Abstract
Mastitis, a highly prevalent disease in dairy cows, is responsible for massive financial losses due to decreased milk yield, milk quality, and costly medication. This research paper investigates antimicrobial susceptibility in cows and the role played by both resistance and virulence gene distribution in bovine mastitis. A total of 984 raw milk samples were collected from five different dairy farms and cultured on sheep blood agar plates. Antimicrobial susceptibility was determined by disc diffusion, and corresponding resistance and virulence genes were detected by PCR. Among the collected milk samples, 73, 32, and 19 isolates of Streptococcus spp., Staphylococcus spp., and coliforms were identified, respectively. The antimicrobial susceptibility results showed that Streptococcus spp. were resistant to tetracycline (86.30%), neomycin (79.45%), and oxacillin (73.97%). Staphylococcus spp. were resistant to tetracycline (59.37%) and oxacillin (53.12%). Lastly, coliforms were resistant to oxacillin (100%) and bacitracin (68.42%). The genotyping results showed that Streptococcus spp. carried the resistance genes tetM (46.57%) against tetracycline, bcrB (41.09%) against bacitracin, and aph(3)-II (39.72%) against neomycin. Staphylococcus spp. carried the resistance genes bcrB (40.62%) and tetM (18.75%), and coliforms carried the resistance genes tetM (42.10%) and bcrB (57.89%). Moreover, 57.53%, 75.0%, and 63.15% of Streptococcus spp., Staphylococcus spp., and coliforms carried lmb, fib, and ompC virulence genes, respectively. All three tested bacterial genera showed no significant association between antimicrobial resistance genes and virulence factors, although they were negatively correlated (p > 0.05). The combination of resistance gene identification and susceptibility tests as components of the diagnosis of bovine mastitis can help in selecting effective antimicrobial agents to treat it.
1. Introduction
Bovine mastitis is a common disease of dairy cattle worldwide that causes considerable economic losses due to decreased milk production, low milk quality, increased therapeutic costs, and early culling. Over 135 types of bacterial species have been recorded from bovine mastitis, but only 20 distinct pathogenic bacteria commonly cause mastitis in dairy animals [1,2]. The most common mastitis-causing etiological agents are bacteria, such as Staphylococcus, Streptococcus, Escherichia coli, mycoplasma, and other coliforms; other nonbacterial microorganisms, such as fungi and algae, can also cause bovine mastitis [3]. Antimicrobial agents are the main approach to treating and preventing bovine mastitis, and they have been considered the first choice against bacterial infection for a long time [4]. The frequent use of antimicrobial agents in food animals can result in the presence of antimicrobial-resistant bacteria in milk, meat, and other dairy products, which may pose food safety hazards to humans [5].
Antimicrobial resistance is a serious issue worldwide. Among the antimicrobial agents used on dairy farms, nearly 60–70% are used only for treating and preventing mastitis [4]. The selection pressure and overuse of antimicrobial agents in animal production might be the main reasons for the development of antimicrobial resistance in microorganisms and the emergence of multidrug-resistant bacteria, a serious threat to public health. Therefore, the World Health Organization has recommended the proper use of antimicrobial agents in the livestock industry [6]. Generally, bacteria evolve resistance through several mechanisms, including gene mutations, horizontal gene transfer, antimicrobial agent inactivation by enzymes, drug target modification, the alteration of membrane permeability, and efflux pumps [7,8]. Many antimicrobial resistance genes have been identified in bovine mastitis: blaZ and blaTEM (β-lactam resistance genes); norA (fluoroquinolone resistance gene); tetM and tetK (tetracycline resistance genes); and ermA, ermB, and ermC (erythromycin resistance genes) [9,10]. The study of gene mutation has become a standard tool to investigate antimicrobial resistance, which allows the investigation of the transmission of bacterial genetic material among host populations in more detail compared to conventional culture-based phenotypic resistance methods.
Bacteria possess several virulence factors that play important roles in pathogenesis in the causative microorganism. Resistance in bacteria may be related to the loss of virulence in different models of infections [11]. In one study, mice intraperitoneally injected with E. coli resistant to more than four types of antimicrobial agents showed higher survival rates than mice injected with the reference strain. Interestingly, E. coli bacteria susceptible to antimicrobial agents and resistant to two classes of antimicrobial agents showed lower survival rates in mice. This phenomenon is explained by the fitness cost of antimicrobial resistance [11].
In Taiwan, it is estimated that every year, approximately 70–76% of all antimicrobial agents are used to treat pets and farm animals [12]. Bovine mastitis is a severe constraint against Taiwanese livestock production because few antimicrobial agent susceptibility reports are available. Studying antimicrobial resistance in dairy cattle is crucial for the proper prevention and cure of bacterial infections. Therefore, the purpose of this study was to identify mastitis-causing bacteria and their antimicrobial resistance patterns and to investigate the association between antimicrobial resistance genes and virulence factors.
2. Results
2.1. Identification of Bacterial Isolates through 16S rRNA Gene Sequencing
Based on 16S rRNA gene sequencing, 73, 32, and 19 isolates were identified as Streptococcus spp., Staphylococcus spp., and coliforms, respectively (Table 1). Among the 73 isolates of Streptococcus spp., the predominant species identified was Strep. uberis (n = 30), followed by Strep. lutetiensis (n = 13) and Strep. dysgalactiae (n = 10). Similarly, among the 32 isolates of Staphylococcus spp., the predominant species was Staph. aureus (n = 14), followed by Staph. epidermidis (n = 7) and Staph. hemolyticus (n = 4). Furthermore, the dominant species among the 19 coliform isolates were Escherichia coli (n = 8), followed by Enterobacter aerogenes (n = 5) and Klebsiella pneumoniae (n = 4).

Table 1.
Identification of bacterial isolates through 16S rRNA gene sequencing.
2.2. Antimicrobial Susceptibility of Streptococcus spp., Staphylococcus spp., and Coliforms
The antimicrobial resistance status of Streptococcus spp., Staphylococcus spp., and coliforms are depicted in Table 2. Most Streptococcus spp. were resistant to tetracycline (86.30%), neomycin (79.45%), and oxacillin (73.97%). In contrast, Streptococcus spp. were susceptible to cephalothin (91.78%), cefuroxime (80.82%), and ceftiofur (73.97%). Among the tested Staphylococcus spp., 59.37% were resistant to tetracycline, followed by oxacillin (53.12%) and ampicillin (43.75%). However, all tested Staphylococcus spp. were susceptible to ceftiofur (100%), cephalothin (100%), and cefuroxime (100%). All tested coliforms bacteria were resistant to oxacillin (100%), and nearly 68% of isolates were resistant to bacitracin. However, coliforms were susceptible to ceftiofur (100%), cefuroxime (84.21%), and neomycin (78.94%)

Table 2.
Antimicrobial susceptibility test results for Streptococcus spp., Staphylococcus spp., and coliforms.
2.3. Comparative Study of Phenotypic and Genotypic Antimicrobial Resistance in Streptococcus spp., Staphylococcus spp., and Coliforms
The results revealed a negative correlation between the phenotypic and genotypic antimicrobial resistance patterns of Streptococcus spp. for bacitracin (p < 0.0234), ampicillin (p < 0.0124), oxacillin (p < 0.0335), cefuroxime (p < 0.0059), and cephalothin (p < 0.0003); however, no significant associations were observed for tetracycline, neomycin, and ceftiofur, although the correlations were negative, as shown in Table 3. The phenotypic and genotypic antimicrobial resistance patterns of Staphylococcus spp. were negatively correlated with tetracycline (p < 0.0239), whereas no significant associations were observed for neomycin, bacitracin, ampicillin, and oxacillin, although they were negatively correlated (Table 4). Lastly, no significant associations were found between the phenotypic and genotypic antimicrobial resistance patterns of coliforms with tetracycline, bacitracin, and ampicillin (Table 5).

Table 3.
Comparative study of phenotypic and genotypic antimicrobial resistance in Streptococcus spp.

Table 4.
Comparative study of phenotypic and genotypic antimicrobial resistance in Staphylococcus spp.

Table 5.
Comparative study of phenotypic and genotypic antimicrobial resistance in coliforms.
2.4. Correlation between Antimicrobial Resistance Genes and the Virulence Factors of Bacterial Isolates
All three bacterial genera, Streptococcus, Staphylococcus, and coliforms, showed no association with antimicrobial resistance genes and virulence factors (Table 6), although they were negatively correlated.

Table 6.
The correlation between antimicrobial resistance genes and virulence factors of bacteria.
2.5. Prevalence of Virulence Genes in Streptococcus spp., Staphylococcus spp., and Coliforms
The prevalence of the virulence genes in the tested bacterial genera is presented in Figure 1. A total of 57.53% of Streptococcus spp. carried the lmb virulence gene, followed by hylB (44.83%), bca (17.80%), and scpB (16.43%); however, none of them were found to be positive for the bac virulence gene. Similarly, 75.0% of Staphylococcus spp. carried the fib virulence gene, followed by hla (71.87%), coa (40.62%), sea (21.87%), and spa (6.25%). Likewise, 63.15% of coliforms harbored the ompC virulence gene, followed by colV (52.63%), fimH (47.36%), Ecs3703 (21.05%), and ompF (5.26%).
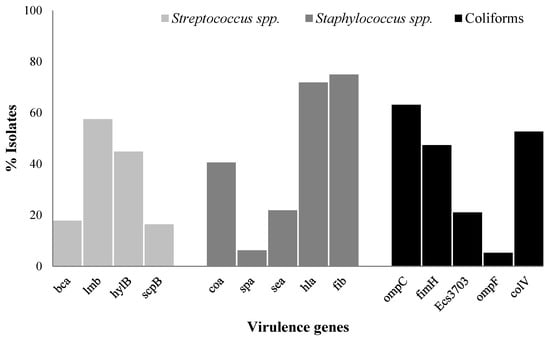
Figure 1.
The prevalence of virulence genes in Streptococcus spp., Staphylococcus spp., and coliforms from bovine mastitis. bca = surface protein ą-C, bac = surface protein ß-C, lmb = laminin-binding protein, hylB = hyaluronidase, scpB = Streptococcal C5a peptidase, coa = coagulase, spa = protein A, sea = enterotoxin A, hla = alpha-hemolysin, and fib = fibrinogen-binding protein, fimH = type 1 fimbriae, Ecs3703 = putative ABC transport protein, ompC and ompF = outer membrane protein, and colV = Colicin V.
3. Discussion
Our study revealed that Strep. uberis was the most frequently identified bacterial species that caused bovine mastitis in Taiwan, whereas Staph. aureus and E. coli were also detected as major bacteria. Previously, Strep. uberis was detected as the most dominant mastitis-causing pathogen in Taiwan [13]. The primary source of Strep. uberis in dairy farms include bedding materials, water, soil, and plant matter; therefore, pathogens may easily enter animal udders and transmit between cows [14]. Staph. aureus has been widely recognized as a common pathogen in intramammary infections in dairy cows due to its high transmissibility and ability to cause chronic infections [15]. The present study identified certain coagulase-negative staphylococci (CoNS) species that cause mastitis in cattle. CoNS are emerging globally as opportunistic pathogens, and their infections are usually self-limiting; however, studies have reported the need for antimicrobial treatment in clinical mastitis cases [16]. Similarly, E. coli is one of the major pathogens that causes bovine mastitis [11]. This study revealed that Klebsiella pneumoniae and Enterobacter aerogenes also caused bovine mastitis, as with other previous findings [17,18]. Therefore, numerous factors may determine bacterial presence in dairy farms, such as environment, management systems, temperature, humidity, and barn design [19].
Determining antimicrobial susceptibility profiles is essential for effective therapy and monitoring the selection and emergence of antimicrobial-resistant microorganisms. Our study found that most Streptococcus spp. were resistant to tetracycline and neomycin, which is similar to previous findings in Taiwan [13] and Northwest China [20]. Tetracycline is the most widely used antibiotic globally to treat various infections in cattle, including Taiwan, due to its broad-spectrum effectiveness, which may be the reason for the widespread resistance against tetracycline. The efflux pump is the most important mechanism of bacterial antimicrobial resistance to tetracycline, although ribosomal protective protein and enzyme inactivation also have a role in resistance development [21]. Aminoglycosides (neomycin) are used for prophylactic purposes in dairy animals, but they are not an effective antimicrobial agent for the treatment of mastitis-causing Streptococcus bacteria because most streptococci have inherited resistance to this class of antimicrobial due to their poor ability to penetrate the cell walls of bacteria [22]. Most of the tested Staphylococcus spp. and coliforms were susceptible to neomycin on our studied farms. The results also showed that 61.64% of Streptococcus spp. were susceptible to bacitracin, which is a much higher percentage than previous findings [23], but lower than the findings of [13]. These differing results might be due to differences in sampling areas or different antimicrobial agent usage histories. The present study’s isolates showed higher susceptibility to cephalothin, cefuroxime, and ceftiofur. This might be due to less exposure in these dairy farm environments, different antimicrobial agents being rotated in the treatment of dairy animals, or the broad-spectrum nature of these antimicrobials. Nearly half of the tested Streptococcus spp. (45.20%) and Staphylococcus spp. (43.75%) and one-third of coliforms (31.57%) developed resistance to ampicillin, as observed in previous findings on Staphylococcus and coliforms [22,24], while contrasting with the findings of [13] regarding Streptococcus. Ampicillin intramammary ointment is highly accessible for mastitis treatments in Taiwan; therefore, bacteria are likely to develop resistance against ampicillin on dairy farms.
In the present study, a high proportion of resistance genes related to tetracycline (tetM), neomycin (aph(3)-II), bacitracin (bcrB), and β-lactam (blaZ) were observed in Streptococcus spp., Staphylococcus spp., as well as coliforms, which have great potential to lead to high resistance rates against these antimicrobial agents. The present study revealed that 15.62% of Staphylococcus spp. carried the mecA gene, but one study found that nearly 2% of Staph. aureus carried the mecA gene [25]. The presence of the beta-lactam resistance gene blaZ varied depending on bacterial type. The present study demonstrated that 79.45% of Streptococcus spp. were resistant to neomycin based on the results of the phenotypical assay, but only 20.54% of Streptococcus spp. carried the resistance gene (aph (3)- I). Very few coliforms showed resistance to tetracycline (31.57%) and neomycin (21.05%) in terms of phenotypic assay, whereas much higher numbers of coliforms harbored the corresponding tetracycline (68.42%) and neomycin (57.89%) resistance genes. Our findings suggest that phenotypic resistance does not necessarily rely on the existence of resistance genes. In the present study, Streptococcus spp. showed no association between the phenotypic and genotypic antimicrobial resistance patterns for some antimicrobial agents, such as tetracycline, neomycin, and ceftiofur. Similarly, a previous study also mentioned that the majority of Streptococcus spp. showed no association between the phenotypic and genotypic characteristics of some antimicrobial agents [26]. These results could be explained by various reasons. Firstly, the majority of bacteria in this study showed a positive resistance phenotype but a negative genotype, which might be explained by the limited number of antimicrobial resistance genes investigated. Therefore, it was necessary for all possible antimicrobial resistance genes to be examined. Secondly, resistance gene expression depends on the existence of a promoter or inducer. Resistance genes distant from a promoter or associated with a weak promoter may lead to hindered gene expression. Thirdly, resistance genes might remain unexpressed due to point mutations [27]. Lastly, the small sample sizes and low number of observations could also contribute to these findings. Staphylococcus spp. showed a negative correlation between phenotypic and genotypic antimicrobial resistance to tetracycline; however, a previous study found a positive correlation in Staph. aureus [24].
In general, increased resistance is linked directly or indirectly to decreased virulence and fitness [28]. This is because developing resistance is a genetic burden and associated with a fitness cost [11,27]. Murine models have shown that penicillin-susceptible Strep. pneumoniae was virulent, although some isolates with low penicillin susceptibility were nonvirulent [29]. Similarly, E. coli resistant to tigecycline showed significantly decreased virulence in a mouse model [30]. In the present study, all three tested bacterial genera showed no association between antimicrobial resistance genes and virulence factors. Perhaps the sample size in the current study was insufficient or under antimicrobial resistance conditions. When bacteria are under the selective pressure of antimicrobials, it weakens the association between antimicrobial resistance and bacterial virulence [31]. Fitness costs in antimicrobial-resistant bacteria should be further studied to elucidate the underlying evolutionary mechanisms for resistance genes’ emergence, stability, and dissemination.
Multiple virulence factors play significant roles in host cell adhesion, invasion, and evasion of the host immune response. Our current findings on virulence factor distribution align with previous studies [32,33,34] but contradict the results reported by [11,35]. Virulence factors have diverse roles in bacterial pathogenesis. For example, laminin-binding protein (lmb) is vital in facilitating adherence to host laminin [36], and fibrinogen-binding protein (fib) is a major plasma protein that is crucial in blood clotting, inflammation, and interactions with cells and the extracellular matrix [37]. Additionally, both the ompF and ompC genes encode major porin proteins that act as passive diffusion channels for nutrients, antimicrobial agents, and small molecules [38]. Hence, these virulence factors might be crucial in mastitis development or persistence.
4. Conclusions
Streptococcus spp. was more dominant than Staphylococcus spp. and coliforms in causing bovine mastitis in Taiwan. These three bacterial genera revealed high-level phenotypic resistance to certain antimicrobial agents. The presence of bacterial resistance and diverse virulence profiles among these pathogens is concerning. Effective antimicrobials, cefuroxime, cephalothin, and ceftiofur, were identified for pathogen treatment, but their usage must be carefully monitored to prevent resistance development. The lack of associations between antimicrobial resistance genes and virulence factors in the tested bacterial genera may be influenced by factors such as bacterial species, host immunity, virulence mechanisms, and environmental conditions. Combining resistance gene identification and susceptibility tests can aid farmers in selecting appropriate chemotherapeutic measures. Regular monitoring of mastitis-causing pathogens is vital to assess antimicrobial resistance patterns in dairy herds.
5. Materials and Methods
5.1. Herd Enrollment Criteria and Milk Sample Collection
A total of 984 raw milk samples were collected from five different commercial dairy farms in Taiwan. Farm details are presented in Figure S1. Farms were required to have at least 200 lactating cows. Herds must participate in regular Dairy Herd Improvement testing or the monthly California Mastitis Test (CMT) must be used for all lactating cows with a yearly farm survey by sending individual cow milk samples to a reference laboratory. Quarter milk samples (including clinical, subclinical, and suspicious mastitis samples) were collected monthly from the one dairy farm in Tainan. For the other four farms, only quarter milk samples from cows diagnosed with clinical mastitis were collected. Veterinarians examined clinical mastitis samples, characterized by observable changes in milk or systemic symptoms. Subclinical mastitis samples, representing intramammary infections lacking clinical symptoms, were identified using the CMT. Raw milk samples were collected using aseptic procedures as described by the National Mastitis Council (https://www.nmconline.org/nmc-protocols-guidelines-and-procedures/) (accessed on 13 March 2020).
5.2. Isolation and Identification of Bacteria
Milk samples (10 µL) were cultured on 5% sheep blood agar and incubated at 37 °C for 20–24 h (Creative Biotechnology Company, New Taipei City, Taiwan). Bacteria were identified based on colonial characteristics and microscopic examination. Although different mastitis-causing bacterial genera were identified, only Staphylococcus spp., Streptococcus spp., Escherichia coli, and other coliforms were selected for further study because they are more common on dairy farms. Bacteria were preserved in tryptic soy broth (Becton, Dickinson and Company, Taipei City) with 20% glycerol (BIONOVAS biotechnology Co., Ltd., Toronto, Canada) and stored at −80 °C for further study.
5.3. Bacterial Species Identification through 16S rRNA Gene Sequencing
Bacterial DNA was extracted using the PureLinkTM Microbiome DNA Purification Kit’s recommended protocol (Invitrogen, Thermo Fisher Scientific, USA, Waltham, MA), and DNA was quantified using a MicroDrop (BIO-DL). Samples were amplified by PCR using 16S rRNA gene targeting primers [39]. PCR was performed on a T100 Thermal Cycler (Bio-Rad) with primers, and PCR conditions are summarized in the Supplementary Materials (Table S1). PCR amplicons were analyzed via electrophoresis in 1.5% agarose gels (w/v) and visualized using a Gel Doc XR+ System. A single discrete PCR amplicon band (458 bp) was purified using a QIAquick PCR Purification kit (QIAGEN, Inc., Toronto, ON, Canada). Purified PCR products were sent for sequencing with forward and reverse primers at National Chung Hsing University biotechnology center in Taichung, Taiwan. The sequencing data were analyzed using the National Center for Biotechnology Information rRNA/ITS nucleotide database.
5.4. Antimicrobial Susceptibility Test
Antimicrobial susceptibility tests were performed using the Kirby–Bauer disc diffusion method on Mueller–Hinton (M-H) agar plates. Eight commercially prepared antimicrobial discs were used: ampicillin (10 μg), oxacillin (1 μg), cephalothin (30 μg), cefuroxime (30 μg), ceftiofur (30 μg), neomycin (30 μg), bacitracin (10 units), and tetracycline (30 μg). Bacterial inoculum (5 × 105 cfu/mL) was inoculated on M-H agar plates according to the Clinical and Laboratory Standards Institute (CLSI) recommendations, and antimicrobial discs were placed and incubated at 37 °C for 20–24 h. The responses of the isolates to various antimicrobial agents were evaluated by measuring the zone of inhibition diameter and interpreting results according to standards recommended by [40], and bacitracin results were interpreted based on [41]’s recommendation. Staphylococcus aureus subsp. aureus Rosenbach ATCC 25923 was used as the quality control strain.
5.5. Identification of Antimicrobial Resistance Genes
Bacterial DNA was used as the template for PCR amplification. All isolates were tested through the PCR amplification of genes that confer resistance to neomycin (aph (3)-I; aph(3)-II), β-lactam (mecA, blaZ, and ampC), bacitracin (bcrA and bcrB), tetracycline (tetM, tetO, tetA, and tetB), and 16S Nossa were used as positive controls, and a PCR mix without a DNA template was used as negative control in all assays. Primers and PCR conditions used in this study are listed in Supplementary Materials (Table S1). Amplicons were analyzed via electrophoresis, as stated before.
5.6. Identification of Virulence Genes
Fifteen virulence genes were selected for Staphylococcus spp. (coa, spa, sea, hla, and fib), Streptococcus spp. (bac, bca, lmb, hylB, and scpB), and coliforms (ompC, fimH, Ecs3703, ompF, and colV). Primers and PCR conditions used in this study are listed in Supplementary Materials (Table S2). Amplicons were analyzed via electrophoresis, as stated before.
5.7. Statistical Analysis
Pearson correlation coefficient values were calculated using SAS 9.4 (SAS Institute Inc., Cary, NC, USA) to determine associations between the phenotypic and genotypic resistance patterns of antimicrobial agents and the relationship between antimicrobial resistance genes and bacterial virulence factors. p ≤ 0.05 was considered statistically significant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13010036/s1, Figure S1: Milk sample collection sites in Taiwan during 2020–2021. Table S1: Oligonucleotide sequences, primers, and targets for Polymerase Chain Reaction amplification of antimicrobial resistance genes. Table S2: Oligonucleotide sequences, primer names, and conditions of Polymerase Chain Reaction amplification for target virulence genes.
Author Contributions
B.D.: methodology, data curation, conducting the experiment, writing the first draft, performing the analysis. S.-T.C.: conceptualization, validation, methodology, writing—review and editing. J.-C.H.: methodology. M.-H.H.: methodology. H.-I.C.: conceptualization, funding acquisition, validation, writing—review and editing, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported (in part) by NSTC 111-2634-F-005-001—project Smart Sustainable New Agriculture Research Center (SMARTer).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We gratefully acknowledge Chih-Feng Chen, Department of Animal Science, NCHU, and Meng-Rong Hsieh for their assistance in the statistical analysis and milk sample collection, respectively.
Conflicts of Interest
We certify that there are no conflicts of interest with any financial organizations regarding the material discussed in this manuscript.
References
- Bradley, A. Bovine mastitis: An evolving disease. Vet. J. 2002, 164, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Barkema, H.W.; Zhang, L.; Liu, G.; Deng, Z.; Cai, L.; Shan, R.; Zhang, S.; Zou, J.; Kastelic, J.P.; et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J. Dairy Sci. 2017, 100, 4797–4806. [Google Scholar] [CrossRef] [PubMed]
- Sztachańska, M.; Barański, W.; Janowski, T.; Pogorzelska, J.; Zduńczyk, S. Prevalence and etiological agents of subclinical mastitis at the end of lactation in nine dairy herds in North-East Poland. Pol. J. Vet. Sci. 2016, 19, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Bergman, P.; Raqib, R.; Rekha, R.S.; Agerberth, B.; Gudmundsson, G.H. Host directed therapy against infection by boosting innate immunity. Front. Immunol. 2020, 11, 1209. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Kong, L.; Gao, H.; Cheng, X.; Wang, X. A review of current bacterial resistance to antibiotics in food animals. Front. Microbiol. 2022, 13, 822689. [Google Scholar] [CrossRef]
- WHO. Guidelines on Use of Medically Important Antimicrobials in Food-Producing Animals. 2017. Available online: http://apps.who.int/iris/bitstream/handle/10665/258970/9789241550130-eng.pdf (accessed on 12 July 2023).
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- McInnes, R.S.; McCallum, G.E.; Lamberte, L.E.; van Schaik, W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 2020, 53, 35–43. [Google Scholar] [CrossRef]
- Monistero, V.; Barberio, A.; Biscarini, F.; Cremonesi, P.; Castiglioni, B.; Graber, H.U.; Bottini, E.; Ceballos-Marquez, A.; Kroemker, V.; Petzer, I.M.; et al. Different distribution of antimicrobial resistance genes and virulence profiles of Staphylococcus aureus strains isolated from clinical mastitis in six countries. J. Dairy Sci. 2020, 103, 3431–3446. [Google Scholar] [CrossRef]
- Patel, K.; Godden, S.M.; Royster, E.E.; Crooker, B.A.; Johnson, T.J.; Smith, E.A.; Sreevatsan, S. Prevalence, antibiotic resistance, virulence and genetic diversity of Staphylococcus aureus isolated from bulk tank milk samples of U.S. dairy herds. BMC Genom. 2021, 22, 367. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Z.; Huang, C.; Gao, X.; Wang, Z.; Liu, Y.; Tian, C.; Hong, W.; Niu, S.; Liu, M. The phylogenetic group, antimicrobial susceptibility, and virulence genes of Escherichia coli from clinical bovine mastitis. J. Dairy Sci. 2018, 101, 572–580. [Google Scholar] [CrossRef]
- Chang, S.C.; Chen, M.W.; Lin, M.C.; Hu, O.Y.P. Antibiotic consumption in human and animals in Taiwan. Infect. Control J. 2003, 13, 334–345. [Google Scholar]
- Hsieh, J.C.; Yen, Y.S.; Chuang, S.T. Identification of Streptococcus spp. isolated from bovine milk and characterization of their antimicrobial susceptibility profiles in Taiwan. Thai J. Vet. Med. 2019, 49, 57–63. [Google Scholar] [CrossRef]
- Abureema, S.; Smooker, P.; Malmo, J.; Deighton, M. Molecular epidemiology of recurrent clinical mastitis due to Streptococcus uberis: Evidence of both an environmental source and recurring infection with the same strain. J. Dairy Sci. 2014, 97, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Monistero, V.; Graber, H.U.; Pollera, C.; Cremonesi, P.; Castiglioni, B.; Bottini, E.; Ceballos-Marquez, A.; Lasso-Rojas, L.; Kroemker, V.; Wente, N.; et al. Staphylococcus aureus isolates from bovine mastitis in eight countries: Genotypes, detection of genes encoding different toxins and other virulence genes. Toxins 2018, 10, 247. [Google Scholar] [CrossRef] [PubMed]
- Fry, P.R.; Middleton, J.R.; Dufour, S. Association of coagulase-negative staphylococcal species, mammary quarter milk somatic cell count, and persistence of intramammary infection in dairy cattle. J. Dairy Sci. 2014, 97, 4876–4885. [Google Scholar] [CrossRef] [PubMed]
- Ngu Ngwa, V.; Cuteri, V.; Awah-Ndukum, J.; Tangwa, B.V.; Manchang, K.T. Bacterial pathogens involved in bovine mastitis and their antibiotic resistance patterns in the Adamawa region of Cameroon. J. Dairy Res. Technol. 2020, 3, 012. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhang, J.; Yang, J.; Yi, B.; Liu, G.; Zhou, M.; Kastelic, J.P.; Han, B.; Gao, J. Klebsiella pneumoniae infection causes mitochondrial damage and dysfunction in bovine mammary epithelial cells. Vet. Res. 2021, 52, 17. [Google Scholar] [CrossRef]
- Quintana, Á.R.; Seseña, S.; Garzón, A.; Arias, R. Factors Affecting Levels of Airborne Bacteria in Dairy Farms: A Review. Animals 2020, 10, 526. [Google Scholar] [CrossRef]
- Shen, J.; Wu, X.; Yang, Y.; Lv, Y.; Li, X.; Ding, X.; Wang, S.; Yan, Z.; Yan, Y.; Yang, F.; et al. Antimicrobial Resistance and Virulence Factor of Streptococcus dysgalactiae Isolated from Clinical Bovine Mastitis Cases in Northwest China. Infect. Drug Resist. 2021, 14, 3519–3530. [Google Scholar] [CrossRef]
- Deng, J.; Liu, K.; Wang, K.; Yang, B.; Xu, H.; Wang, J.; Dai, F.; Xiao, X.; Gu, X.; Zhang, L.; et al. The prevalence of coagulase-negative staphylococcus associated with bovine mastitis in China and its antimicrobial resistance rate: A meta-analysis. J. Dairy Res. 2023, 90, 158–163. [Google Scholar] [CrossRef]
- Giguère, S. Principles of antimicrobial drug selection and Use. In Antimicrobial Therapy in Veterinary Medicine, 5th ed.; Giguère, S., Prescott, J.F., Dowling, P.M., Eds.; John Wiley & Sons: London, UK, 2013; pp. 105–115. [Google Scholar]
- Pascu, C.; Herman, V.; Iancu, I.; Costinar, L. Etiology of mastitis and antimicrobial resistance in dairy cattle farms in the western part of Romania. Antibiotics 2022, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Rana, E.A.; Fazal, M.A.; Alim, M.A. Frequently used therapeutic antimicrobials and their resistance patterns on Staphylococcus aureus and Escherichia coli in mastitis affected lactating cows. Int. J. Vet. Sci. 2022, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Qi, W.; Wang, X.; Ling, W.; Li, X.; Luo, J.; Zhang, S.; Li, H. Genetic characterization of antimicrobial resistance in Staphylococcus aureus isolated from bovine mastitis cases in Northwest China. J. Integr. Agric. 2016, 15, 2842–2847. [Google Scholar]
- Vélez, J.R.; Cameron, M.; Rodríguez-Lecompte, J.C.; Xia, F.; Heider, L.C.; Saab, M.; McClure, J.T.; Sánchez, J. Whole-genome sequence analysis of antimicrobial resistance genes in Streptococcus uberis and Streptococcus dysgalactiae isolates from Canadian Dairy Herds. Front. Vet. Sci. 2017, 4, 63. [Google Scholar] [CrossRef]
- Gao, J.; Yu, F.Q.; Luo, L.P.; He, J.Z.; Hou, R.G.; Zhang, H.Q.; Li, S.M.; Su, J.L.; Han, B. Antibiotic resistance of Streptococcus agalactiae from cows with mastitis. Vet. J. 2012, 194, 423–424. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [PubMed]
- Azoulay-Dupuis, E.; Rieux, V.; Muffat-Joly, M.; Bédos, J.P.; Vallée, E.; Rivier, C.; Isturiz, R.; Carbon, C.; Moine, P. Relationship between capsular type, penicillin susceptibility, and virulence of human Streptococcus pneumoniae isolates in mice. Antimicrob. Agents Chemother. 2000, 44, 1575–1577. [Google Scholar] [CrossRef]
- Linkevicius, M.; Sandegren, L.; Andersson, D.I. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. J. Antimicrob. Chemother. 2013, 68, 2809–2819. [Google Scholar] [CrossRef]
- Darmancier, H.; Domingues, C.P.F.; Rebelo, J.S.; Amaro, A.; Dionísio, F.; Pothier, J.; Serra, O.; Nogueira, T. Are virulence and antibiotic resistance genes linked? A comprehensive analysis of bacterial chromosomes and plasmids. Antibiotics 2022, 11, 706. [Google Scholar] [CrossRef]
- Ding, Y.; Zhao, J.; He, X.; Li, M.; Guan, H.; Zhang, Z.; Li, P. Antimicrobial resistance and virulence-related genes of Streptococcus obtained from dairy cows with mastitis in Inner Mongolia, China. Pharm. Biol. 2016, 54, 162–167. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, L.; Zhou, X.; He, Y.; Yong, C.; Shen, M.; Szenci, O.; Han, B. Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J. Dairy Sci. 2016, 99, 9560–9569. [Google Scholar] [CrossRef] [PubMed]
- Kaczorek, E.; Małaczewska, J.; Wójcik, R.; Siwicki, A.K. Biofilm production and other virulence factors in Streptococcus spp. isolated from clinical cases of bovine mastitis in Poland. BMC Vet. Res. 2017, 13, 398. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tan, X.; Zhang, X.; Xia, X.; Sun, H. The diversities of staphylococcal species, virulence and antibiotic resistance genes in the subclinical mastitis milk from a single Chinese cow herd. Microb. Pathog. 2015, 88, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Pires, R.; Rolo, D.; Gama-Norton, L. Group A Streptococci from carriage and disease in Portugal: Evolution of antimicrobial resistance and T antigenic types during 2000–2002. Microb. Drug Resist. 2005, 11, 360–370. [Google Scholar] [CrossRef]
- Meh, D.A.; Siebenlist, K.R.; Brennan, S.O.; Holyst, T.; Mosesson, M.W. The amino acid sequence in fibrin responsible for high affinity thrombin binding. Thromb. Haemost. 2001, 85, 470–474. [Google Scholar]
- Hejair, H.M.A.; Zhu, Y.; Ma, J.; Zhang, Y.; Pan, Z.; Zhang, W.; Yao, H. Functional role of ompF and ompC porins in pathogenesis of avian pathogenic Escherichia coli. Microb. Pathog. 2017, 107, 29–37. [Google Scholar] [CrossRef]
- Nossa, C.W.; Oberdorf, W.E.; Yang, L.; Aas, J.A.; Paster, B.J.; Desantis, T.Z.; Brodie, E.L.; Malamud, D.; Poles, M.A.; Pei, Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010, 16, 4135–4144. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. 2018. Available online: https://clsi.org/media/2321/vet08ed4_sample.pdf (accessed on 15 July 2023).
- FDA. BD BBL™ Sensi-Disc™ Antimicrobial Susceptibility Test Discs. 2007. Available online: https://legacy.bd.com/europe/regulatory/Assets/IFU/US/8840621(1107)_No.pdf (accessed on 5 July 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).