Widespread Multidrug Resistance of Arcobacter butzleri Isolated from Clinical and Food Sources in Central Italy
Abstract
1. Introduction
2. Results
2.1. Molecular Identification of Arcobacter Species
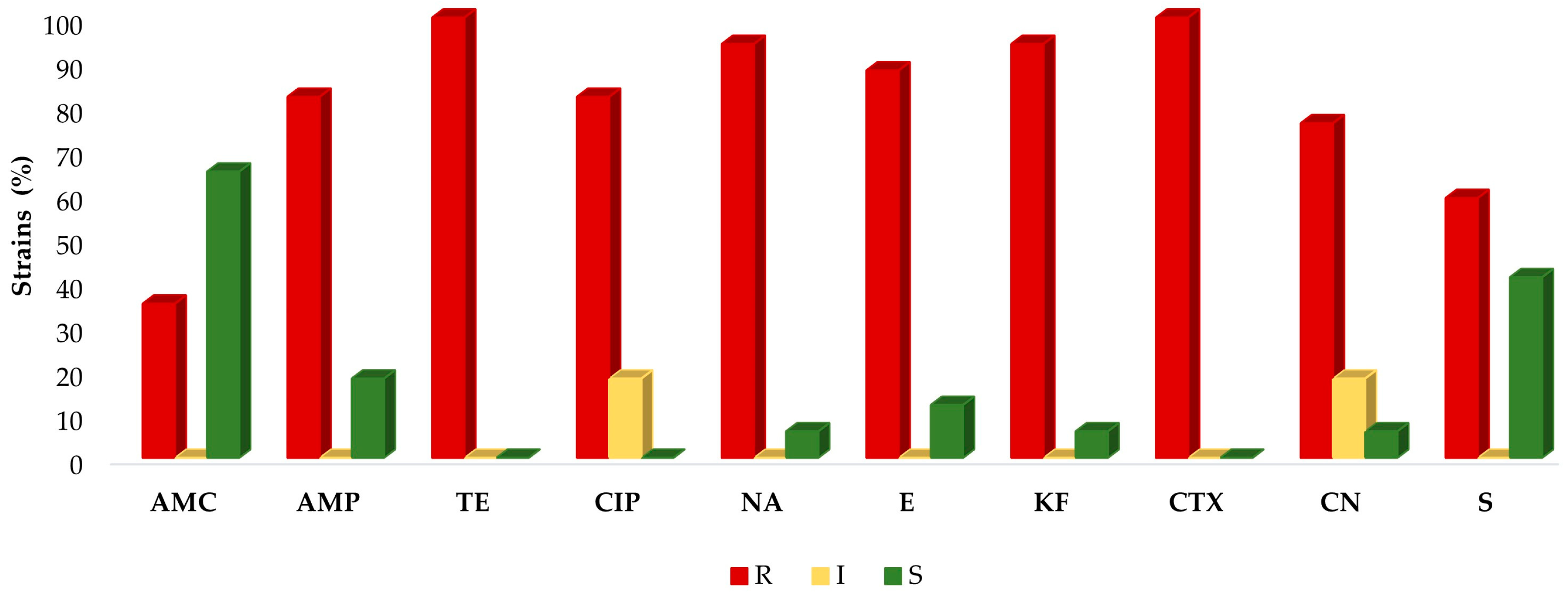
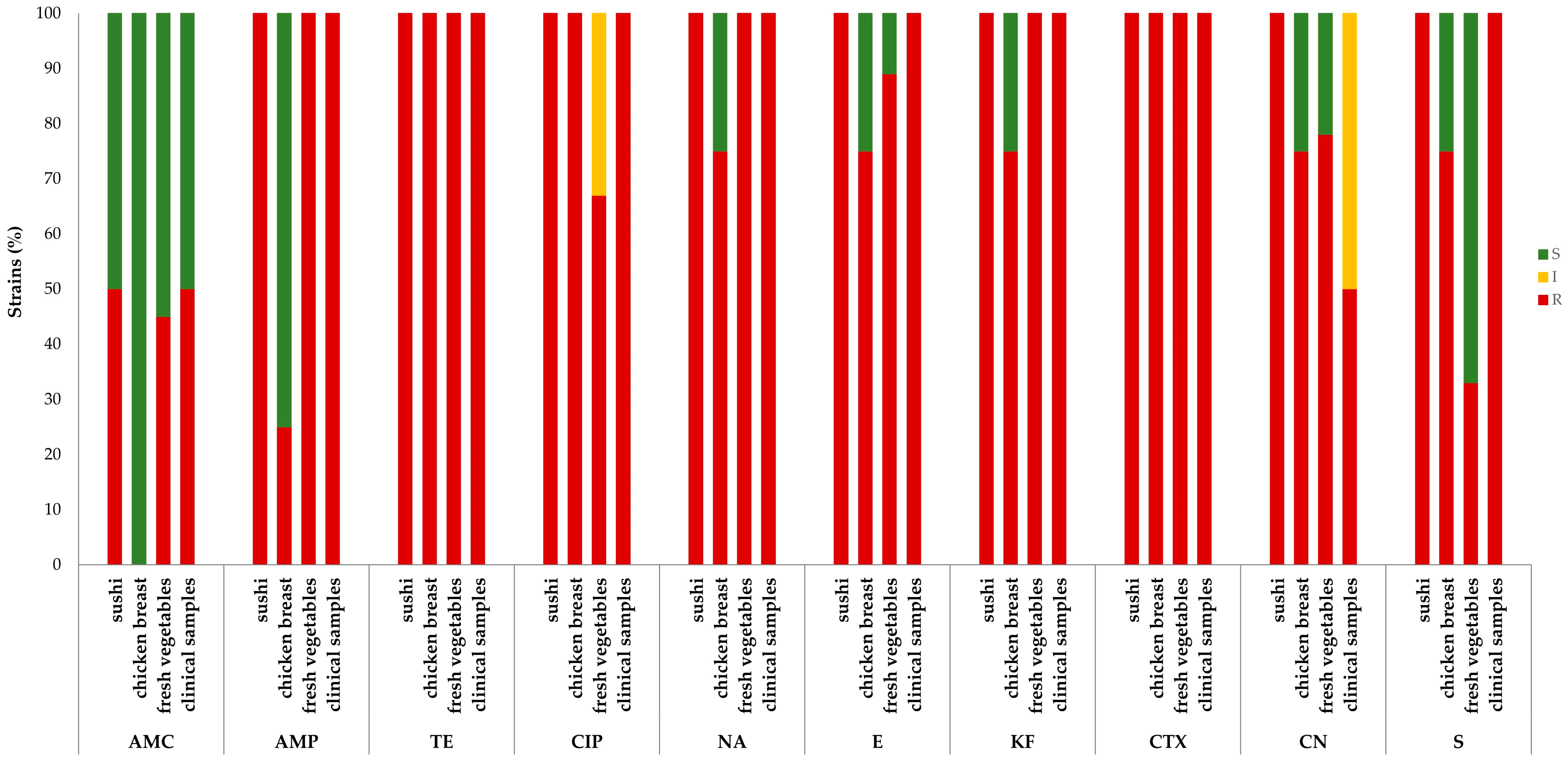
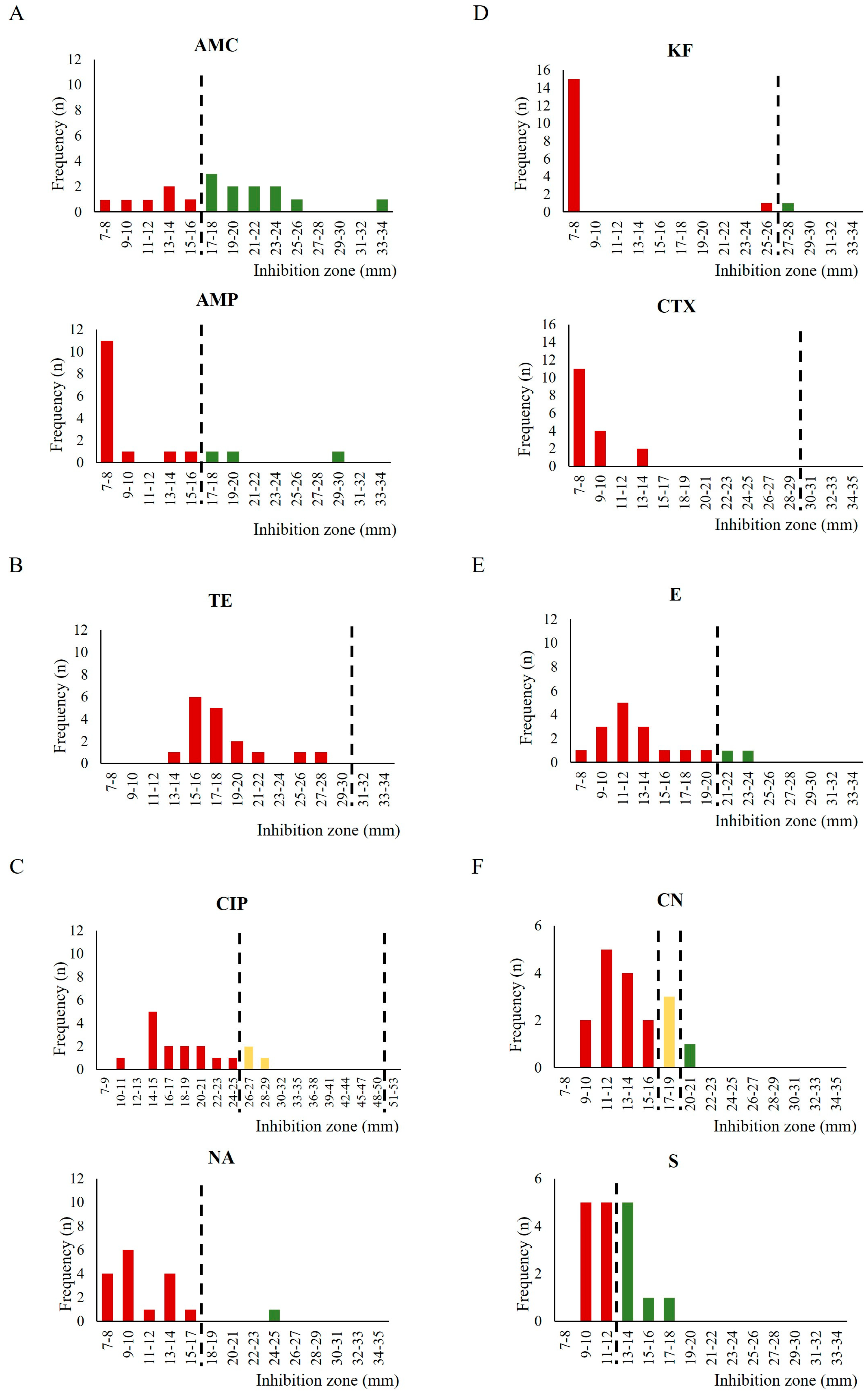
2.2. Antimicrobial Susceptibility Testing
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Multiplex PCR-Based Species Identification
4.3. Antibiotic Susceptibility Tests
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferreira, S.; Oleastro, M.; Domingues, F. Current insights on Arcobacter butzleri in food chain. Curr. Opin. Food Sci. 2019, 26, 9–17. [Google Scholar] [CrossRef]
- Figueras, M.J.; Pérez-Cataluña, A.; Salas-Massó, N.; Levican, A.; Collado, L. ‘Arcobacter porcinus’ sp. nov., a novel Arcobacter species uncovered by Arcobacter thereius. New Microbes New Infect. 2017, 15, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cataluña, A.; Salas-Massó, N.; Figueras, M.J. Arcobacter canalis sp. nov., isolated from a water canal contaminated with urban sewage. Int. J. Syst. Evol. Microbiol. 2018, 68, 1258–1264. [Google Scholar] [CrossRef]
- Callbeck, C.M.; Pelzer, C.; Lavik, G.; Ferdelman, T.G.; Graf, J.S.; Vekeman, B.; Schunck, H.; Littmann, S.; Fuchs, B.M.; Hach, P.F.; et al. Arcobacter peruensis sp. nov., a Chemolithoheterotroph Isolated from Sulfide- and Organic-Rich Coastal Waters off Peru. Appl. Environ. Microbiol. 2019, 85, e01344-19. [Google Scholar] [CrossRef]
- Kerkhof, P.-J.; On, S.L.W.; Houf, K. Arcobacter vandammei sp. nov., isolated from the rectal mucus of a healthy pig. Int. J. Syst. Evol. Microbiol. 2021, 71, 005113. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jung, Y.-T.; Kim, S.; Yoon, J.-H. Arcobacter acticola sp. nov., isolated from seawater on the East Sea in South Korea. J. Microbiol. 2016, 54, 655–659. [Google Scholar] [CrossRef]
- Martins, I.; Mateus, C.; Domingues, F.; Oleastro, M.; Ferreira, S. Putative Role of an ABC Efflux System in Aliarcobacter butzleri Resistance and Virulence. Antibiotics 2023, 12, 339. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Anderson, W.; Anelich, L.; Cordier, J.L.; Dewanti-Hariyadi, R.; Ross, T.; Zwietering, M.H. Microorganisms in Foods 7. Microbiological Testing in Food Safety Management; Springer: Cham, Switzerland, 2018; Volume 7. [Google Scholar]
- Chieffi, D.; Fanelli, F.; Fusco, V. Arcobacter butzleri: Up-to-date taxonomy, ecology, and pathogenicity of an emerging pathogen. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2071–2109. [Google Scholar] [CrossRef]
- Mottola, A.; Ciccarese, G.; Sinisi, C.; Savarino, A.E.; Marchetti, P.; Terio, V.; Tantillo, G.; Barrasso, R.; Di Pinto, A. Occurrence and characterization of Arcobacter spp. from ready-to-eat vegetables produced in Southern Italy. Ital. J. Food Saf. 2021, 10, 8585. [Google Scholar] [CrossRef]
- Kim, N.H.; Park, S.M.; Kim, H.W.; Cho, T.J.; Kim, S.H.; Choi, C.; Rhee, M.S. Prevalence of pathogenic Arcobacter species in South Korea: Comparison of two protocols for isolating the bacteria from foods and examination of nine putative virulence genes. Food Microbiol. 2019, 78, 18–24. [Google Scholar] [CrossRef]
- Ramees, T.p.; Rathore, R.; Kumar, A.; Remesh, A.; Kumar, R.; Karthik, K.; Malik, Y.; Dhama, K.; Singh, R. Phylogenetic analysis of Arcobacter butzleri and Arcobacter skirrowii isolates and their detection from contaminated vegetables by multiplex PCR. J. Exp. Biol. Agric. Sci. 2018, 6, 307–314. [Google Scholar] [CrossRef]
- González, A.; Bayas Morejón, I.F.; Ferrús, M.A. Isolation, molecular identification and quinolone-susceptibility testing of Arcobacter spp. isolated from fresh vegetables in Spain. Food Microbiol. 2017, 65, 279–283. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Ferrús, M.A. Study of Arcobacter spp. contamination in fresh lettuces detected by different cultural and molecular methods. Int. J. Food Microbiol. 2011, 145, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Choi, C. Survival of Arcobacter butzleri in Apple and Pear Purees. J. Food Saf. 2013, 33, 333–339. [Google Scholar] [CrossRef]
- Laishram, M.; Rathlavath, S.; Lekshmi, M.; Kumar, S.; Nayak, B.B. Isolation and characterization of Arcobacter spp. from fresh seafood and the aquatic environment. Int. J. Food Microbiol. 2016, 232, 87–89. [Google Scholar] [CrossRef]
- Leoni, F.; Chierichetti, S.; Santarelli, S.; Talevi, G.; Masini, L.; Bartolini, C.; Rocchegiani, E.; Naceur Haouet, M.; Ottaviani, D. Occurrence of Arcobacter spp. and correlation with the bacterial indicator of faecal contamination Escherichia coli in bivalve molluscs from the Central Adriatic, Italy. Int. J. Food Microbiol. 2017, 245, 6–12. [Google Scholar] [CrossRef]
- Barel, M.; Yildirim, Y. Arcobacter species isolated from various seafood and water sources; virulence genes, antibiotic resistance genes and molecular. World J. Microbiol. Biotechnol. 2023, 183, 12. [Google Scholar] [CrossRef]
- Figueras, M.J.; Levican, A.; Pujol, I.; Ballester, F.; Rabada Quilez, M.J.; Gomez-Bertomeu, F. A severe case of persistent diarrhoea associated with Arcobacter cryaerophilus but attributed to Campylobacter sp. and a review of the clinical incidence of Arcobacter spp. New Microbes New Infect. 2014, 2, 31–37. [Google Scholar] [CrossRef]
- Van den Abeele, A.M.; Vogelaers, D.; Van Hende, J.; Houf, K. Prevalence of Arcobacter species among humans, Belgium, 2008–2013. Emerg. Infect. Dis. 2014, 20, 1731–1734. [Google Scholar] [CrossRef]
- Prouzet-Mauléon, V.; Labadi, L.; Bouges, N.; Ménard, A.; Mégraud, F. Arcobacter butzleri: Underestimated enteropathogen. Emerg. Infect. Dis. 2006, 12, 307–309. [Google Scholar] [CrossRef]
- Samie, A.; Obi, C.L.; Barrett, L.J.; Powell, S.M.; Guerrant, R.L. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool samples from the Venda region, Limpopo, South Africa: Studies using molecular diagnostic methods. J. Infect. 2007, 54, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Švarcová, K.; Pejchalová, M.; Šilha, D. The Effect of Antibiotics on Planktonic Cells and Biofilm Formation Ability of Collected Arcobacter-like Strains and Strains Isolated within the Czech Republic. Antibiotics 2022, 11, 87. [Google Scholar] [CrossRef]
- Yan, J.J.; Ko, W.C.; Huang, A.H.; Chen, H.M.; Jin, Y.T.; Wu, J.J. Arcobacter butzleri bacteremia in a patient with liver cirrhosis. J. Formos. Med. Assoc. 2000, 99, 166–169. [Google Scholar] [PubMed]
- Lau, S.K.; Woo, P.C.; Teng, J.L.; Leung, K.W.; Yuen, K.Y. Identification by 16S ribosomal RNA gene sequencing of Arcobacter butzleri bacteraemia in a patient with acute gangrenous appendicitis. Mol. Pathol. 2002, 55, 182–185. [Google Scholar] [CrossRef]
- Collado, L.; Figueras, M.J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 2011, 24, 174–192. [Google Scholar] [CrossRef]
- Vandenberg, O.; Dediste, A.; Houf, K.; Ibekwem, S.; Souayah, H.; Cadranel, S.; Douat, N.; Zissis, G.; Butzler, J.P.; Vandamme, P. Arcobacter species in humans. Emerg. Infect. Dis. 2004, 10, 1863–1867. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.H.; Saleha, A.; Zakaria, Z.; Marimuthu, M. Arcobacter—An emerging threat to animals and animal origin food products? Trends Food Sci. Technol. 2011, 22, 225–236. [Google Scholar] [CrossRef]
- McGregor, A.C.; Wright, S.G. Gastrointestinal symptoms in travellers. Clin. Med. 2015, 15, 93–95. [Google Scholar] [CrossRef]
- Arguello, E.; Otto, C.C.; Mead, P.; Babady, N.E. Bacteremia caused by Arcobacter butzleri in an immunocompromised host. J. Clin. Microbiol. 2015, 53, 1448–1451. [Google Scholar] [CrossRef]
- Soelberg, K.K.; Danielsen, T.K.L.; Martin-Iguacel, R.; Justesen, U.S. Arcobacter butzleri is an opportunistic pathogen: Recurrent bacteraemia in an immunocompromised patient without diarrhoea. Access Microbiol. 2020, 2, acmi000145. [Google Scholar] [CrossRef]
- Gölz, G.; Karadas, G.; Alutis, M.; Fischer, A.; Kühl, A.; Breithaupt, A.; Göbel, U.; Alter, T.; Bereswill, S.; Heimesaat, M. Arcobacter butzleri Induce Colonic, Extra-Intestinal and Systemic Inflammatory Responses in Gnotobiotic IL-10 Deficient Mice in a Strain-Dependent Manner. PLoS ONE 2015, 10, e0139402. [Google Scholar] [CrossRef] [PubMed]
- Isidro, J.; Ferreira, S.; Pinto, M.; Domingues, F.; Oleastro, M.; Gomes, J.P.; Borges, V. Virulence and antibiotic resistance plasticity of Arcobacter butzleri: Insights on the genomic diversity of an emerging human pathogen. Infect. Genet. Evol. 2020, 80, 104213. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Sánchez, A.; Baztarrika, I.; Alonso, R.; Fernández-Astorga, A.; Martínez-Ballesteros, I.; Martinez-Malaxetxebarria, I. Arcobacter butzleri Biofilms: Insights into the Genes Beneath Their Formation. Microorganisms 2022, 10, 1280. [Google Scholar] [CrossRef]
- Martinez-Malaxetxebarria, I.; Girbau, C.; Salazar-Sánchez, A.; Baztarrika, I.; Martínez-Ballesteros, I.; Laorden, L.; Alonso, R.; Fernández-Astorga, A. Genetic characterization and biofilm formation of potentially pathogenic foodborne Arcobacter isolates. Int. J. Food Microbiol. 2022, 373, 109712. [Google Scholar] [CrossRef] [PubMed]
- Girbau, C.; Martinez-Malaxetxebarria, I.; Muruaga, G.; Carmona, S.; Alonso, R.; Fernandez-Astorga, A. Study of Biofilm Formation Ability of Foodborne Arcobacter butzleri under Different Conditions. J. Food Prot. 2017, 80, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Mudadu, A.G.; Melillo, R.; Salza, S.; Mara, L.; Marongiu, L.; Piras, G.; Spanu, C.; Tedde, T.; Fadda, A.; Virgilio, S.; et al. Detection of Arcobacter spp. in environmental and food samples collected in industrial and artisanal sheep’s milk cheese-making plants. Food Control 2021, 126, 108100. [Google Scholar] [CrossRef]
- Šilha, D.; Hrušková, L.; Brožková, I.; Moťková, P.; Vytřasová, J. Survival of selected bacteria from the genus Arcobacter on various metallic surfaces. J. Food Nutr. Res. 2014, 53, 217–223. [Google Scholar]
- Falahchai, S.; Bahador, N.; Ghane, M. Isolation and Characterization of Arcobacter butzleri from Environmental Samples and Determination of their Pathogenic Gene Expression under Different Physicochemical Conditions. Pol. J. Environ. Stud. 2021, 30, 3963–3974. [Google Scholar] [CrossRef]
- Serraino, A.; Giacometti, F. Short communication: Occurrence of Arcobacter species in industrial dairy plants. J. Dairy Sci. 2014, 97, 2061–2065. [Google Scholar] [CrossRef]
- Šilha, D.; Šilhová, L.; Vytřasová, J.; Brožková, I.; Pejchalova, M. Survival of selected bacteria of Arcobacter genus in disinfectants and possibility of acquired secondary resistance to disinfectants. J. Microbiol. Biotechnol. Food Sci. 2016, 5, 326–329. [Google Scholar] [CrossRef]
- Rasmussen, L.H.; Kjeldgaard, J.; Christensen, J.P.; Ingmer, H. Multilocus sequence typing and biocide tolerance of Arcobacter butzleri from Danish broiler carcasses. BMC Res. Notes 2013, 6, 322. [Google Scholar] [CrossRef]
- Uljanovas, D.; Gölz, G.; Brückner, V.; Grineviciene, A.; Tamuleviciene, E.; Alter, T.; Malakauskas, M. Prevalence, antimicrobial susceptibility and virulence gene profiles of Arcobacter species isolated from human stool samples, foods of animal origin, ready-to-eat salad mixes and environmental water. Gut Pathog. 2021, 13, 76. [Google Scholar] [CrossRef] [PubMed]
- Ramees, T.P.; Dhama, K.; Karthik, K.; Rathore, R.S.; Kumar, A.; Saminathan, M.; Tiwari, R.; Malik, Y.S.; Singh, R.K. Arcobacter: An emerging food-borne zoonotic pathogen, its public health concerns and advances in diagnosis and control—A comprehensive review. Vet. Q. 2017, 37, 136–161. [Google Scholar] [CrossRef] [PubMed]
- Brückner, V.; Fiebiger, U.; Ignatius, R.; Friesen, J.; Eisenblätter, M.; Höck, M.; Alter, T.; Bereswill, S.; Gölz, G.; Heimesaat, M.M. Prevalence and antimicrobial susceptibility of Arcobacter species in human stool samples derived from out- and inpatients: The prospective German Arcobacter prevalence study Arcopath. Gut Pathog. 2020, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Sciortino, S.; Arculeo, P.; Alio, V.; Cardamone, C.; Nicastro, L.; Arculeo, M.; Alduina, R.; Costa, A. Occurrence and Antimicrobial Resistance of Arcobacter spp. Recovered from Aquatic Environments. Antibiotics 2021, 10, 288. [Google Scholar] [CrossRef]
- Khodamoradi, S.; Abiri, R. The incidence and antimicrobial resistance of Arcobacter species in animal and poultry meat samples at slaughterhouses in Iran. Iran. J. Microbiol. 2020, 12, 531–536. [Google Scholar] [CrossRef]
- Jribi, H.; Sellami, H.; Amor, S.B.; Ducournau, A.; Sifré, E.; Benejat, L.; Mégraud, F.; Gdoura, R. Occurrence and Antibiotic Resistance of Arcobacter Species Isolates from Poultry in Tunisia. J. Food Prot. 2020, 83, 2080–2086. [Google Scholar] [CrossRef]
- Fanelli, F.; Chieffi, D.; Di Pinto, A.; Mottola, A.; Baruzzi, F.; Fusco, V. Phenotype and genomic background of Arcobacter butzleri strains and taxogenomic assessment of the species. Food Microbiol. 2020, 89, 103416. [Google Scholar] [CrossRef]
- Zautner, A.E.; Riedel, T.; Bunk, B.; Spröer, C.; Boahen, K.G.; Akenten, C.W.; Dreyer, A.; Färber, J.; Kaasch, A.J.; Overmann, J.; et al. Molecular characterization of Arcobacter butzleri isolates from poultry in rural Ghana. Front. Cell. Infect. Microbiol. 2023, 13, 1094067. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. In Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 13.0; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2023.
- BSAC. BSAC Methods for Antimicrobial Susceptibility Testing. Version 14.0, 05-01-2015. Available online: https://www.bsac.org.uk/wp-content/uploads/2012/02/BSAC-Susceptibility-testing-version-14.pdf (accessed on 10 June 2023).
- Çelik, C.; Pınar, O.; Sipahi, N. The Prevalence of Aliarcobacter Species in the Fecal Microbiota of Farm Animals and Potential Effective Agents for Their Treatment: A Review of the Past Decade. Microorganisms 2022, 10, 2430. [Google Scholar] [CrossRef]
- Collado, L.; Guarro, J.; Figueras, M.J. Prevalence of Arcobacter in meat and shellfish. J. Food Prot. 2009, 72, 1102–1106. [Google Scholar] [CrossRef]
- Ferreira, S.; Queiroz, J.A.; Oleastro, M.; Domingues, F.C. Insights in the pathogenesis and resistance of Arcobacter: A review. Crit. Rev. Microbiol. 2016, 42, 364–383. [Google Scholar] [CrossRef] [PubMed]
- Dekker, D.; Eibach, D.; Boahen, K.G.; Akenten, C.W.; Pfeifer, Y.; Zautner, A.E.; Mertens, E.; Krumkamp, R.; Jaeger, A.; Flieger, A.; et al. Fluoroquinolone-Resistant Salmonella enterica, Campylobacter spp., and Arcobacter butzleri from Local and Imported Poultry Meat in Kumasi, Ghana. Foodborne Pathog. Dis. 2019, 16, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, H.; Maruyama, S.; Morita, Y.; Ohsuga, T.; Ozawa, S.; Kobayashi, Y.; Abe, M.; Katsube, Y.; Mikami, T. Prevalence of Arcobacter species in retail meats and antimicrobial susceptibility of the isolates in Japan. Int. J. Food Microbiol. 2004, 90, 303–308. [Google Scholar] [CrossRef]
- Rathlavath, S.; Kohli, V.; Singh, A.S.; Lekshmi, M.; Tripathi, G.; Kumar, S.; Nayak, B.B. Virulence genotypes and antimicrobial susceptibility patterns of Arcobacter butzleri isolated from seafood and its environment. Int. J. Food Microbiol. 2017, 263, 32–37. [Google Scholar] [CrossRef]
- Shah, A.H.; Saleha, A.A.; Zunita, Z.; Murugaiyah, M.; Aliyu, A.B.; Jafri, N. Prevalence, distribution and antibiotic resistance of emergent Arcobacter spp. from clinically healthy cattle and goats. Transbound. Emerg. Dis. 2013, 60, 9–16. [Google Scholar] [CrossRef][Green Version]
- Šilha, D.; Pejchalová, M.; Šilhová, L. Susceptibility to 18 drugs and multidrug resistance of Arcobacter isolates from different sources within the Czech Republic. J. Glob. Antimicrob. Resist. 2017, 9, 74–77. [Google Scholar] [CrossRef]
- Vicente-Martins, S.; Oleastro, M.; Domingues, F.C.; Ferreira, S. Arcobacter spp. at retail food from Portugal: Prevalence, genotyping and antibiotics resistance. Food Control 2018, 85, 107–112. [Google Scholar] [CrossRef]
- Elmali, M.; Yeşim Can, H. Occurence and antimicrobial resistance of Arcobacter species in food and slaughterhouse samples. Food Sci. Technol. 2017, 37, 280–285. [Google Scholar] [CrossRef]
- Rahimi, E. Prevalence and antimicrobial resistance of Arcobacter species isolated from poultry meat in Iran. Br. Poult. Sci. 2014, 55, 174–180. [Google Scholar] [CrossRef]
- Debelo, M.; Mohammed, N.; Tiruneh, A.; Tolosa, T. Isolation, identification and antibiotic resistance profile of thermophilic Campylobacter species from Bovine, Knives and personnel at Jimma Town Abattoir, Ethiopia. PLoS ONE 2022, 17, e0276625. [Google Scholar] [CrossRef]
- Brown, D.F.; Wootton, M.; Howe, R.A. Antimicrobial susceptibility testing breakpoints and methods from BSAC to EUCAST. J. Antimicrob. Chemother. 2016, 71, 3–5. [Google Scholar] [CrossRef]
- EUCAST. MIC and Zone Diameter Distributions and ECOFFs. Available online: https://www.eucast.org/mic_and_zone_distributions_and_ecoffs (accessed on 10 June 2023).
- Sousa, V.; Luís, Â.; Oleastro, M.; Domingues, F.; Ferreira, S. Polyphenols as resistance modulators in Arcobacter butzleri. Folia Microbiol. 2019, 64, 547–554. [Google Scholar] [CrossRef]
- Collado, L.; Inza, I.; Guarro, J.; Figueras, M.J. Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ. Microbiol. 2008, 10, 1635–1640. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.U.H.; Cloutier, M.; Libby, M.; Lapen, D.R.; Wilkes, G.; Topp, E. Enhanced Single-tube Multiplex PCR Assay for Detection and Identification of Six Arcobacter Species. J. Appl. Microbiol. 2017, 123, 1522–1532. [Google Scholar] [CrossRef] [PubMed]
- Matuschek, E.; Brown, D.F.J.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef] [PubMed]
| Strain ID | Species | Isolation Source | Isolation Date |
|---|---|---|---|
| 31164/3 | A. butzleri | Sushi | July 2022 |
| 35709/3 | A. butzleri | Sushi | August 2022 |
| 35683/1 | A. butzleri | Chicken breast | August 2022 |
| 36981/2 | A. butzleri | Chicken breast | August 2022 |
| 37809/1 | A. butzleri | Chicken breast | August 2022 |
| 39884/1 | A. butzleri | Chicken breast | August 2022 |
| 25176/2 | A. butzleri | Fresh vegetables, curly endive | June 2022 |
| 29991/1 | A. butzleri | Fresh vegetables, escarole | June 2022 |
| 32455/2 | A. butzleri | Fresh vegetables, curly endive | July 2022 |
| 35638/2 | A. butzleri | Fresh vegetables, curly endive | August 2022 |
| 40619/1 | A. butzleri | Fresh vegetables, escarole | September 2022 |
| 43130/1 | A. butzleri | Fresh vegetables, escarole | September 2022 |
| 43130/2 | A. butzleri | Fresh vegetables, curly endive | September 2022 |
| 43130/3 | A. butzleri | Fresh vegetables, radicchio | September 2022 |
| 45224/3 | A. butzleri | Fresh vegetables, chicory | October 2022 |
| 8722325 | A. butzleri | Clinical isolate | September 2018 |
| 9291368 | A. butzleri | Clinical isolate | August 2022 |
| Penicillins | Tetracyclines | Fluoroquinolones | Macrolides | Cephalosporins | Aminoglycosides | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain ID | AMC | AMP | TE | CIP | NA | E | KF | CTX | CN | S | ||
| Food matrices | Sushi | 31164/3 | R | R | R | R | R | R | R | R | R | R |
| 35709/3 | S | R | R | R | R | R | R | R | R | R | ||
| Chicken breast | 35683/1 | S | S | R | R | R | R | R | R | R | R | |
| 36981/2 | S | R | R | R | R | R | R | R | R | R | ||
| 37809/1 | S | S | R | R | S | S | S | R | S | S | ||
| 39884/1 | S | S | R | R | R | R | R | R | R | R | ||
| Fresh vegetables | 25176/2 | S | R | R | R | R | R | R | R | I | S | |
| 29991/1 | R | R | R | I | R | S | R | R | R | S | ||
| 32455/2 | S | R | R | I | R | R | R | R | R | S | ||
| 35638/2 | R | R | R | R | R | R | R | R | R | R | ||
| 40619/1 | S | R | R | R | R | R | R | R | I | S | ||
| 43130/1 | R | R | R | I | R | R | R | R | R | R | ||
| 43130/2 | S | R | R | R | R | R | R | R | R | S | ||
| 43130/3 | R | R | R | R | R | R | R | R | R | S | ||
| 45224/3 | S | R | R | R | R | R | R | R | R | R | ||
| Clinical | 8722325 | S | R | R | R | R | R | R | R | I | R | |
| 9291368 | R | R | R | R | R | R | R | R | R | R | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabucci, C.; Baldelli, G.; Amagliani, G.; Schiavano, G.F.; Savelli, D.; Russo, I.; Di Lullo, S.; Blasi, G.; Napoleoni, M.; Leoni, F.; et al. Widespread Multidrug Resistance of Arcobacter butzleri Isolated from Clinical and Food Sources in Central Italy. Antibiotics 2023, 12, 1292. https://doi.org/10.3390/antibiotics12081292
Gabucci C, Baldelli G, Amagliani G, Schiavano GF, Savelli D, Russo I, Di Lullo S, Blasi G, Napoleoni M, Leoni F, et al. Widespread Multidrug Resistance of Arcobacter butzleri Isolated from Clinical and Food Sources in Central Italy. Antibiotics. 2023; 12(8):1292. https://doi.org/10.3390/antibiotics12081292
Chicago/Turabian StyleGabucci, Claudia, Giulia Baldelli, Giulia Amagliani, Giuditta Fiorella Schiavano, David Savelli, Ilaria Russo, Stefania Di Lullo, Giuliana Blasi, Maira Napoleoni, Francesca Leoni, and et al. 2023. "Widespread Multidrug Resistance of Arcobacter butzleri Isolated from Clinical and Food Sources in Central Italy" Antibiotics 12, no. 8: 1292. https://doi.org/10.3390/antibiotics12081292
APA StyleGabucci, C., Baldelli, G., Amagliani, G., Schiavano, G. F., Savelli, D., Russo, I., Di Lullo, S., Blasi, G., Napoleoni, M., Leoni, F., Primavilla, S., Massacci, F. R., Garofolo, G., & Petruzzelli, A. (2023). Widespread Multidrug Resistance of Arcobacter butzleri Isolated from Clinical and Food Sources in Central Italy. Antibiotics, 12(8), 1292. https://doi.org/10.3390/antibiotics12081292







