Structures, Interactions and Activity of the N-Terminal Truncated Variants of Antimicrobial Peptide Thanatin
Abstract
1. Introduction
2. Results
2.1. Thanatin Peptide Fragments and Antibacterial Activity
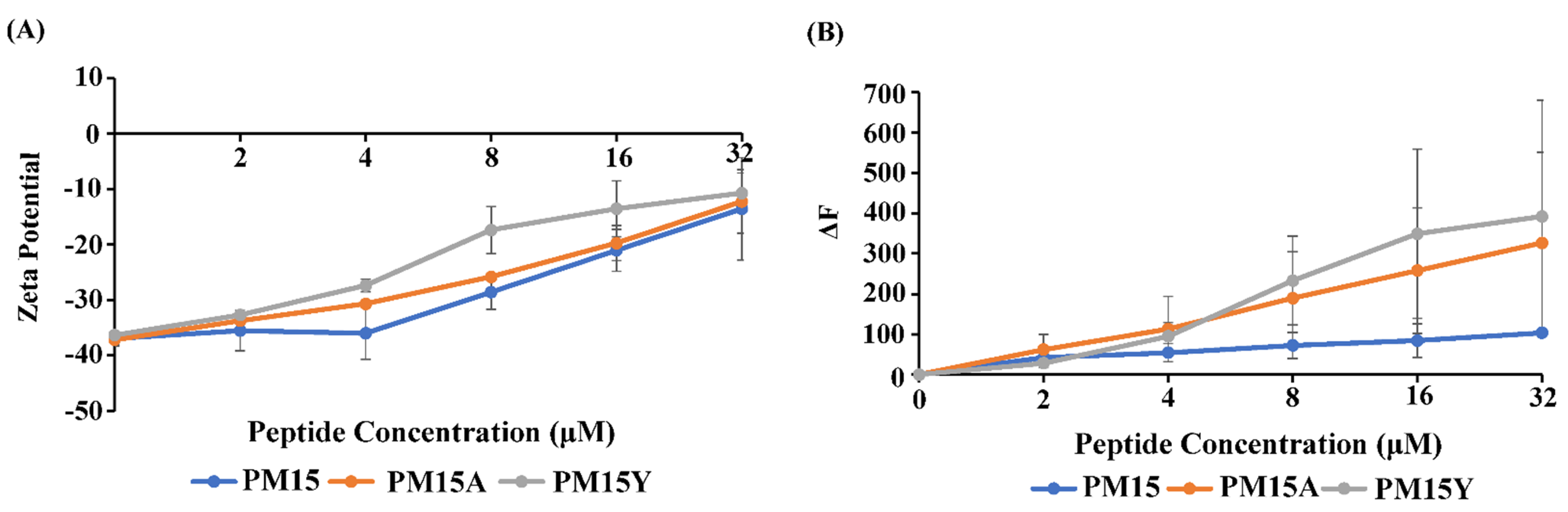
2.2. Surface Charge Neutralization, Outer-Membrane Permeabilization and LPS Interactions
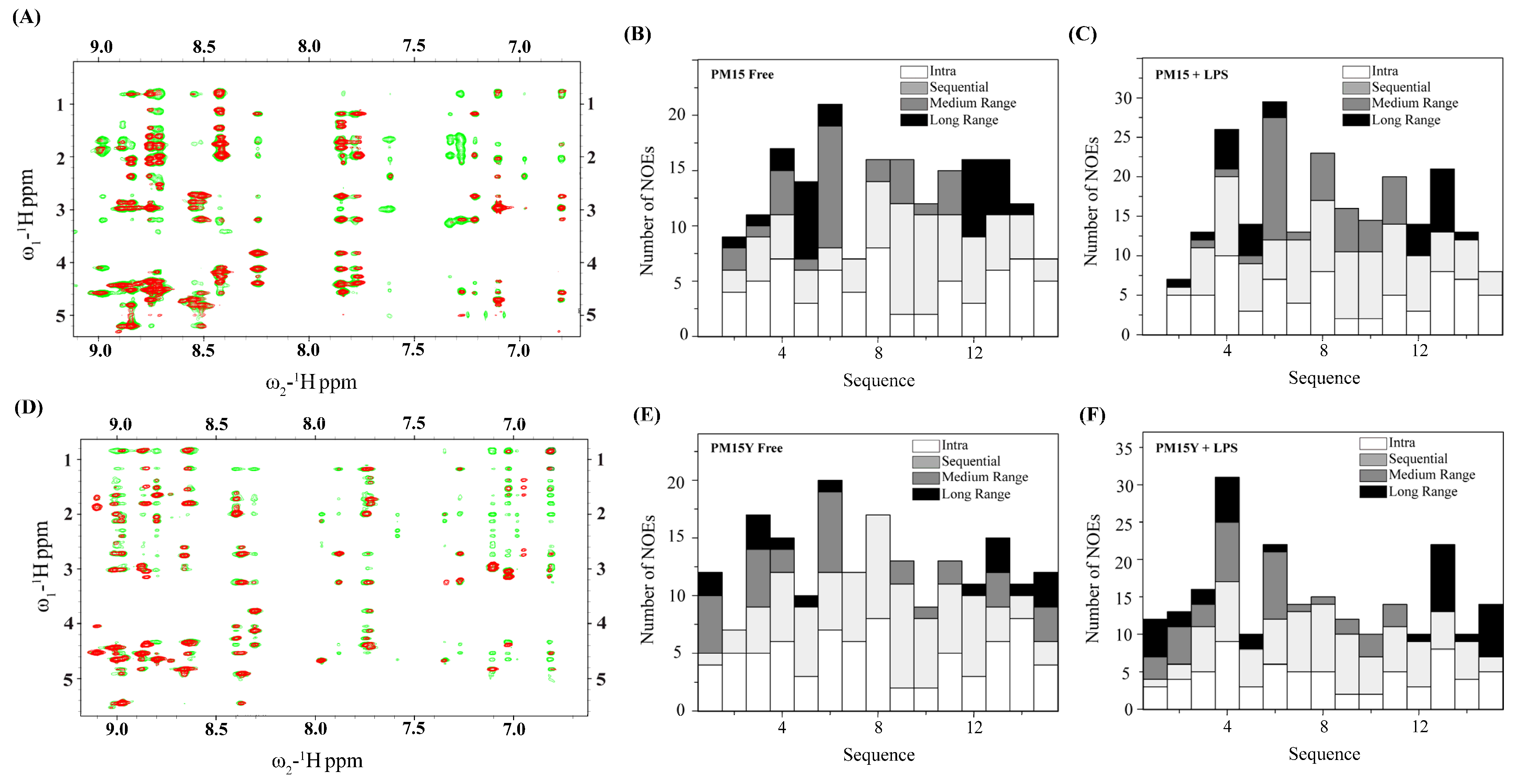
2.3. NMR Analyses of PM15 and PM15Y Peptides
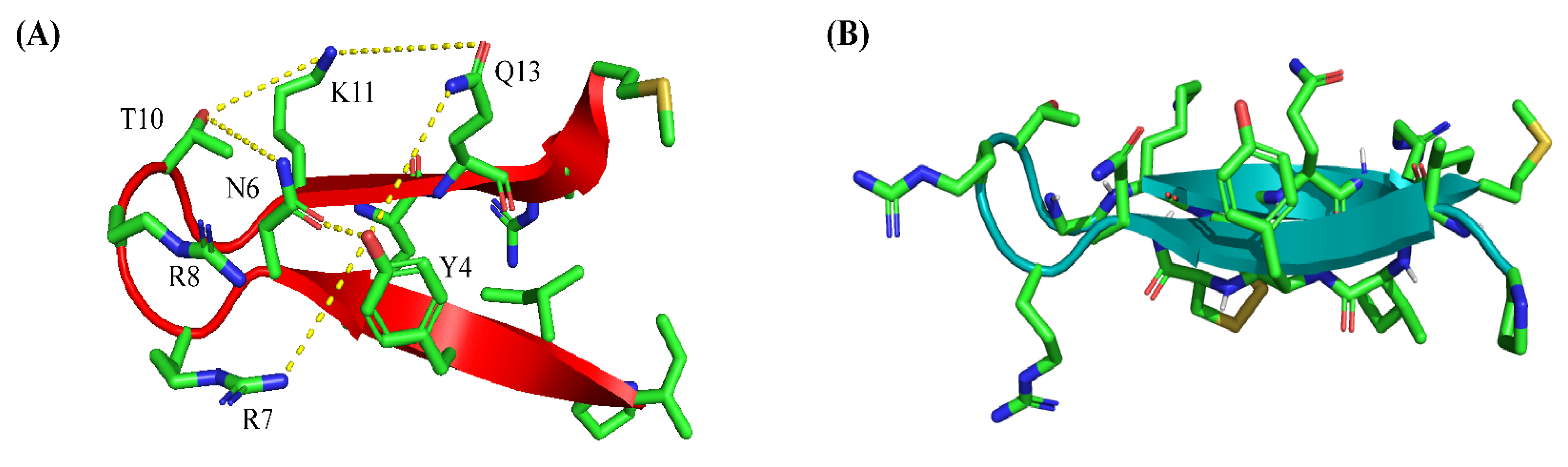
2.4. Atomic-Resolution Structures of PM15 and PM15Y in Free Solution and in Complex with LPS Micelle
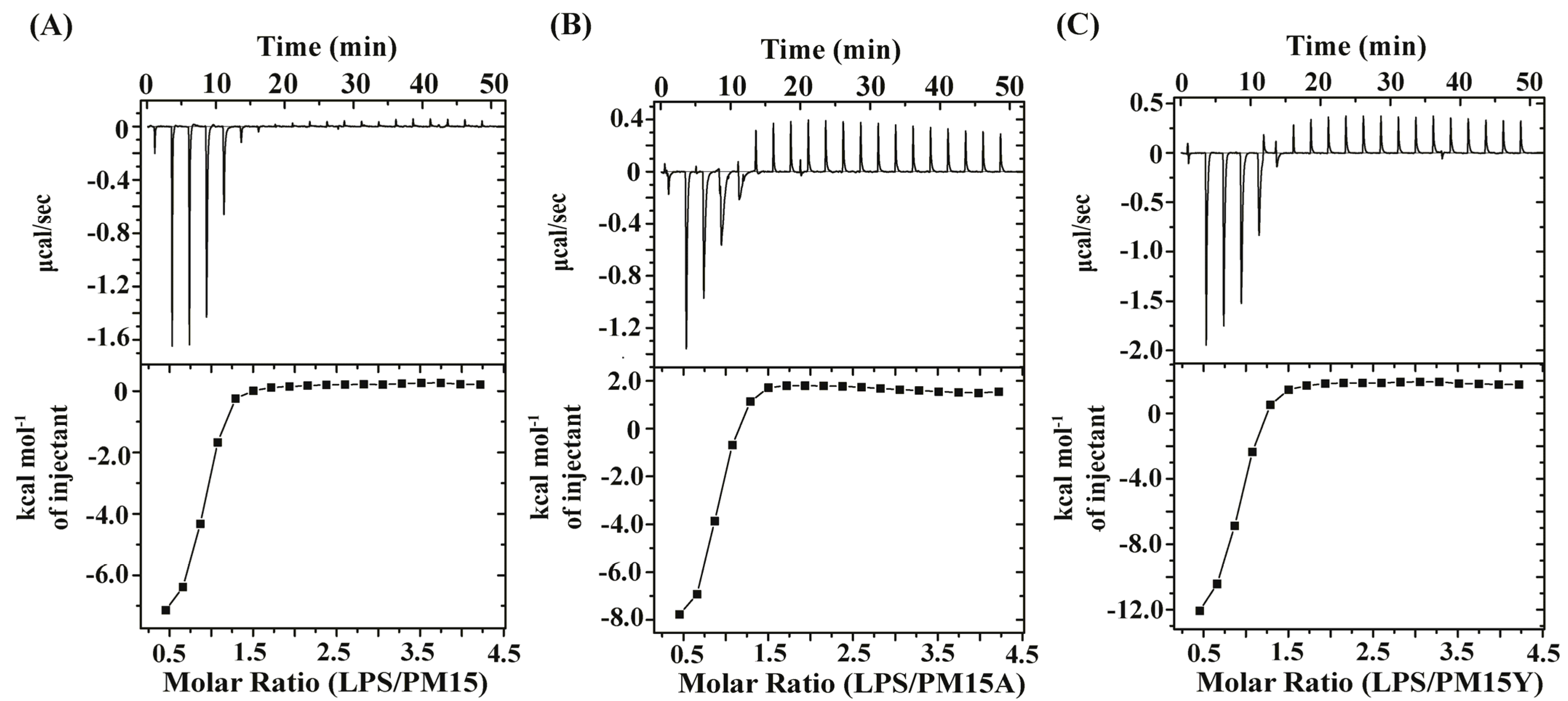
2.5. Thermodynamics of Interactions of PM15 and PM15Y Peptides with LptAm
3. Discussion
4. Materials and Methods
4.1. Peptides, Bacterial Strains and Media
4.2. Determination of Minimal Inhibitory Concentration (MIC) of the Peptides
4.3. NPN Fluorescence Assay
4.4. Zeta Potential Measurements
4.5. Isothermal Titration Calorimetry (ITC) Studies
4.6. Purification of LptAm protein
4.7. NMR Experiments, Structure Determination of PM15, PM15Y Peptides and LPS-PM15 Docking
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vaara, M.; Siikanen, O.; Apajalahti, J.; Fox, J.; Frimodt-Møller, N.; He, H.; Poudyal, A.; Li, J.; Nation, R.L.; Vaara, T. A Novel Polymyxin Derivative That Lacks the Fatty Acid Tail and Carries Only Three Positive Charges Has Strong Synergism with Agents Excluded by the Intact Outer Membrane. Antimicrob. Agents Chemother. 2010, 54, 3341–3346. [Google Scholar] [CrossRef]
- Morita, Y.; Tomida, J.; Kawamura, Y. Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 2014, 4, 422. [Google Scholar] [CrossRef]
- Umstätter, F.; Domhan, C.; Hertlein, T.; Ohlsen, K.; Mühlberg, E.; Kleist, C.; Zimmermann, S.; Beijer, B.; Klika, K.D.; Haberkorn, U.; et al. Vancomycin Resistance Is Overcome by Conjugation of Polycationic Peptides. Angew. Chem. Int. Ed. 2020, 59, 8823–8827. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992, 56, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Collaborators, A.R. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling drug-resistant infections globally: Final report and recommendations. In The Review on Antimicrobial Resistance (AMR); Wellcome Trust; HM Government: London, UK, 2016. [Google Scholar]
- Morrison, L.; Zembower, T.R. Antimicrobial Resistance. Gastrointest. Endosc. Clin. N. Am. 2020, 30, 619–635. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Adams-Sapper, S.; Nolen, S.; Donzelli, G.F.; Lal, M.; Chen, K.; da Silva, L.H.J.; Moreira, B.M.; Riley, L.W. Rapid Induction of High-Level Carbapenem Resistance in Heteroresistant KPC-Producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2015, 59, 3281–3289. [Google Scholar] [CrossRef] [PubMed]
- Rabanal, F.; Cajal, Y. Recent advances and perspectives in the design and development of polymyxins. Nat. Prod. Rep. 2017, 34, 886–908. [Google Scholar] [CrossRef]
- Brown, P.; Dawson, M.J. Development of new polymyxin derivatives for multi-drug resistant Gram-negative infections. J. Antibiot. 2017, 70, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms: My Perspective. Adv. Exp. Med. Biol. 2019, 1117, 3–6. [Google Scholar] [CrossRef]
- MacNair, C.R.; Rutherford, S.T.; Tan, M.-W. Alternative therapeutic strategies to treat antibiotic-resistant pathogens. Nat. Rev. Microbiol. 2023. [Google Scholar] [CrossRef]
- Boman, H.G. Antibacterial peptides: Basic facts and emerging concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Vitale, M. Antibiotic Resistance: Do We Need Only Cutting-Edge Methods, or Can New Visions Such as One Health Be More Useful for Learning from Nature? Antibiotics 2023, 12, 1694. [Google Scholar] [CrossRef]
- Wei, L.; Yang, J.; He, X.; Mo, G.; Hong, J.; Yan, X.; Lin, D.; Lai, R. Structure and Function of a Potent Lipopolysaccharide-Binding Antimicrobial and Anti-inflammatory Peptide. J. Med. Chem. 2013, 56, 3546–3556. [Google Scholar] [CrossRef] [PubMed]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef]
- Huynh, K.; Kibrom, A.; Donald, B.R.; Zhou, P. Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA. J. Struct. Biol. X 2023, 8, 100091. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cha, W.H.; Lee, D.-W. Multiple Precursor Proteins of Thanatin Isoforms, an Antimicrobial Peptide Associated with the Gut Symbiont of Riptortus pedestris. Front. Microbiol. 2022, 12, 796548. [Google Scholar] [CrossRef] [PubMed]
- Dash, R.; Bhattacharjya, S. Thanatin: An Emerging Host Defense Antimicrobial Peptide with Multiple Modes of Action. Int. J. Mol. Sci. 2021, 22, 1522. [Google Scholar] [CrossRef]
- Sinha, S.; Dhanabal, V.B.; Sperandeo, P.; Polissi, A.; Bhattacharjya, S. Linking dual mode of action of host defense antimicrobial peptide thanatin: Structures, lipopolysaccharide and LptAm binding of designed analogs. Biochim. Biophys. Acta (BBA)-Biomembr. 2022, 1864, 183839. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef]
- Wu, G.; Fan, X.; Li, L.; Wang, H.; Ding, J.; Hongbin, W.; Zhao, R.; Gou, L.; Shen, Z.; Xi, T. Interaction of antimicrobial peptide s-thanatin with lipopolysaccharide in vitro and in an experimental mouse model of septic shock caused by a multidrug-resistant clinical isolate of Escherichia coli. Int. J. Antimicrob. Agents 2010, 35, 250–254. [Google Scholar] [CrossRef]
- Pagès, J.-M.; Dimarcq, J.-L.; Quenin, S.; Hetru, C. Thanatin activity on multidrug resistant clinical isolates of Enterobacter aerogenes and Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2003, 22, 265–269. [Google Scholar] [CrossRef]
- Schuster, M.; Brabet, E.; Oi, K.K.; Desjonquères, N.; Moehle, K.; Le Poupon, K.; Hell, S.; Gable, S.; Rithié, V.; Dillinger, S.; et al. Peptidomimetic antibiotics disrupt the lipopolysaccharide transport bridge of drug-resistant Enterobacteriaceae. Sci. Adv. 2023, 9, eadg3683. [Google Scholar] [CrossRef]
- Sinha, S.; Zheng, L.; Mu, Y.; Ng, W.J.; Bhattacharjya, S. Structure and interactions of a host defense antimicrobial peptide thanatin in lipopolysaccharide micelles reveal mechanism of bacterial cell agglutination. Sci. Rep. 2017, 7, 17795. [Google Scholar] [CrossRef]
- Vetterli, S.U.; Zerbe, K.; Müller, M.; Urfer, M.; Mondal, M.; Wang, S.-Y.; Moehle, K.; Zerbe, O.; Vitale, A.; Pessi, G.; et al. Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv. 2018, 4, eaau2634. [Google Scholar] [CrossRef] [PubMed]
- Moura, E.C.C.M.; Baeta, T.; Romanelli, A.; Laguri, C.; Martorana, A.M.; Erba, E.; Simorre, J.-P.; Sperandeo, P.; Polissi, A. Thanatin Impairs Lipopolysaccharide Transport Complex Assembly by Targeting LptC–LptA Interaction and Decreasing LptA Stability. Front. Microbiol. 2020, 11, 909. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, F.; Sauer, J.B.; Qiu, X.; Corey, R.A.; Cassidy, C.K.; Mynors-Wallis, B.; Mehmood, S.; Bolla, J.R.; Stansfeld, P.J.; Robinson, C.V. Dynamics of an LPS translocon induced by substrate and an antimicrobial peptide. Nat. Chem. Biol. 2021, 17, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.J.; Yan, B.; Palanivelu, N.; Dhanabal, V.; Bifani, J.; Bhattacharjya, S. Outer-membrane Permeabilization, LPS Transport Inhibition: Activity, Interactions and Structures of Thanatin Derived Antimicrobial Peptides. Int. J. Mol. Sci. 2024. Submitted. [Google Scholar]
- Ön, A.; Vejzovic, D.; Jennings, J.; Parigger, L.; Cordfunke, R.A.; Drijfhout, J.W.; Lohner, K.; Malanovic, N. Bactericidal Activity to Escherichia coli: Different Modes of Action of Two 24-Mer Peptides SAAP-148 and OP-145, Both Derived from Human Cathelicidine LL-37. Antibiotics 2023, 12, 1163. [Google Scholar] [CrossRef] [PubMed]
- Maillard, A.P.F.; Espeche, J.C.; Maturana, P.; Cutro, A.C.; Hollmann, A. Zeta potential beyond materials science: Applications to bacterial systems and to the development of novel antimicrobials. Biochim. Biophys. Acta (BBA)-Biomembr. 2021, 1863, 183597. [Google Scholar] [CrossRef]
- Alves, C.S.; Melo, M.N.; Franquelim, H.G.; Ferre, R.; Planas, M.; Feliu, L.; Bardají, E.; Kowalczyk, W.; Andreu, D.; Santos, N.C.; et al. Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J. Biol. Chem. 2010, 285, 27536–27544. [Google Scholar] [CrossRef]
- Akhoundsadegh, N.; Belanger, C.R.; Hancock, R.E.W. Outer Membrane Interaction Kinetics of New Polymyxin B Analogs in Gram-Negative Bacilli. Antimicrob. Agents Chemother. 2019, 63, e00935-19. [Google Scholar] [CrossRef]
- Sinha, S.; Dhanabal, V.B.; Manivannen, V.L.; Cappiello, F.; Tan, S.-M.; Bhattacharjya, S. Ultra-Short Cyclized β-Boomerang Peptides: Structures, Interactions with Lipopolysaccharide, Antibiotic Potentiator and Wound Healing. Int. J. Mol. Sci. 2022, 24, 263. [Google Scholar] [CrossRef]
- Hafsa, N.E.; Wishart, D.S. CSI 2.0: A significantly improved version of the Chemical Shift Index. J. Biomol. NMR 2014, 60, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Güntert, P. Automated NMR protein structure calculation with CYANA, Methods. Mol. Biol. 2004, 278, 353–378. [Google Scholar] [CrossRef]
- Mandard, N.; Sodano, P.; Labbe, H.; Bonmatin, J.; Bulet, P.; Hetru, C.; Ptak, M.; Vovelle, F. Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data. JBIC J. Biol. Inorg. Chem. 1998, 256, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Ng, W.J.; Bhattacharjya, S. NMR structure and localization of the host defense antimicrobial peptide thanatin in zwit-terionic dodecylphosphocholine micelle: Implications in antimicrobial activity. Biochim. Biophys. Acta. Biomembr. 2020, 1862, 183432. [Google Scholar] [CrossRef] [PubMed]
- Luther, A.; Moehle, K.; Chevalier, E.; Dale, G.; Obrecht, D. Protein epitope mimetic macrocycles as biopharmaceuticals. Curr. Opin. Chem. Biol. 2017, 38, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Pipiya, S.O.; Kudzhaev, A.M.; Mirzoeva, N.Z.; Mokrushina, Y.A.; Ziganshin, R.H.; Komlev, A.S.; Petrova, P.E.; Smirnov, I.V.; Gabibov, A.G.; Shamova, O.V.; et al. Bioengineering the Antimicrobial Activity of Yeast by Recombinant Thanatin Production. Antibiotics 2023, 12, 1719. [Google Scholar] [CrossRef]
- Sperandeo, P.; Martorana, A.M.; Zaccaria, M.; Polissi, A. Targeting the LPS export pathway for the development of novel therapeutics. Biochim. Biophys. Acta Mol. Cell. Res. 2023, 1870, 119406. [Google Scholar] [CrossRef]
- Yu, L.; Tan, M.; Ho, B.; Ding, J.L.; Wohland, T. Determination of critical micelle concentrations and aggregation numbers by fluorescence correlation spectroscopy: Aggregation of a lipopolysaccharide. Anal. Chim. Acta 2006, 556, 216–225. [Google Scholar] [CrossRef]
- Laguri, C.; Sperandeo, P.; Pounot, K.; Ayala, I.; Silipo, A.; Bougault, C.M.; Molinaro, A.; Polissi, A.; Simorre, J.-P. Interaction of lipopolysaccharides at intermolecular sites of the periplasmic Lpt transport assembly. Sci. Rep. 2017, 7, 9715. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]





| Peptides | Gram-Negative | Gram-Positive | ||||
|---|---|---|---|---|---|---|
| EC | KP | SE | AB | SP | EF | |
| PM15 | 4 (>16) | 4 (>16) | 4–8 (>16) | >16 (>16) | 8 (>16) | 8 (>16) |
| PM15A | 8 (>16) | 4 (>16) | 8 (>16) | >16 (>16) | 8 (>16) | 8 (>16) |
| PM15Y | 4 (>16) | 4 (>16) | >16(>16) | >16 (>16) | 8 (>16) | 8(>16) |
| Peptides | Kd (μM) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|
| PM15 | 0.57 | −7.33 | 1.54 | −8.87 |
| PM15A | 0.33 | −8.74 | 0.47 | −9.21 |
| PM15Y | 0.28 | −11.79 | −2.49 | −9.30 |
| Free PM15 | PM15 in LPS Micelle |
|---|---|
| 2 ILE δH3–14 ARG εH 3 ILE δH3–13 GLN H 5 CYS αH–13 GLN H 5 CYS αH–12 CYS αH 5 CYS αH–12 CYS H 6 ASN H–11 LYS H 6 ASN H–13 GLN H 6 ASN βH2–11 LYS H 6 ASN βH3–11 LYS H 12 CYS αH–6 ASN H 12 CYS βH–5 CYS αH 13 GLN βHs–4 TYR δHs 13 GLN γHs–4 TYR δHs | 2 ILE δH3–14 ARG εH 3 ILE δH3–13 GLN H 4 TYR H–13 GLN H 5 CYS αH–13 GLN H 5 CYS αH–12 CYS H 5 CYS βH2–12 CYS H 6 ASN H–11 LYS H 6 ASN H–13 GLN H 6 ASN βH2–11 LYS H 6 ASN βH3–11 LYS H 11 LYS H–6 ASN δHs 11 LYS βHs–6 ASN H 12 CYS αH–6 ASN H 12 CYS βHs–5 CYS H 12 CYS βHs–5 CYS αH 13 GLN βHs–4 TYR δHs 13 GLN γHs–4 TYR δHs 13 GLN γHs–4 TYR εHs |
| Free PM15Y | PM15Y in LPS Micelle |
|---|---|
| 3 ILE αH–14 ARG H 3 ILE αH–15 MET H 3 ILE δH3–13 GLN αH 5 CYS αH–13 GLN H 6 ASN βH2–11 LYS H 6 ASN βH3–11 LYS H 13 CYS αH–6 ASN αH 13 GLN γH2–4 TYR δHs 15 MET γH2–1 TYR δHs 15 MET βH2–1 TYR δHs | 3 ILE αH–14 ARG H 3 ILE αH–15 MET H 2 ILE γH2–14 ARG H 3 ILE δH3–13 GLN H 3 ILE δH3–15 MET H 4 ILE δH3–13 GLN H 4 TYR H–13 GLN H 4 TYR αH–13 GLN H 5 CYS H–13 GLN H 5 CYS αH–13 GLN H 6 ASN H–11 LYS H 6 ASN βH2–11 LYS H 6 ASN βH3–11 LYS H 12 CYS αH–6 ASN αH 13 GLN γH2–4 TYR δHs 13 GLN γH3–4 TYR δHs 15 MET βH2–1 TYR δHs 15 MET βH3–1 TYR δHs 15 MET βH2–1 TYR ƐHs 15 MET γH2–1 TYR ƐHs 15 MET γH2–4 TYR δHs |
| Free PM15 | PM15 in LPS | Free PM15Y | PM15Y in LPS | |
|---|---|---|---|---|
| Distance constraints | ||||
| Intra-residue [|i − j| = 0] | 67 | 73 | 87 | 69 |
| Sequential [|i − j| = 1] | 34 | 50 | 32 | 45 |
| Medium range [1 < |i − j| < 4] | 11 | 12 | 5 | 12 |
| Long range [|i − j| ≥ 4] | 13 | 18 | 10 | 21 |
| Total NOE | 128 | 156 | 134 | 147 |
| Dihedral angle constraints (φ, ψ) | 24 | 24 | 24 | 24 |
| Deviation from mean structure | ||||
| All backbone atoms (Å) | 0.58 | 0.44 | 0.40 | 0.55 |
| All heavy atoms (Å) | 1.51 | 1.27 | 1.14 | 1.45 |
| Ramachandran plot for the mean structure | ||||
| % of residues in most favored region and additional allowed region | 100 | 100 | 100 | 100 |
| % of residues in generously allowed region | 0 | 0 | 0 | 0 |
| % of residues in disallowed region | 0 | 0 | 0 | 0 |
| Kd (μM) | ΔH (Kcal/mol) | TΔS (Kcal/mol) | ΔG (Kcal/mol) | |
|---|---|---|---|---|
| PM15 | 0.60 | −2.93 | 6.23 | −9.16 |
| PM15Y | 0.40 | −11.79 | −2.49 | −9.30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah, S.J.; Mu, Y.; Bhattacharjya, S. Structures, Interactions and Activity of the N-Terminal Truncated Variants of Antimicrobial Peptide Thanatin. Antibiotics 2024, 13, 74. https://doi.org/10.3390/antibiotics13010074
Abdullah SJ, Mu Y, Bhattacharjya S. Structures, Interactions and Activity of the N-Terminal Truncated Variants of Antimicrobial Peptide Thanatin. Antibiotics. 2024; 13(1):74. https://doi.org/10.3390/antibiotics13010074
Chicago/Turabian StyleAbdullah, Swaleeha Jaan, Yuguang Mu, and Surajit Bhattacharjya. 2024. "Structures, Interactions and Activity of the N-Terminal Truncated Variants of Antimicrobial Peptide Thanatin" Antibiotics 13, no. 1: 74. https://doi.org/10.3390/antibiotics13010074
APA StyleAbdullah, S. J., Mu, Y., & Bhattacharjya, S. (2024). Structures, Interactions and Activity of the N-Terminal Truncated Variants of Antimicrobial Peptide Thanatin. Antibiotics, 13(1), 74. https://doi.org/10.3390/antibiotics13010074









