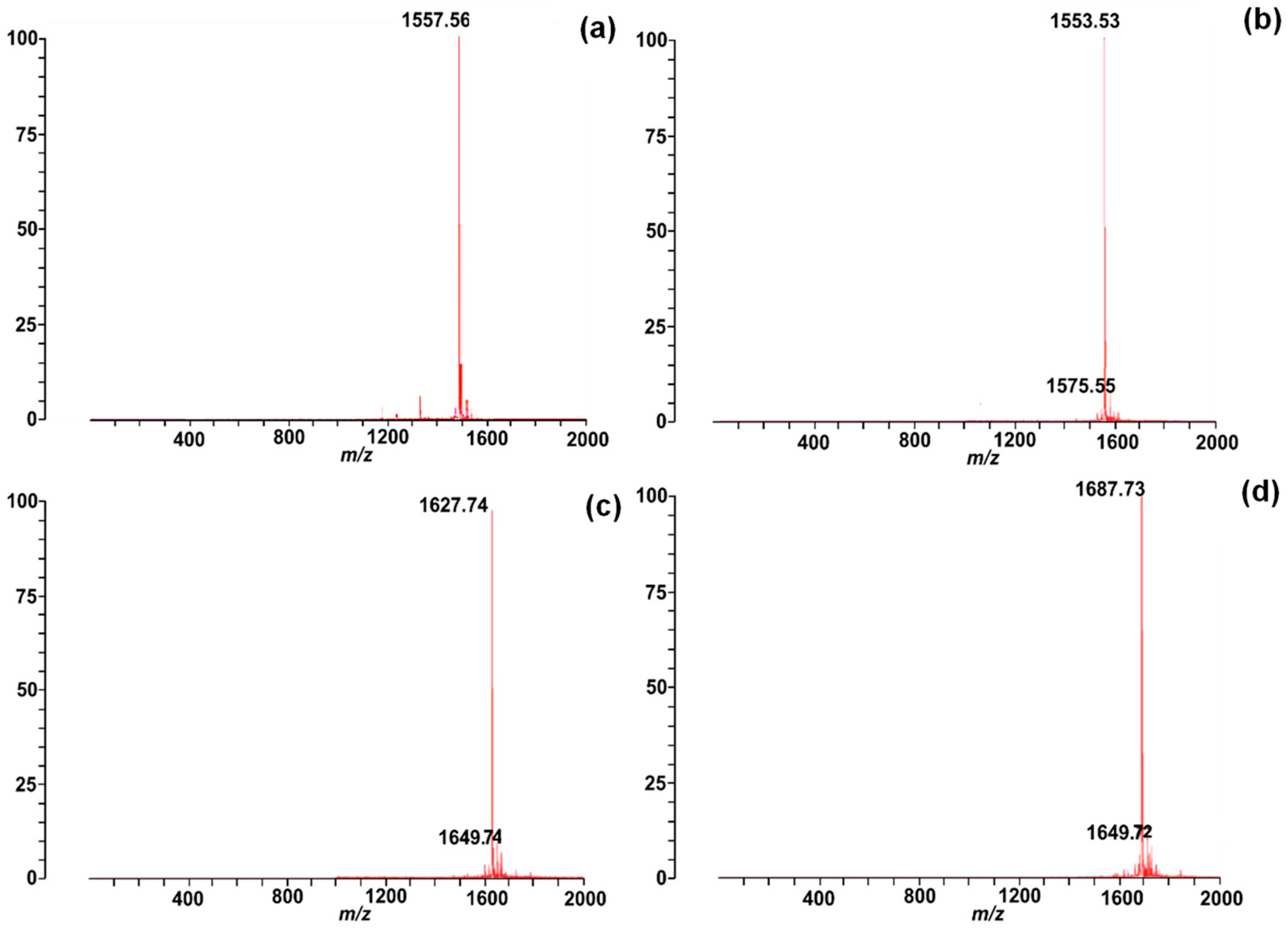
2.2. Synthesis and Characterisation of Chenopodin-Derived Peptides
2.3. Chenopodin-Derived Peptides Exhibit Antibacterial Activities
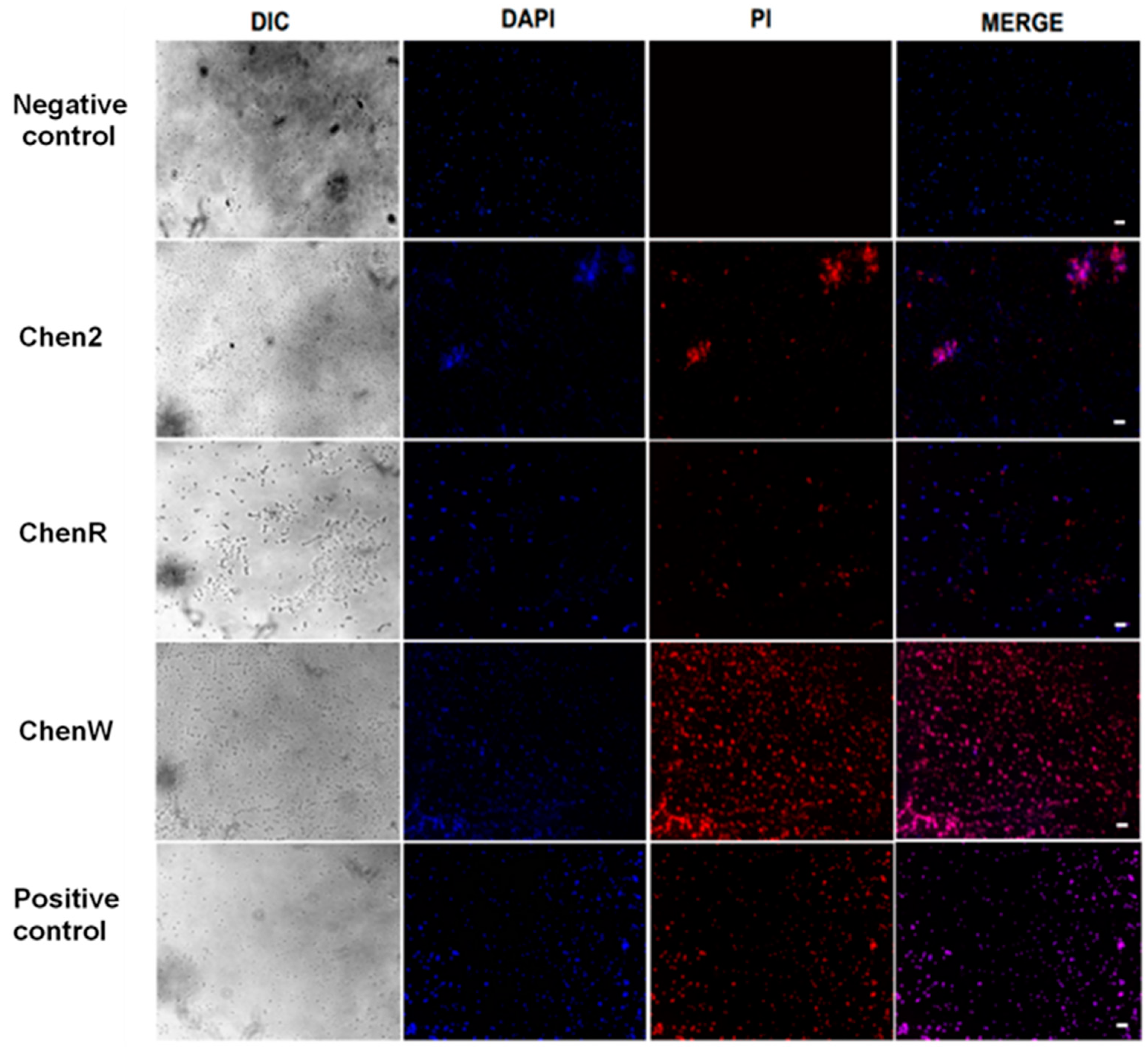
2.4. Chenopodin-Derived Peptides Damage the Bacterial Cell Membrane
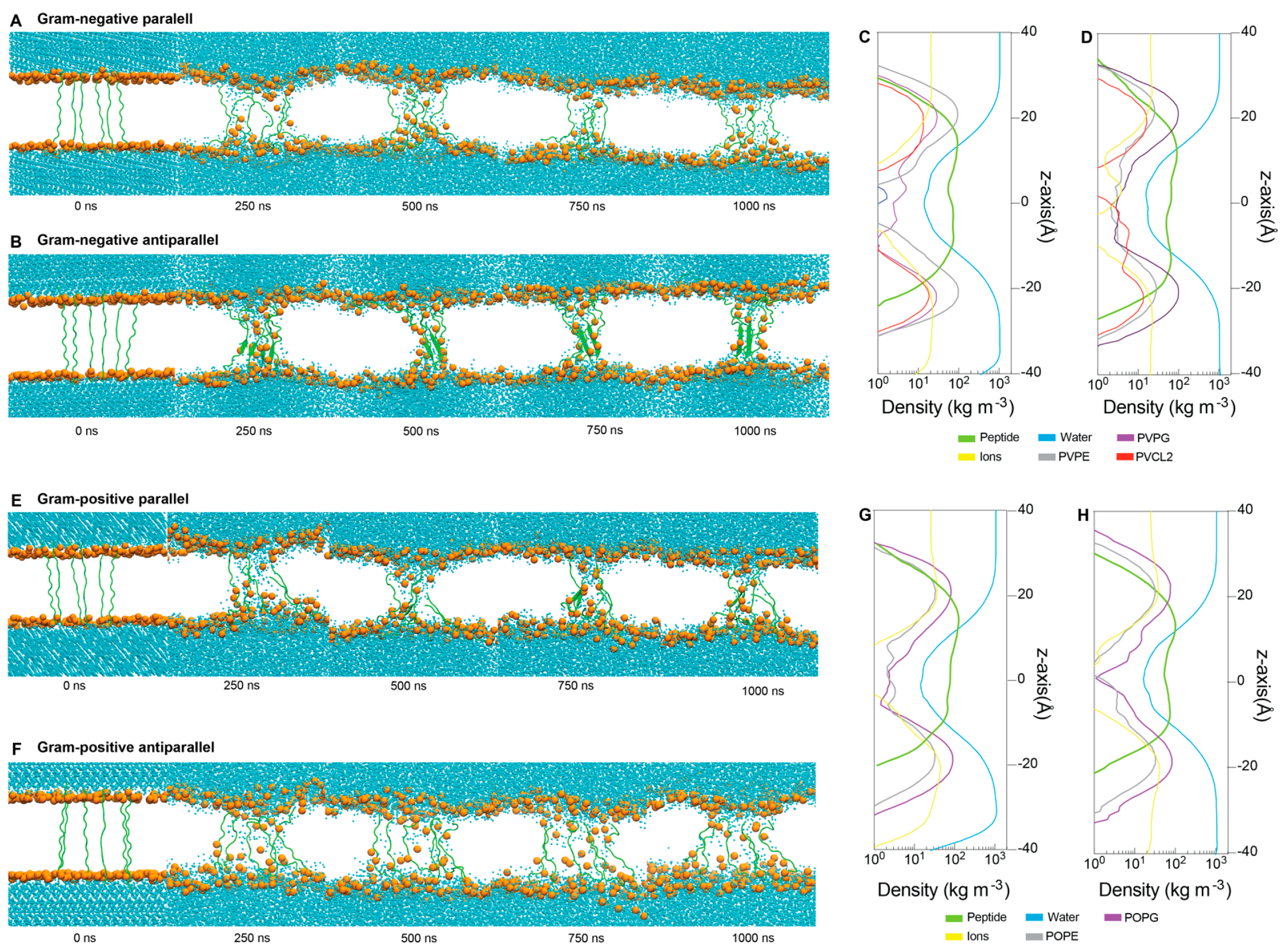
2.5. Interactions between ChenW and Bacterial Membranes
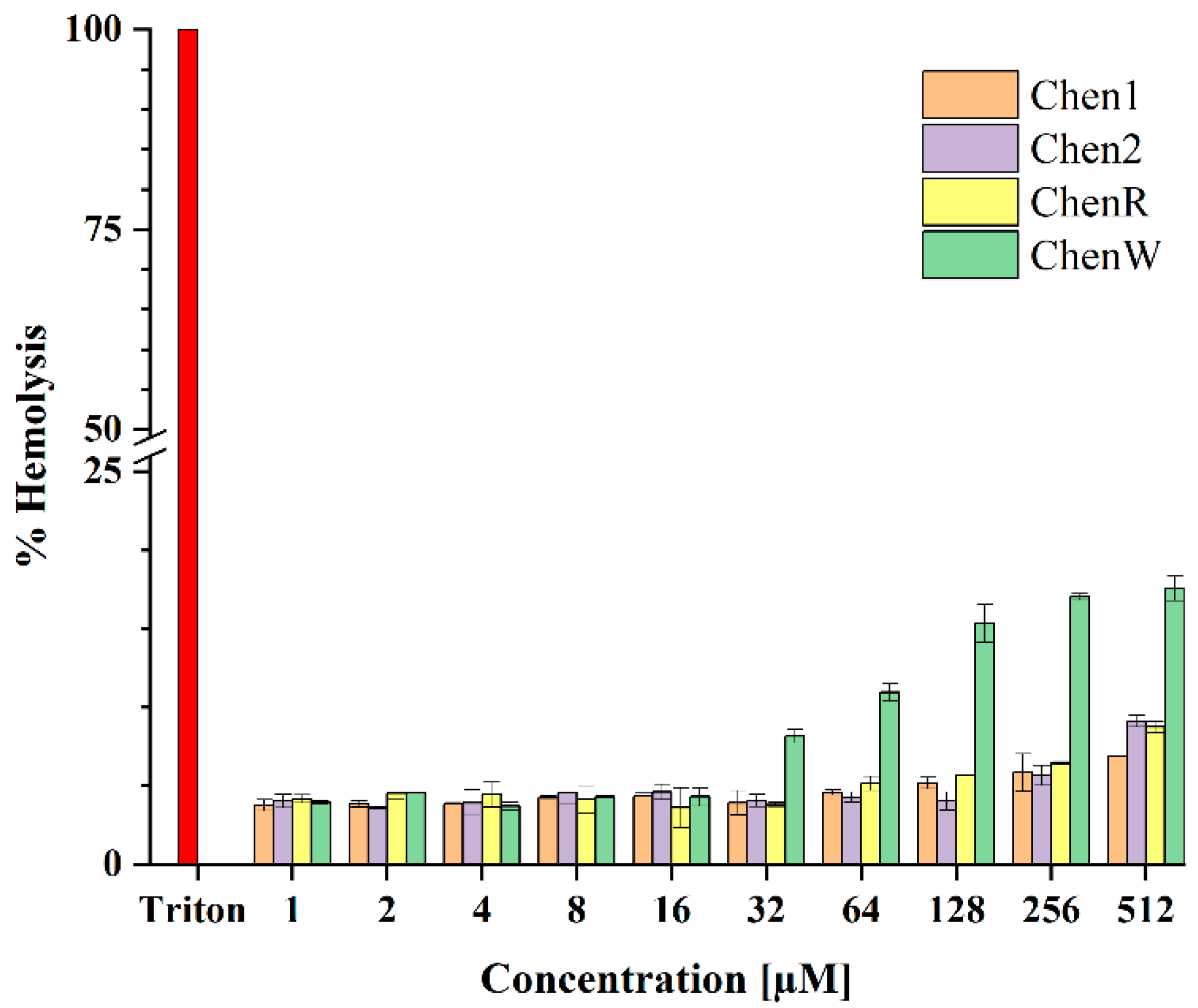
2.6. ChenW Peptide Exhibits Slight Haemolytic Activity
2.7. Chen2 and ChenR Peptides Display Antiviral Activities
3. Discussion
4. Materials and Methods
4.1. Computer-Aided Design of Cationic Peptides Based on Chenopodin
Screening of Chenopodin Sequence: Prediction of Toxicity, Structure and Biological Activity
4.2. Chenopodin-Directed Peptide Synthesis, Purification, and Molecular Mass Determination
4.3. Biological Evaluation
4.3.1. Peptide Bactericidality
4.3.2. Antiviral Activity and MTT Viability Assay
4.3.3. Hemolytic Activity
4.4. Insights into the Mechanism of Action
4.4.1. Fluorescence Microscopy to Assess Membranolytic Properties
4.4.2. Molecular Dynamic Simulations
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Monetary Fund (IMF). World Economic Outlook: Recovery during a Pandemic-Health Concerns, Supply Disruptions, Price Pressures; International Monetary Fund: Washington, DC, USA, 2021; ISBN 9781513577524.
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.-F.; et al. Infectious Disease in an Era of Global Change. Nat. Rev. Microbiol. 2022, 20, 193–205. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). WHO Coronavirus (COVID-19) Dashboard. Available online: https://data.who.int/dashboards/covid19/cases?n=c (accessed on 31 December 2023).
- Lai, C.-C.; Chen, S.-Y.; Ko, W.-C.; Hsueh, P.-R. Increased Antimicrobial Resistance during the COVID-19 Pandemic. Int. J. Antimicrob. Agents 2021, 57, 106324. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Bornman, C.; Zafer, M.M. Antimicrobial Resistance Threats in the Emerging COVID-19 Pandemic: Where Do We Stand? J. Infect. Public Health 2021, 14, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Westwood, D.; MacFadden, D.R.; Soucy, J.-P.R.; Daneman, N. Bacterial Co-Infection and Secondary Infection in Patients with COVID-19: A Living Rapid Review and Meta-Analysis. Clin. Microbiol. Infect. 2020, 26, 1622–1629. [Google Scholar] [CrossRef]
- Liu, C.; Wen, Y.; Wan, W.; Lei, J.; Jiang, X. Clinical Characteristics and Antibiotics Treatment in Suspected Bacterial Infection Patients with COVID-19. Int. Immunopharmacol. 2021, 90, 107157. [Google Scholar] [CrossRef]
- Kolář, M. Bacterial Infections, Antimicrobial Resistance and Antibiotic Therapy. Life 2022, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Pavlicevic, M.; Marmiroli, N.; Maestri, E. Immunomodulatory Peptides—A Promising Source for Novel Functional Food Production and Drug Discovery. Peptides 2022, 148, 170696. [Google Scholar] [CrossRef]
- Singh, B.P.; Bangar, S.P.; Alblooshi, M.; Ajayi, F.F.; Mudgil, P.; Maqsood, S. Plant-Derived Proteins as a Sustainable Source of Bioactive Peptides: Recent Research Updates on Emerging Production Methods, Bioactivities, and Potential Application. Crit. Rev. Food Sci. Nutr. 2022, 63, 9539–9560. [Google Scholar] [CrossRef]
- Graf, B.L.; Rojas-Silva, P.; Rojo, L.E.; Delatorre-Herrera, J.; Baldeón, M.E.; Raskin, I. Innovations in Health Value and Functional Food Development of Quinoa (Chenopodium quinoa Willd.). Compr. Rev. Food Sci. Food Saf. 2015, 14, 431–445. [Google Scholar] [CrossRef]
- Elam, E.; Feng, J.; Lv, Y.-M.; Ni, Z.-J.; Sun, P.; Thakur, K.; Zhang, J.-G.; Ma, Y.-L.; Wei, Z.-J. Recent Advances on Bioactive Food Derived Anti-Diabetic Hydrolysates and Peptides from Natural Resources. J. Funct. Foods 2021, 86, 104674. [Google Scholar] [CrossRef]
- Mudgil, P.; Omar, L.S.; Kamal, H.; Kilari, B.P.; Maqsood, S. Multi-Functional Bioactive Properties of Intact and Enzymatically Hydrolysed Quinoa and Amaranth Proteins. LWT 2019, 110, 207–213. [Google Scholar] [CrossRef]
- Abbasi, S.; Moslehishad, M.; Salami, M. Antioxidant and Alpha-Glucosidase Enzyme Inhibitory Properties of Hydrolyzed Protein and Bioactive Peptides of Quinoa. Int. J. Biol. Macromol. 2022, 213, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, F.; Segura, M.R. Amaranth, Quinoa and Chia Bioactive Peptides: A Comprehensive Review on Three Ancient Grains and Their Potential Role in Management and Prevention of Type 2 Diabetes. Crit. Rev. Food Sci. Nutr. 2022, 62, 2707–2721. [Google Scholar] [CrossRef] [PubMed]
- Capraro, J.; De Benedetti, S.; Di Dio, M.; Bona, E.; Abate, A.; Corsetto, P.A.; Scarafoni, A. Characterization of Chenopodin Isoforms from Quinoa Seeds and Assessment of Their Potential Anti-Inflammatory Activity in Caco-2 Cells. Biomolecules 2020, 10, 795. [Google Scholar] [CrossRef] [PubMed]
- Pathan, S.; Siddiqui, R.A. Nutritional Composition and Bioactive Components in Quinoa (Chenopodium quinoa Willd.) Greens: A Review. Nutrients 2022, 14, 558. [Google Scholar] [CrossRef] [PubMed]
- Chris, B.; Saraswathi, G. Isolation and Characterization of Chenopodin, the 11S Seed Storage Protein of Quinoa (Chenopodium quinoa). J. Agric. Food Chem. 1993, 41, 182–185. [Google Scholar] [CrossRef]
- Pompeu, D.G.; Mattioli, M.A.; Ribeiro, R.I.M.A.; Gonçalves, D.B.; De Magalhães, J.T.; Marangoni, S.; da Silva, J.A.; Granjero, P.A. Purification, partial characterization and antimicrobial activity of lectin from Chenopodium Quinoa seeds. Food Sci. Technol. 2015, 35, 696–703. [Google Scholar] [CrossRef]
- Singh, A.; Selvakumar, P.; Saraswat, A.; Tomar, P.; Mishra, M.; Singh, P.; Sharma, A. Characterization and Cloning of an 11S Globulin with Hemagglutination Activity from Murraya paniculata. Protein Pept. Lett. 2015, 22, 750–761. [Google Scholar] [CrossRef]
- Pompeu, D.G.; Cordeiro, H.G.; Tonelli, F.C.P.; Godin, A.M.; de Melo, I.S.F.; Matsui, T.C.; Rodrigues, F.F.; da Silva, J.A.; Coelho, M.d.M.; Machado, R.R.; et al. Chenopodin as an Anti-Inflammatory Compound. Nat. Prod. Res. 2021, 36, 4429–4432. [Google Scholar] [CrossRef]
- Dakhili, S.; Abdolalizadeh, L.; Hosseini, S.M.; Shojaee-Aliabadi, S.; Mirmoghtadaie, L. Quinoa Protein: Composition, Structure and Functional Properties. Food Chem. 2019, 299, 125161. [Google Scholar] [CrossRef]
- Rastogi, S.; Shukla, S.; Kalaivani, M.; Singh, G.N. Peptide-Based Therapeutics: Quality Specifications, Regulatory Considerations, and Prospects. Drug Discov. Today 2019, 24, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Lopez Cascales, J.J.; Zenak, S.; García de La Torre, J.; Lezama, O.G.; Garro, A.; Enriz, R.D. Small Cationic Peptides: Influence of Charge on Their Antimicrobial Activity. ACS Omega 2018, 3, 5390–5398. [Google Scholar] [CrossRef] [PubMed]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef] [PubMed]
- Robles-Loaiza, A.A.; Pinos-Tamayo, E.A.; Mendes, B.; Ortega-Pila, J.A.; Proaño-Bolaños, C.; Plisson, F.; Teixeira, C.; Gomes, P.; Almeida, J.R. Traditional and Computational Screening of Non-Toxic Peptides and Approaches to Improving Selectivity. Pharmaceuticals 2022, 15, 323. [Google Scholar] [CrossRef] [PubMed]
- Amit, S.; Irfan, A.M.; Gaurav, S.; Nalini, T.; Sarmad, M. A Review on Medicinal and Pharmaceutical Importance of Quinoa (Chenopodium quinoa). Res. J. Pharm. Technol. 2021, 14, 1779–1784. [Google Scholar] [CrossRef]
- Guo, H.; Hao, Y.; Yang, X.; Ren, G.; Richel, A. Exploration on Bioactive Properties of Quinoa Protein Hydrolysate and Peptides: A Review. Crit. Rev. Food Sci. Nutr. 2021, 63, 2896–2909. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Wang, C.; Ren, Y.; Yang, C.; Tian, F. Computational Peptidology: A New and Promising Approach to Therapeutic Peptide Design. Curr. Med. Chem. 2013, 20, 1985–1996. [Google Scholar] [CrossRef]
- Basith, S.; Manavalan, B.; Hwan Shin, T.; Lee, G. Machine Intelligence in Peptide Therapeutics: A Next-generation Tool for Rapid Disease Screening. Med. Res. Rev. 2020, 40, 1276–1314. [Google Scholar] [CrossRef]
- Chen, C.H.; Starr, C.G.; Troendle, E.; Wiedman, G.; Wimley, W.C.; Ulmschneider, J.P.; Ulmschneider, M.B. Simulation-Guided Rational De Novo Design of a Small Pore-Forming Antimicrobial Peptide. J. Am. Chem. Soc. 2019, 141, 4839–4848. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, C.H.; Hu, D.; Ulmschneider, M.B.; Ulmschneider, J.P. Spontaneous Formation of Structurally Diverse Membrane Channel Architectures from a Single Antimicrobial Peptide. Nat. Commun. 2016, 7, 13535. [Google Scholar] [CrossRef]
- Ulmschneider, J.P.; Smith, J.C.; White, S.H.; Ulmschneider, M.B. In Silico Partitioning and Transmembrane Insertion of Hydrophobic Peptides under Equilibrium Conditions. J. Am. Chem. Soc. 2011, 133, 15487–15495. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Wiedman, G.; Khan, A.; Ulmschneider, M.B. Absorption and Folding of Melittin onto Lipid Bilayer Membranes via Unbiased Atomic Detail Microsecond Molecular Dynamics Simulation. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2014, 1838, 2243–2249. [Google Scholar] [CrossRef] [PubMed]
- Matthes, D.; de Groot, B.L. Molecular Dynamics Simulations Reveal the Importance of Amyloid-Beta Oligomer β-Sheet Edge Conformations in Membrane Permeabilization. J. Biol. Chem. 2023, 299, 103034. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial Peptides: Do They Have a Future as Therapeutics? In Antimicrobial Peptides; Springer International Publishing: Cham, Switzerland, 2016; pp. 147–154. [Google Scholar]
- Lakshmaiah, J.; Chen, J.-Y. Antimicrobial Peptides: Possible Anti-Infective Agents. Peptides 2015, 72, 88–94. [Google Scholar] [CrossRef]
- Barashkova, A.S.; Rogozhin, E.A. Isolation of Antimicrobial Peptides from Different Plant Sources: Does a General Extraction Method Exist? Plant Methods 2020, 16, 143. [Google Scholar] [CrossRef]
- Simoes-Silva, R.; Alfonso, J.; Gomez, A.; Holanda, R.J.; Sobrinho, J.C.; Zaqueo, K.D.; Moreira-Dill, L.S.; Kayano, A.M.; Grabner, F.P.; da Silva, S.L.; et al. Snake Venom, A Natural Library of New Potential Therapeutic Molecules: Challenges and Current Perspectives. Curr. Pharm. Biotechnol. 2018, 19, 308–335. [Google Scholar] [CrossRef]
- Harvey, A.L. Snake Venom Peptides. In Handbook of Biologically Active Peptides; Academic Press: Cambridge, MA, USA, 2006; pp. 355–362. [Google Scholar]
- Furman, B.L. The Development of Byetta (Exenatide) from the Venom of the Gila Monster as an Anti-Diabetic Agent. Toxicon 2012, 59, 464–471. [Google Scholar] [CrossRef]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in Peptide Drug Discovery. Nat Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]
- Porto, W.F.; Irazazabal, L.; Alves, E.S.F.; Ribeiro, S.M.; Matos, C.O.; Pires, Á.S.; Fensterseifer, I.C.M.; Miranda, V.J.; Haney, E.F.; Humblot, V.; et al. In Silico Optimization of a Guava Antimicrobial Peptide Enables Combinatorial Exploration for Peptide Design. Nat. Commun. 2018, 9, 1490. [Google Scholar] [CrossRef]
- Wang, G. Antimicrobial Peptides Discovery, Design and Novel Therapeutic Strategies; CABI: Wallingford, UK, 2010. [Google Scholar]
- Chen, C.H.; Lu, T.K. Development and Challenges of Antimicrobial Peptides for Therapeutic Applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef]
- Fathi, F.; Ghobeh, M.; Mahboubi, A.; Tabarzad, M. Controversy Between In Vitro Biological Activities of a Novel Designed Antimicrobial Peptide and Its In Silico Predicted Activities: Novel In Silico Designed AMP. Trends Pept. Protein Sci. 2022, 7, 1–12. [Google Scholar]
- Hoyt, D.W.; Gierasch, L.M. A Peptide Corresponding to an Export-Defective Mutant OmpA Signal Sequence with Asparagine in the Hydrophobic Core Is Unable to Insert into Model Membranes. J. Biol. Chem. 1991, 266, 14406–14412. [Google Scholar] [CrossRef] [PubMed]
- Protopapa, E.; Ringstad, L.; Aggeli, A.; Nelson, A. Interaction of Self-Assembling β-Sheet Peptides with Phospholipid Monolayers: The Effect of Serine, Threonine, Glutamine and Asparagine Amino Acid Side Chains. Electrochim. Acta 2010, 55, 3368–3375. [Google Scholar] [CrossRef]
- Suarez, M.; Haenni, M.; Canarelli, S.; Fisch, F.; Chodanowski, P.; Servis, C.; Michielin, O.; Freitag, R.; Moreillon, P.; Mermod, N. Structure-Function Characterization and Optimization of a Plant-Derived Antibacterial Peptide. Antimicrob. Agents Chemother. 2005, 49, 3847–3857. [Google Scholar] [CrossRef]
- Li, S.; Shui, Y.; Ma, J.; Yuan, Y.; Jiang, W.; Xu, C.; Wang, L.; Ren, Y.; Deng, B.; Zhang, W.; et al. Preliminary Antimicrobial Activity of CT-K3K7, a Modified Peptide by Lysine Substitutions from Scorpion Venom Peptide. Res. Sq. 2022, 218, 88–98. [Google Scholar] [CrossRef]
- Peña-Carrillo, M.S.; Pinos-Tamayo, E.A.; Mendes, B.; Domínguez-Borbor, C.; Proaño-Bolaños, C.; Miguel, D.C.; Almeida, J.R. Dissection of Phospholipases A2 Reveals Multifaceted Peptides Targeting Cancer Cells, Leishmania and Bacteria. Bioorg. Chem. 2021, 114, 105041. [Google Scholar] [CrossRef]
- Oh, H.Y.; Go, H.-J.; Park, N.G. Identification and Characterization of SaRpAMP, a 60S Ribosomal Protein L27-Derived Antimicrobial Peptide from Amur Catfish, Silurus Asotus. Fish Shellfish Immunol. 2020, 106, 480–490. [Google Scholar] [CrossRef]
- Shagaghi, N.; Palombo, E.A.; Clayton, A.H.A.; Bhave, M. Archetypal Tryptophan-Rich Antimicrobial Peptides: Properties and Applications. World. J. Microbiol. Biotechnol. 2016, 32, 31. [Google Scholar] [CrossRef]
- Mishra, A.; Choi, J.; Moon, E.; Baek, K.-H. Tryptophan-Rich and Proline-Rich Antimicrobial Peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef]
- Xiang, W.; Clemenza, P.; Klousnitzer, J.; Chen, J.; Qin, W.; Tristram-Nagle, S.; Doi, Y.; Di, Y.P.; Deslouches, B. Rational Framework for the Design of Trp- and Arg-Rich Peptide Antibiotics Against Multidrug-Resistant Bacteria. Front. Microbiol. 2022, 13, 889791. [Google Scholar] [CrossRef]
- Mendes, B.; Almeida, J.R.; Vale, N.; Gomes, P.; Gadelha, F.R.; Da Silva, S.L.; Miguel, D.C. Potential Use of 13-mer Peptides Based on Phospholipase and Oligoarginine as Leishmanicidal Agents. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2019, 226, 108612. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Jin, S.; Wang, M.; Pang, Q.; Liu, C.; Liu, R.; Wang, Y.; Yang, H.; Liu, F.; Liu, Y. The Critical Role of Tryptophan in the Antimicrobial Activity and Cell Toxicity of the Duck Antimicrobial Peptide DCATH. Front. Microbiol. 2020, 11, 1146. [Google Scholar] [CrossRef] [PubMed]
- Staubitz, P.; Peschel, A.; Nieuwenhuizen, W.F.; Otto, M.; Götz, F.; Jung, G.; Jack, R.W. Structure-Function Relationships in the Tryptophan-Rich, Antimicrobial Peptide Indolicidin. J. Pept. Sci. 2001, 7, 552–564. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef] [PubMed]
- Vanzolini, T.; Bruschi, M.; Rinaldi, A.C.; Magnani, M.; Fraternale, A. Multitalented Synthetic Antimicrobial Peptides and Their Antibacterial, Antifungal and Antiviral Mechanisms. Int. J. Mol. Sci. 2022, 23, 545. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.R.; Mendes, B.; Lancellotti, M.; Franchi, G.C.; Passos, Ó.; Ramos, M.J.; Fernandes, P.A.; Alves, C.; Vale, N.; Gomes, P.; et al. Lessons from a Single Amino Acid Substitution: Anticancer and Antibacterial Properties of Two Phospholipase A2-Derived Peptides. Curr. Issues Mol. Biol. 2021, 44, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Zanfardino, A.; Di Giuseppe, A.M.A.; Bosso, A.; Landi, N.; Ragucci, S.; Varcamonti, M.; Notomista, E.; Di Maro, A. A New Active Antimicrobial Peptide from PD-L4, a Type 1 Ribosome Inactivating Protein of Phytolacca Dioica L.: A New Function of RIPs for Plant Defence? FEBS Lett. 2015, 589, 2812–2818. [Google Scholar] [CrossRef]
- Liu, S.; Lin, Y.; Liu, J.; Chen, X.; Ma, C.; Xi, X.; Zhou, M.; Chen, T.; Burrows, J.F.; Wang, L. Targeted Modification and Structure-Activity Study of GL-29, an Analogue of the Antimicrobial Peptide Palustrin-2ISb. Antibiotics 2022, 11, 1048. [Google Scholar] [CrossRef]
- Chen, C.; Pan, F.; Zhang, S.; Hu, J.; Cao, M.; Wang, J.; Xu, H.; Zhao, X.; Lu, J.R. Antibacterial Activities of Short Designer Peptides: A Link between Propensity for Nanostructuring and Capacity for Membrane Destabilization. Biomacromolecules 2010, 11, 402–411. [Google Scholar] [CrossRef]
- Zhao, H.; To, K.K.W.; Sze, K.-H.; Yung, T.T.-M.; Bian, M.; Lam, H.; Yeung, M.L.; Li, C.; Chu, H.; Yuen, K.-Y. A Broad-Spectrum Virus- and Host-Targeting Peptide against Respiratory Viruses Including Influenza Virus and SARS-CoV-2. Nat. Commun. 2020, 11, 4252. [Google Scholar] [CrossRef]
- Kurpe, S.R.; Grishin, S.Y.; Surin, A.K.; Panfilov, A.V.; Slizen, M.V.; Chowdhury, S.D.; Galzitskaya, O.V. Antimicrobial and Amyloidogenic Activity of Peptides. Can Antimicrobial Peptides Be Used against SARS-CoV-2? Int. J. Mol. Sci. 2020, 21, 9552. [Google Scholar] [CrossRef] [PubMed]
- Zannella, C.; Chianese, A.; Palomba, L.; Marcocci, M.E.; Bellavita, R.; Merlino, F.; Grieco, P.; Folliero, V.; De Filippis, A.; Mangoni, M.; et al. Broad-Spectrum Antiviral Activity of the Amphibian Antimicrobial Peptide Temporin L and Its Analogs. Int. J. Mol. Sci. 2022, 23, 2060. [Google Scholar] [CrossRef] [PubMed]
- Bruno, F.; Malvaso, A.; Canterini, S.; Bruni, A.C. Antimicrobial Peptides (AMPs) in the Pathogenesis of Alzheimer’s Disease: Implications for Diagnosis and Treatment. Antibiotics 2022, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- Mousa, S.; Ayoub, B. Repositioning of Dipeptidyl Peptidase-4 Inhibitors and Glucagon like Peptide-1 Agonists as Potential Neuroprotective Agents. Neural Regen Res. 2019, 14, 745. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, W.-H.; Chen, H.-F.; Huang, L.-M.; Gao, J.-Y.; Lin, C.-W.; Wang, Y.-C.; Yang, C.-S.; Liu, Y.-L.; Hou, M.-H.; et al. Tafenoquine and Its Derivatives as Inhibitors for the Severe Acute Respiratory Syndrome Coronavirus 2. J. Biol. Chem. 2022, 298, 101658. [Google Scholar] [CrossRef]
- Zhao, H.; To, K.K.W.; Lam, H.; Zhou, X.; Chan, J.F.-W.; Peng, Z.; Lee, A.C.Y.; Cai, J.; Chan, W.-M.; Ip, J.D.; et al. Cross-Linking Peptide and Repurposed Drugs Inhibit Both Entry Pathways of SARS-CoV-2. Nat. Commun. 2021, 12, 1517. [Google Scholar] [CrossRef]
- Choi, Y.-M.; Jeong, H.; Park, U.; Cho, N.-H.; Kim, B.-J. A Hepatitis B Virus-Derived Peptide Can Inhibit Infection of Human Lung Cells with SARS-CoV-2 in a Type-1 Interferon-Dependent Manner. Viruses 2021, 13, 1227. [Google Scholar] [CrossRef]
- Bakovic, A.; Risner, K.; Bhalla, N.; Alem, F.; Chang, T.L.; Weston, W.K.; Harness, J.A.; Narayanan, A. Brilacidin Demonstrates Inhibition of SARS-CoV-2 in Cell Culture. Viruses 2021, 13, 271. [Google Scholar] [CrossRef]
- Enayathullah, M.G.; Parekh, Y.; Banu, S.; Ram, S.; Nagaraj, R.; Kumar, B.K.; Idris, M.M. Gramicidin S and Melittin: Potential Anti-Viral Therapeutic Peptides to Treat SARS-CoV-2 Infection. Sci. Rep. 2022, 12, 3446. [Google Scholar] [CrossRef]
- Torrent, M.; Di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An Automated Web Server for Prediction of Protein Antimicrobial Regions. Bioinformatics 2012, 28, 130–131. [Google Scholar] [CrossRef]
- Chung, C.R.; Kuo, T.R.; Wu, L.C.; Lee, T.Y.; Horng, J.T. Characterization and Identification of Antimicrobial Peptides with Different Functional Activities. Brief. Bioinform. 2020, 21, 1098–1114. [Google Scholar] [CrossRef] [PubMed]
- Veltri, D.; Kamath, U.; Shehu, A. Deep Learning Improves Antimicrobial Peptide Recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Schaduangrat, N.; Basith, S.; Lee, G.; Shoombuatong, W.; Manavalan, B. HLPpred-Fuse: Improved and Robust Prediction of Hemolytic Peptide and its Activity by Fusing Multiple Feature Representation. Bioinformatics 2020, 36, 3350–3356. [Google Scholar] [CrossRef] [PubMed]
- Timmons, P.B.; Hewage, C.M. HAPPENN is a Novel Tool for Hemolytic Activity Prediction for Therapeutic Peptides which Employs Neural Networks. Sci. Rep. 2020, 10, 10869. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, R.; Agrawal, P.; Patiyal, S.; Raghava, G.P. A Method for Predicting Hemolytic Potency of Chemically Modified Peptides from its Structure. Front. Pharmacol. 2020, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.T.; Sun, Y.Y.; Wang, C.T.; Cheng, W.C.; Lu, I.H.; Lin, C.Y.; Chen, S.H. AI4AVP: An Antiviral Peptides Predictor in Deep Learning Approach with Generative Adversarial Network Data Augmentation. Bioinform Adv. 2022, 26, vbac080. [Google Scholar] [CrossRef]
- Dittmar, M.; Lee, J.S.; Whig, K.; Segrist, E.; Li, M.; Kamalia, B.; Castellana, L.; Ayyanathan, K.; Cardenas-Diaz, F.L.; Morrisey, E.E.; et al. Drug Repurposing Screens Reveal Cell-Type-Specific Entry Pathways and FDA-Approved Drugs Active against SARS-Cov-2. Cell Rep. 2021, 35, 108959. [Google Scholar] [CrossRef]
- Valdivieso-Rivera, F.; Bermúdez-Puga, S.; Proaño-Bolaños, C.; Almeida, J.R. Deciphering the Limitations and Antibacterial Mechanism of Cruzioseptins. Int. J. Pept. Res. Ther. 2022, 28, 73. [Google Scholar] [CrossRef]


| Peptides | Length (aa) | pI | Charge | Hydrophobicity (Kcal∙mol−1) | Mass (Da) |
|---|---|---|---|---|---|
| Chen1 | 14 | 12.79 | +4 | +15.03 | 1556.89 |
| Chen2 | 14 | 12.79 | +4 | +15.54 | 1552.90 |
| ChenR | 14 | 13.09 | +6 | +17.03 | 1626.99 |
| ChenW | 14 | 12.79 | +4 | +9.23 | 1686.95 |
| Peptides/Antibiotic | MIC (µM) | MBC (µM) | ||
|---|---|---|---|---|
| E. coli | S. aureus | E. coli | S. aureus | |
| Chen1 | - | - | - | - |
| Chen2 | 64 | 128 | 128 | 128 |
| ChenR | 16 | 128 | 32 | 256 |
| ChenW | 8 | 8 | 8 | 8 |
| Ampicillin | 16 | 2 | 64 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feijoo-Coronel, M.L.; Mendes, B.; Ramírez, D.; Peña-Varas, C.; de los Monteros-Silva, N.Q.E.; Proaño-Bolaños, C.; de Oliveira, L.C.; Lívio, D.F.; da Silva, J.A.; da Silva, J.M.S.F.; et al. Antibacterial and Antiviral Properties of Chenopodin-Derived Synthetic Peptides. Antibiotics 2024, 13, 78. https://doi.org/10.3390/antibiotics13010078
Feijoo-Coronel ML, Mendes B, Ramírez D, Peña-Varas C, de los Monteros-Silva NQE, Proaño-Bolaños C, de Oliveira LC, Lívio DF, da Silva JA, da Silva JMSF, et al. Antibacterial and Antiviral Properties of Chenopodin-Derived Synthetic Peptides. Antibiotics. 2024; 13(1):78. https://doi.org/10.3390/antibiotics13010078
Chicago/Turabian StyleFeijoo-Coronel, Marcia L., Bruno Mendes, David Ramírez, Carlos Peña-Varas, Nina Q. E. de los Monteros-Silva, Carolina Proaño-Bolaños, Leonardo Camilo de Oliveira, Diego Fernandes Lívio, José Antônio da Silva, José Maurício S. F. da Silva, and et al. 2024. "Antibacterial and Antiviral Properties of Chenopodin-Derived Synthetic Peptides" Antibiotics 13, no. 1: 78. https://doi.org/10.3390/antibiotics13010078
APA StyleFeijoo-Coronel, M. L., Mendes, B., Ramírez, D., Peña-Varas, C., de los Monteros-Silva, N. Q. E., Proaño-Bolaños, C., de Oliveira, L. C., Lívio, D. F., da Silva, J. A., da Silva, J. M. S. F., Pereira, M. G. A. G., Rodrigues, M. Q. R. B., Teixeira, M. M., Granjeiro, P. A., Patel, K., Vaiyapuri, S., & Almeida, J. R. (2024). Antibacterial and Antiviral Properties of Chenopodin-Derived Synthetic Peptides. Antibiotics, 13(1), 78. https://doi.org/10.3390/antibiotics13010078












