Comparison of Cefotaxime-Resistant Escherichia coli and sul1 and intI1 by qPCR for Monitoring of Antibiotic Resistance of Wastewater, Surface Water, and Recycled Water
Abstract
1. Introduction
2. Results
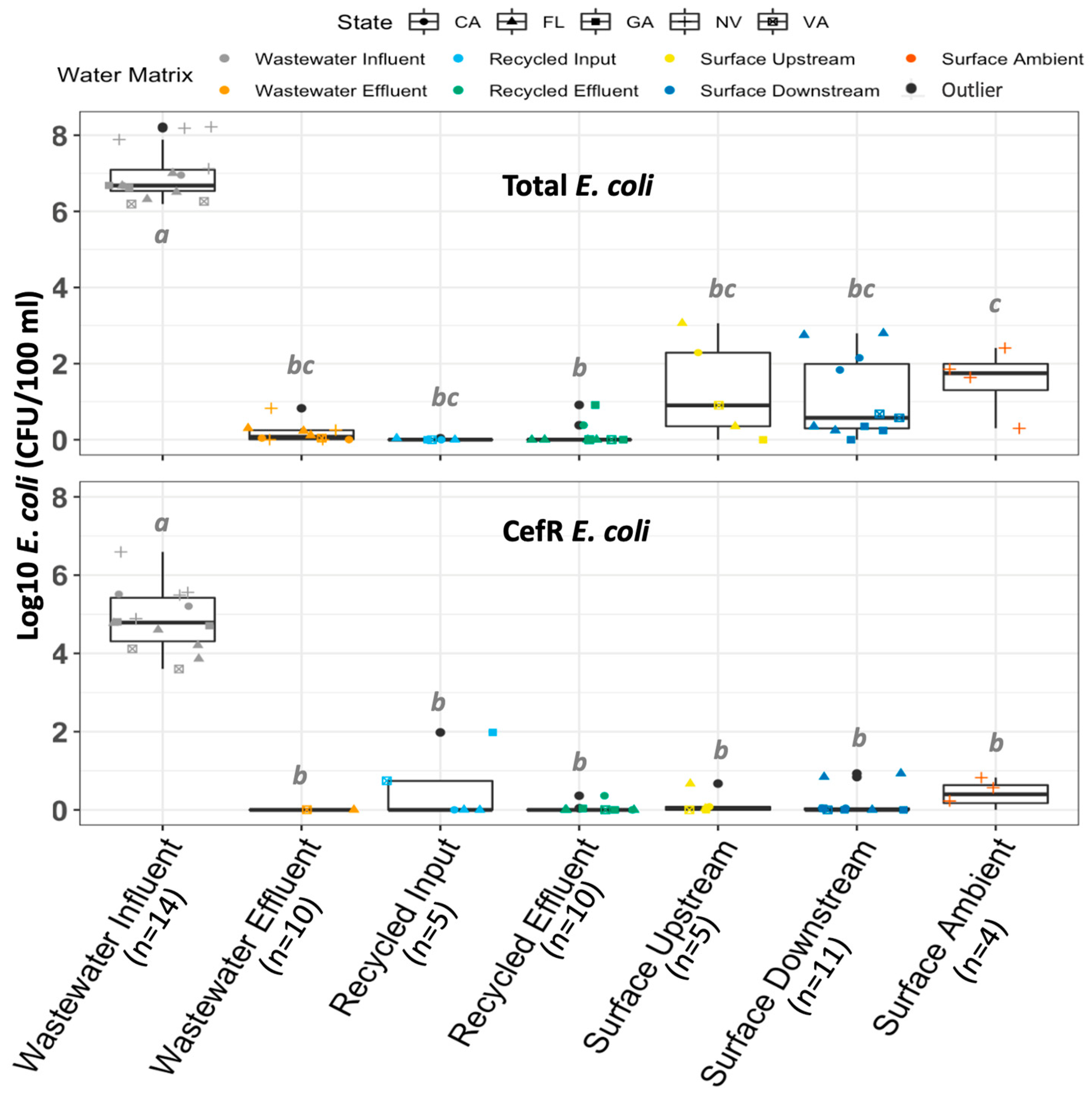
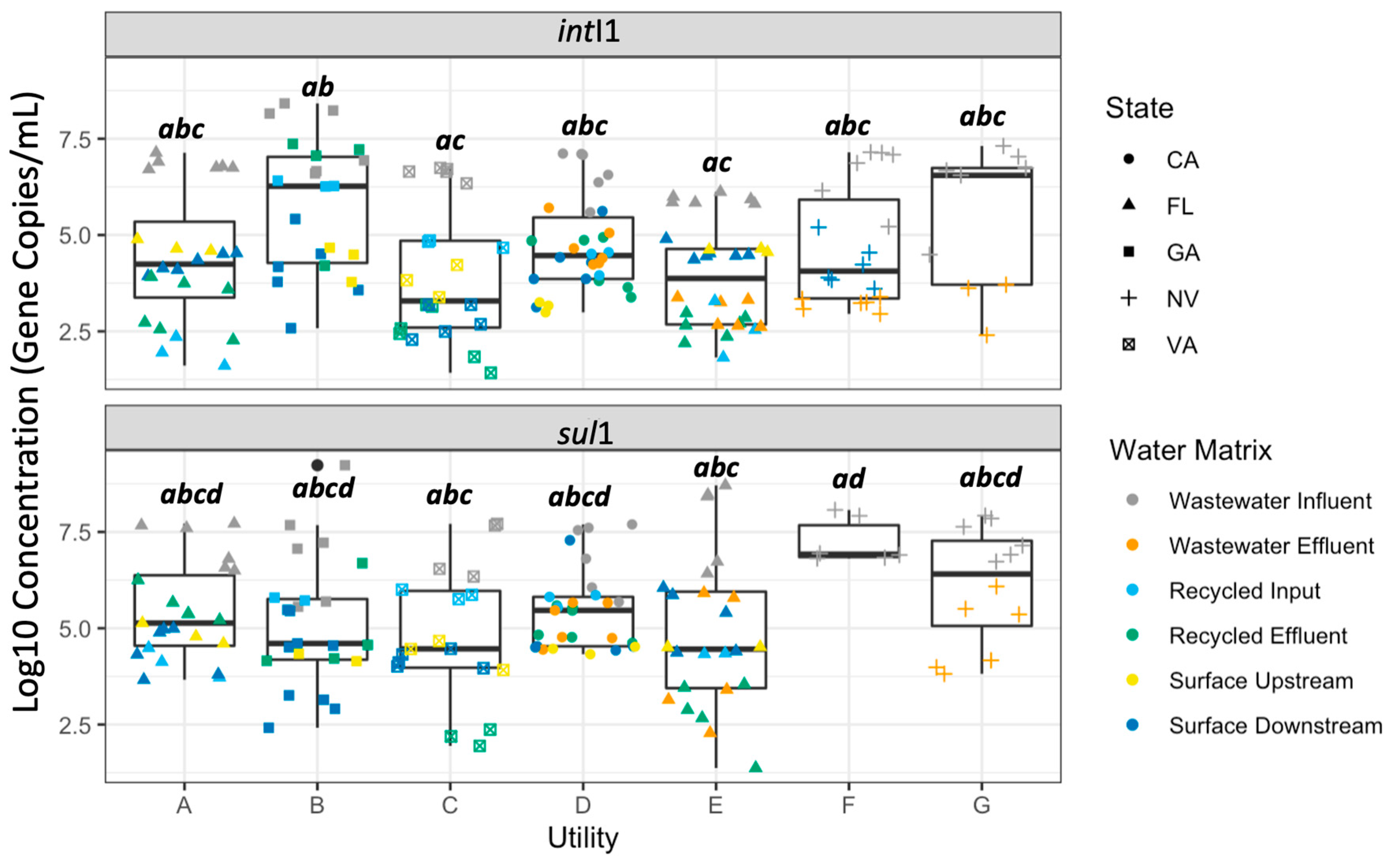
2.1. Comparison of AMR Measures across Water Matrixes, Geographic Locations, and Water Utilities
2.2. Correlation of sul1 and intI1 with cefR E. coli
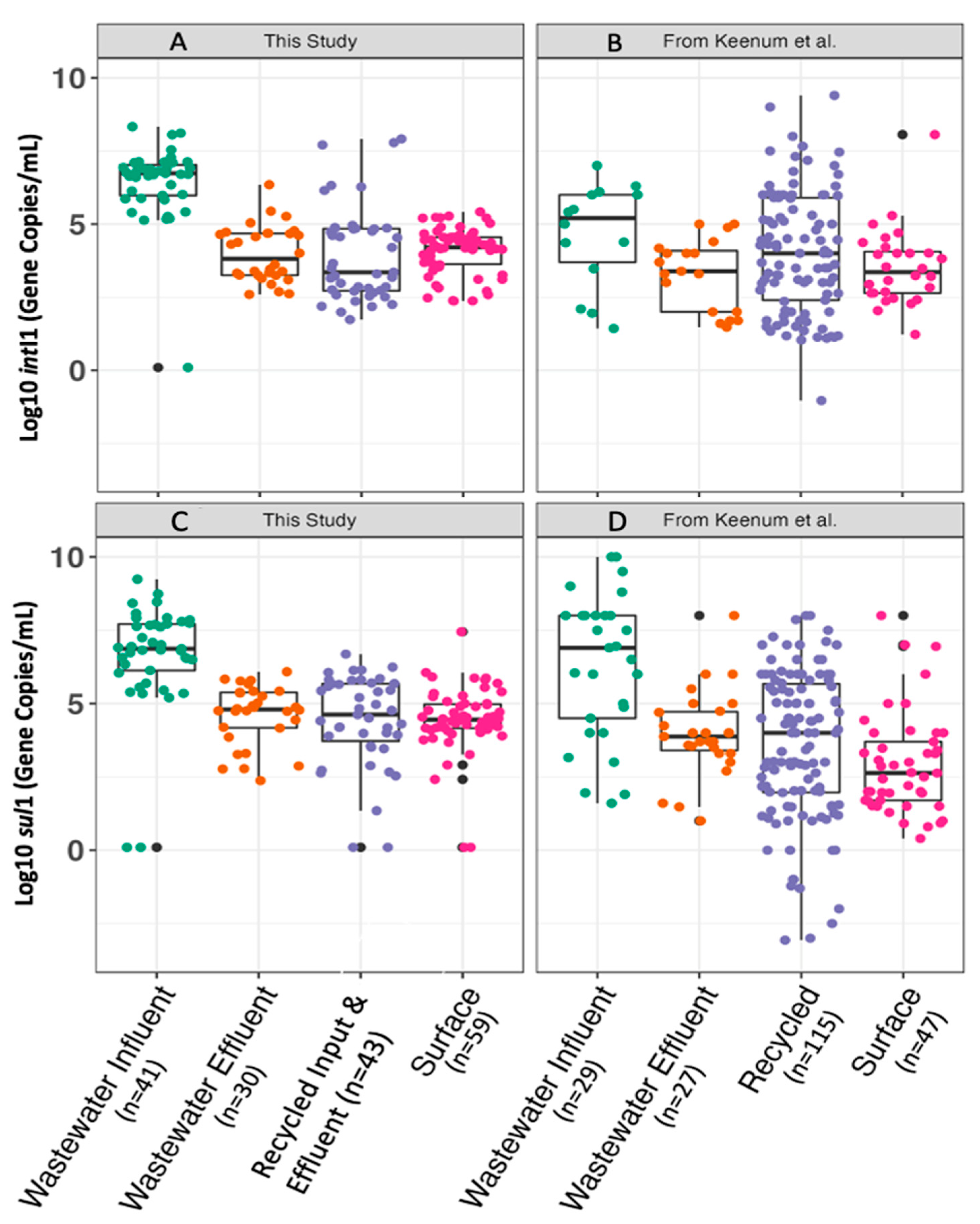
2.3. Interlaboratory Comparison
2.4. Repeatability of Technical and Biological Replicates
3. Discussion
3.1. Limitations of This Study
3.2. E. coli, sul1, and intI1 Measures Correlate as Indicators of Water Quality
3.3. Linking Measurements to Human Health Risk
3.4. Applying Measurements for Assessing Removal by Treatment Processes
3.5. Assessing Ecological Hot Spots for AMR Selection and Evolution
3.6. Wastewater-Based Surveillance and Epidemiology
3.7. Practical Considerations
3.8. QA/QC Recommendations
4. Materials and Methods
4.1. Sampling and Interlaboratory Comparison
4.2. CefR- E. coli Culture
4.3. qPCR Enumeration of sul1 and intI1
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef] [PubMed]
- United Nations Environment Programme. Bracing for Superbugs: Strengthening Environmental Action in the One Health Response to Antimicrobial Resistance. Geneva. 2023. Available online: https://www.unep.org/resources/superbugs/environmental-action (accessed on 3 April 2023).
- National Academies of Sciences, Engineering, and Medicine. Combating Antimicrobial Resistance and Protecting the Miracle of Modern Medicine; The National Academies Press: Washington, DC, USA, 2022. [Google Scholar] [CrossRef]
- Pruden, A.; Vikesland, P.J.; Davis, B.C.; de Roda Husman, A.M. Seizing the moment: Now is the time for integrated global surveillance of antimicrobial resistance in wastewater environments. Curr. Opin. Microbiol. 2021, 64, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Anjum, M.F.; Schmitt, H.; Börjesson, S.; Berendonk, T.U.; Donner, E.; Stehling, E.G.; Boerlin, P.; Topp, E.; Jardine, C.; Li, X.; et al. The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opin. Microbiol. 2021, 64, 152–158. [Google Scholar] [CrossRef]
- Wuijts, S.; van den Berg, H.H.; Miller, J.; Abebe, L.; Sobsey, M.; Andremont, A.; Medlicott, K.O.; van Passel, M.W.; de Roda Husman, A.M. Towards a research agenda for water, sanitation and antimicrobial resistance. J. Water Health 2017, 15, 175–184. [Google Scholar] [CrossRef]
- Huijbers, P.M.; Flach, C.F.; Larsson, D.J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019, 130, 104880. [Google Scholar] [CrossRef]
- Baquero, F.; Martinez, J.L.; Canton, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Lupo, A.; Coyne, S.; Berendonk, T.U. Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front. Microbiol. 2012, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Nappier, S.P.; Liguori, K.; Ichida, A.M.; Stewart, J.R.; Jones, K.R. Antibiotic resistance in recreational waters: State of the science. Int. J. Environ. Res. Public Health 2020, 17, 8034. [Google Scholar] [CrossRef]
- Leonard, A.F.; Zhang, L.; Balfour, A.J.; Garside, R.; Gaze, W.H. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ. Int. 2015, 82, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, S.B.; Søraas, A.V.; Arnesen, L.S.; Leegaard, T.M.; Sundsfjord, A.; Jenum, P.A. A comparison of extended spectrum β-lactamase producing Escherichia coli from clinical, recreational water and wastewater samples associated in time and location. PLoS ONE 2017, 12, e0186576. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Liguori, K.; Keenum, I.; Davis, B.C.; Calarco, J.; Milligan, E.; Hardwood, V.J.; Pruden, A. Antimicrobial Resistance Monitoring of Water Environments: A Framework for Standardized Methods and Quality Control. Environ. Sci. Technol. 2022, 56, 9149–9160. [Google Scholar] [CrossRef] [PubMed]
- Marano, R.B.; Fernandes, T.; Manaia, C.M.; Nunes, O.; Morrison, D.; Berendonk, T.U.; Kreuzinger, N.; Tenson, T.; Corno, G.; Fatta-Kassinos, D.; et al. A global multinational survey of cefotaxime-resistant coliforms in urban wastewater treatment plants. Environ. Int. 2020, 144, 106035. [Google Scholar] [CrossRef]
- World Health Organization. WHO Integrated Global Surveillance on ESBL-Producing E. coli Using a “One Health” Approach: Implementation and Opportunities. 2021. Available online: https://www.who.int/publications/i/item/9789240021402 (accessed on 29 December 2022).
- Adegoke, A.A.; Madu, C.E.; Aiyegoro, O.A.; Stenström, T.A.; Okoh, A.I. Antibiogram and beta-lactamase genes among cefotaxime resistant E. coli from wastewater treatment plant. Antimicrob. Resist. Infect. Control. 2020, 9, 1–12. [Google Scholar] [CrossRef]
- Melzer, M.; Petersen, I. Mortality following bacteraemic infection caused by extended spectrum beta-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J. Infect. 2007, 55, 254–259. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Korzeniewska, A.; Harnisz, M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013, 91, 96–102. [Google Scholar] [CrossRef]
- Mir, R.A.; Weppelmann, T.A.; Johnson, J.A.; Archer, D.; Morris, J.G., Jr.; Jeong, K.C. Identification and characterization of cefotaxime resistant bacteria in beef cattle. PLoS ONE 2016, 11, e0163279. [Google Scholar] [CrossRef]
- Wiegand, I.; Geiss, H.K.; Mack, D.; Sturenburg, E.; Seifert, H. Detection of extended-spectrum beta-lactamases among Enterobacteriaceae by use of semiautomated microbiology systems and manual detection procedures. J. Clin. Microbiol. 2007, 45, 1167–1174. [Google Scholar] [CrossRef][Green Version]
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- CDC. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022. Available online: https://www.cdc.gov/drugresistance/covid19.html (accessed on 20 January 2023).
- Larcher, R.; Laffont-Lozes, P.; Roger, C.; Doncesco, R.; Groul-Viaud, C.; Martin, A.; Loubet, P.; Lavigne, J.P.; Pantel, A.; Sotto, A. Last resort beta-lactam antibiotics for treatment of New-Delhi Metallo-Beta-Lactamase producing Enterobacterales and other Difficult-to-Treat Resistance in Gram-negative bacteria: A real-life study. Front. Cell. Infect. Microbiol. 2022, 12, 1048633. [Google Scholar] [CrossRef] [PubMed]
- Høg, B.B.; Bager, F.; Korsgaard, H.B.; Ellis-Iversen, J.; Pedersen, K.; Jensen, L.B.; Hendriksen, R.S.; Bortolaia, V.; Larsen, A.R.; Petersen, A. DANMAP 2017-Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark; DTU: Lyngby, Denmark, 2018. [Google Scholar]
- Schechner, V.; Temkin, E.; Harbarth, S.; Carmeli, Y.; Schwaber, M.J. Epidemiological interpretation of studies examining the effect of antibiotic usage on resistance. Clin. Microbiol. Rev. 2013, 26, 289–307. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.C.; Keenum, I.; Calarco, J.; Liguori, K.; Milligan, E.; Pruden, A.; Harwood, V.J. Towards the standardization of Enterococcus culture methods for waterborne antibiotic resistance monitoring: A critical review of trends across studies. Water Res. X 2022, 17, 100161. [Google Scholar] [CrossRef]
- Keenum, I.; Liguori, K.; Calarco, J.; Davis, B.C.; Milligan, E.; Harwood, V.J.; Pruden, A. A framework for standardized qPCR-targets and protocols for quantifying antibiotic resistance in surface water, recycled water and wastewater. Crit. Rev. Environ. Sci. Technol. 2022, 52, 4395–4419. [Google Scholar] [CrossRef]
- Milligan, E.G.; Calarco, J.; Davis, B.C.; Keenum, I.M.; Liguori, K.; Pruden, A.; Harwood, V.J. A Systematic Review of Culture-Based Methods for Monitoring Antibiotic-Resistant Acinetobacter, Aeromonas, and Pseudomonas as Environmentally Relevant Pathogens in Wastewater and Surface Water. Curr. Environ. Health Rep. 2023, 10, 1–18. [Google Scholar] [CrossRef]
- Sköld, O. Sulfonamide resistance: Mechanisms and trends. Drug Resist. Updates 2000, 3, 155–160. [Google Scholar] [CrossRef]
- Pei, R.; Kim, S.C.; Carlson, K.H.; Pruden, A. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef]
- Nardelli, M.; Scalzo, P.M.; Ramírez, M.S.; Quiroga, M.P.; Cassini, M.H.; Centrón, D. Class 1 integrons in environments with different degrees of urbanization. PLoS ONE 2012, 7, e39223. [Google Scholar] [CrossRef]
- Czekalski, N.; Sigdel, R.; Birtel, J.; Matthews, B.; Bürgmann, H. Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environ. Int. 2015, 81, 45–55. [Google Scholar] [CrossRef]
- Gillings, M.R. Integrons: Past, present, and future. Microbiol. Mol. Biol. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Ku, J. Using the Class 1 Integron-Integrase Gene as a Proxy for Anthropogenic Pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef]
- Hardwick, S.A.; Stokes, H.W.; Findlay, S.; Taylor, M.; Gillings, M.R. Quantification of class 1 integron abundance in natural environments using real-time quantitative PCR. FEMS Microbiol. Lett. 2008, 278, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P. Antibiotic-resistance genes in waste water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Tarek, M.H.; Garner, E. A proposed framework for the identification of indicator genes for monitoring antibiotic resistance in wastewater: Insights from metagenomic sequencing. Sci. Total Environ. 2023, 854, 158698. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.C.; Riquelme, M.V.; Ramirez-Toro, G.; Bandaragoda, C.; Garner, E.; Rhoads, W.J.; Vikesland, P.; Pruden, A. Demonstrating an integrated antibiotic resistance gene surveillance approach in Puerto Rican watersheds Post-Hurricane Maria. Environ. Sci. Technol. 2020, 54, 15108–15119. [Google Scholar] [CrossRef]
- CARD; Andrew, G.; McArthur, G.D.W. SUL1. The Comprehensive Antibiotic Resistance Database. Available online: https://card.mcmaster.ca/ontology/36549 (accessed on 14 March 2023).
- Makkaew, P.; Kongprajug, A.; Chyerochana, N.; Sresung, M.; Precha, N.; Mongkolsuk, S.; Sirikanchana, K. Persisting antibiotic resistance gene pollution and its association with human sewage sources in tropical marine beach waters. Int. J. Hyg. Environ. Health 2021, 238, 113859. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, W.; Xu, T.; Zheng, B.; Yin, D. Occurrence and distribution of antibiotic resistance genes in the water and sediments of Qingcaosha Reservoir, Shanghai, China. Environ. Sci. Eur. 2019, 31, 1–9. [Google Scholar] [CrossRef]
- Adelowo, O.O.; Helbig, T.; Knecht, C.; Reincke, F.; Mäusezahl, I.; Müller, J.A. High abundances of class 1 integrase and sulfonamide resistance genes, and characterisation of class 1 integron gene cassettes in four urban wetlands in Nigeria. PLoS ONE 2018, 13, e0208269. [Google Scholar] [CrossRef]
- Branger, C.; Zamfir, O.; Geoffroy, S.; Laurans, G.; Arlet, G.; Thien, H.V.; Gouriou, S.; Picard, B.; Denamur, E. Genetic background of Escherichia coli and extended-spectrum β-lactamase type. Emerg. Infect. Dis. 2005, 11, 54. [Google Scholar] [CrossRef]
- Franz, E.; Veenman, C.; van Hoek, A.H.; Husman, A.D.R.; Blaak, H. Pathogenic Escherichia coli producing Extended-Spectrum β-Lactamases isolated from surface water and wastewater. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef]
- US EPA. FAQ: NPDES Water-Quality Based Permit Limits for Recreational. 2014. Available online: https://www3.epa.gov/npdes/pubs/npdes_pathogen_faq.pdf (accessed on 5 April 2023).
- WHO. WHO Guidelines for the Safe Use of Wastewater, Excreta and Greywater (Volume IV: Excreta and Greywater Use in Agriculture); World Health Organization (WHO): Geneva, Switzerland, 2006; ISBN 9241546859. [Google Scholar]
- Bengtsson-Palme, J.; Larsson, D.J. Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environ. Int. 2016, 86, 140–149. [Google Scholar] [CrossRef]
- Manaia, C.M.; Aga, D.S.; Cytryn, E.; Gaze, W.H.; Graham, D.W.; Guo, J.; Leonard, A.F.; Li, L.; Murray, A.K.; Nunes, O.C. The Complex Interplay Between Antibiotic Resistance and Pharmaceutical and Personal Care Products in the Environment. Environ. Toxicol. Chem. 2022; ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Bréchet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater treatment plants release large amounts of extended-spectrum β-lactamase–producing Escherichia coli into the environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.C.; Brown, C.; Gupta, S.; Calarco, J.; Liguori, K.; Milligan, E.; Harwood, V.J.; Pruden, A.; Keenum, I. Recommendations for the use of metagenomics for routine monitoring of antibiotic resistance in wastewater and impacted aquatic environments. Crit. Rev. Environ. Sci. Technol. 2023, 53, 1–26. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef]
- Oh, M.; Pruden, A.; Chen, C.; Heath, L.S.; Xia, K.; Zhang, L. MetaCompare: A computational pipeline for prioritizing environmental resistome risk. FEMS Microbiol. Ecol. 2018, 94, fiy079. [Google Scholar] [CrossRef]
- Larsen, D.A.; Green, H.; Collins, M.B.; Kmush, B.L. Wastewater monitoring, surveillance and epidemiology: A review of terminology for a common understanding. FEMS Microbes 2021, 2, xtab011. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Woolhouse, M.E. Using sewage for surveillance of antimicrobial resistance. Science 2020, 367, 630–632. [Google Scholar] [CrossRef]
- Adhikari, S.; Halden, R.U. Opportunities and limits of wastewater-based epidemiology for tracking global health and attainment of UN sustainable development goals. Environ. Int. 2022, 163, 107217. [Google Scholar] [CrossRef]
- Miłobedzka, A.; Ferreira, C.; Vaz-Moreira, I.; Calderón-Franco, D.; Gorecki, A.; Purkrtova, S.; Bartacek, J.; Dziewit, L.; Singleton, C.M.; Nielsen, P.H. Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the one-health cycle. J. Hazard. Mater. 2022, 424, 127407. [Google Scholar] [CrossRef] [PubMed]
- Prieto Riquelme, M.V.; Garner, E.; Gupta, S.; Metch, J.; Zhu, N.; Blair, M.F.; Arango-Argoty, G.; Maile-Moskowitz, A.; Li, A.D.; Flach, C.F. Demonstrating a Comprehensive Wastewater-Based Surveillance Approach That Differentiates Globally Sourced Resistomes. Environ. Sci. Technol. 2022, 56, 14982–14993. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency (USEPA). Method 1603: E. coli in Water by Membrane Filtration Using Modified mTEC. In Standard Methods (Issue September); EPA: Washington, DC, USA, 2014. [Google Scholar]
- Calarco, J.; Liguori, K.; Keenum, I.; Davis, B.C.; Milligan, E.; Pruden, A.; Harwood, V.J. Effect of Methodological Rigor on Trends in Antibiotic Resistance of E. coli in Built and Natural Aquatic Habitats; University of South Florida: Tampa, FL, USA, 2023; manuscript in preparation. [Google Scholar]
- Rocha, J.; Fernandes, T.; Riquelme, M.V.; Zhu, N.; Pruden, A.; Manaia, C.M. Comparison of culture-and quantitative PCR-based indicators of antibiotic resistance in wastewater, recycled water, and tap water. Int. J. Environ. Res. Public Health 2019, 16, 4217. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The ultimate qPCR experiment: Producing publication quality, reproducible data the first time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Sanders, R.; Mason, D.J.; Foy, C.A.; Huggett, J.F. Considerations for accurate gene expression measurement by reverse transcription quantitative PCR when analysing clinical samples. Anal. Bioanal. Chem. 2014, 406, 6471–6483. [Google Scholar] [CrossRef]
- Grgicak, C.M.; Urban, Z.M.; Cotton, R.W. Investigation of reproducibility and error associated with qPCR methods using Quantifiler® Duo DNA quantification kit. J. Forensic Sci. 2010, 55, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Kokkoris, V.; Vukicevich, E.; Richards, A.; Thomsen, C.; Hart, M.M. Challenges using droplet digital PCR for environmental samples. Appl. Microbiol. 2021, 1, 74–88. [Google Scholar] [CrossRef]
- Chern, E.C.; Brenner, K.P.; Wymer, L.; Haugland, R.A. Comparison of Fecal Indicator Bacteria Densities in Marine Recreational Waters by QPCR. Water Expo. Health 2009, 1, 203–214. [Google Scholar] [CrossRef]
- Li, A.D.; Metch, J.W.; Wang, Y.; Garner, E.; Zhang, A.N.; Riquelme, M.V.; Vikesland, P.J.; Pruden, A.; Zhang, T. Effects of sample preservation and DNA extraction on enumeration of antibiotic resistance genes in wastewater. FEMS Microbiol. Ecol. 2018, 94, 189. [Google Scholar] [CrossRef]
- Pope, M.L.; Bussen, M.; Feige, M.A.; Shadix, L.; Gonder, S.; Rodgers, C.; Chambers, Y.; Pulz, J.; Miller, K.; Connell, K.; et al. Assessment of the effects of holding time and temperature on Escherichia coli densities in surface water samples. Appl. Environ. Microbiol. 2003, 69, 6201–6207. [Google Scholar] [CrossRef]
- Borchardt, M.A.; Boehm, A.B.; Salit, M.; Spencer, S.K.; Wigginton, K.R.; Noble, R.T. The environmental microbiology minimum information (EMMI) guidelines: qPCR and dPCR quality and reporting for environmental microbiology. Environ. Sci. Technol. 2021, 55, 10210–10223. [Google Scholar] [CrossRef] [PubMed]
- Webb, D.; Hamilton, M.A.; Harkin, G.J.; Lawrence, S.; Camper, A.K.; Lewandowski, Z. Assessing technician effects when extracting quantities from microscope images. J. Microbiol. Methods 2003, 53, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, Z.; Zhu, W. Variability: Human nature and its impact on measurement and statistical analysis. J. Sport Health Sci. 2019, 8, 527. [Google Scholar] [PubMed]
- Liguori, K.; Keenum, I.; Davis, B.; Milligan, E.; Heath, L.S.; Pruden, A.; Calarco, J.; Harwood, V.J. Standardizing Methods with QA/QC Standards for Investigating the Occurrence and Removal of Antibiotic Resistant Bacteria/Antibiotic Resistance Genes (ARB/ARGs) in Surface Water, Wastewater, and Recycled Water. Project 5052; The Water Research Foundation: Denver, CO, USA, 2023; p. 239. [Google Scholar]
- Barraud, O.; Baclet, M.-C.; Denis, F.; Ploy, M.-C. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 2010, 65, 1642–1645. [Google Scholar] [CrossRef] [PubMed]

| Method | Target | Rationale | Details | Assay Reference |
|---|---|---|---|---|
| Culture | Total E. coli | Comparison point for cefR E. coli and allows percent resistance calculations | Modified mTEC | EPA Method 1603 [62] |
| cefR E. coli | Can cause difficult-to-treat resistant infections | Modified mTEC with cefotaxime | Modified from EPA Method 1603 [62] | |
| qPCR | intI1 | Indicator of mobile, anthropogenic sources of multi-antibiotic resistance | TaqMan probe assay (5′-3′): F: GCCTTGATGTTACCCGAGAG; R: GATCGGTCGAATGCGTGT; P: (6-FAM) ATTCCTGGCCGTGGTTCTGGGTTTT (BHQ1) | Barraud et al., 2010 [76] |
| sul1 | ARG that encodes resistance to sulfonamides and correlates with anthropogenic inputs | F (5′-3′): CGCACCGGAAACATCGCTGCAC; R (5′-3′): TGAAGTTCCGCCGCAAGGCTCG | Pei et al., 2006 [33] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liguori, K.; Calarco, J.; Maldonado Rivera, G.; Kurowski, A.; Keenum, I.; Davis, B.C.; Harwood, V.J.; Pruden, A. Comparison of Cefotaxime-Resistant Escherichia coli and sul1 and intI1 by qPCR for Monitoring of Antibiotic Resistance of Wastewater, Surface Water, and Recycled Water. Antibiotics 2023, 12, 1252. https://doi.org/10.3390/antibiotics12081252
Liguori K, Calarco J, Maldonado Rivera G, Kurowski A, Keenum I, Davis BC, Harwood VJ, Pruden A. Comparison of Cefotaxime-Resistant Escherichia coli and sul1 and intI1 by qPCR for Monitoring of Antibiotic Resistance of Wastewater, Surface Water, and Recycled Water. Antibiotics. 2023; 12(8):1252. https://doi.org/10.3390/antibiotics12081252
Chicago/Turabian StyleLiguori, Krista, Jeanette Calarco, Gabriel Maldonado Rivera, Anna Kurowski, Ishi Keenum, Benjamin C. Davis, Valerie J. Harwood, and Amy Pruden. 2023. "Comparison of Cefotaxime-Resistant Escherichia coli and sul1 and intI1 by qPCR for Monitoring of Antibiotic Resistance of Wastewater, Surface Water, and Recycled Water" Antibiotics 12, no. 8: 1252. https://doi.org/10.3390/antibiotics12081252
APA StyleLiguori, K., Calarco, J., Maldonado Rivera, G., Kurowski, A., Keenum, I., Davis, B. C., Harwood, V. J., & Pruden, A. (2023). Comparison of Cefotaxime-Resistant Escherichia coli and sul1 and intI1 by qPCR for Monitoring of Antibiotic Resistance of Wastewater, Surface Water, and Recycled Water. Antibiotics, 12(8), 1252. https://doi.org/10.3390/antibiotics12081252






