Abstract
Hospital wastewater is a recognized reservoir for resistant Gram-negative bacteria. This study aimed to screen for carbapenemase-producing Escherichia coli and Klebsiella pneumoniae and their resistance determinants in two hospital effluents of Ouagadougou. Carbapenem-resistant E. coli and K. pneumoniae were selectively isolated from wastewater collected from two public hospitals in Ouagadougou, Burkina Faso. Bacterial species were identified via MALDI-TOF mass spectrometry. Carbapenemase production was studied phenotypically using antibiotic susceptibility testing via the disk diffusion method. The presence of carbapenemases was further characterized by PCR. A total of 14 E. coli (13.59%) and 19 K. pneumoniae (17.92%) carbapenemase-producing isolates were identified with different distributions. They were, respectively, blaNDM (71.43%), blaVIM (42.86%), blaIMP (28.57%), blaKPC (14.29%), blaOXA-48 (14.29%); and blaKPC (68.42%), blaNDM (68.42%), blaIMP (10.53%), blaVIM (10.53%), and blaOXA-48 (5.26%). In addition, eight (57.14%) E. coli and eleven (57.89%) K. pneumoniae isolates exhibited more than one carbapenemase, KPC and NDM being the most prevalent combination. Our results highlight the presence of clinically relevant carbapenemase-producing isolates in hospital effluents, suggesting their presence also in hospitals. Their spread into the environment via hospital effluents calls for intensive antimicrobial resistance (AMR) surveillance.
1. Introduction
Antimicrobial resistance (AMR) is a growing public health problem worldwide due to the presence of multidrug-resistant (MDR) bacterial pathogens in healthcare settings [1] and their prevalence and persistence in all natural and man-made environments [2]. In 2017, the World Health Organization published a list of pathogens of priority attention, among which are carbapenem-resistant Enterobacterales [3]. Carbapenemase-producing Enterobacterales (CPE) remain one of the most pressing threats to healthcare [4], as carbapenems are the last resort antimicrobials in clinical settings. Several studies have reported the presence of carbapenemase-producing bacteria in Africa [5], where low- and middle-income countries face a disproportionate burden due to several factors (lack of epidemiological studies and poor diagnostic) that characterize AMR differently than in different countries [5]. Carbapenemases are widespread in many parts of the world [6], and NDM-, KPC-, and OXA-48-like enzymes have become the major mechanism of carbapenem resistance [7], and the most prevalent carbapenemases in the world [8]. Enterobacterales such as Escherichia coli and Klebsiella pneumoniae are common human pathogens and asymptomatic colonizers of the human gastrointestinal tract and environmental niches [9]. K. pneumoniae and E. coli are responsible for various types of infections in humans, including pneumonia, septicemia, and urinary tract infections, in both community and hospital settings [10].
In Burkina Faso, recent surveillance data from the Ministry of Health revealed that in 2018 and 2019, the most frequently isolated bacteria were highly resistant. In E. coli, resistance levels to penicillin and sulfonamide groups were 90% and over 80%, respectively, in both years. In Klebsiella spp., resistance to quinolones was approximately 50%, whereas resistance to third-generation cephalosporins rose from 50% in 2018 to 60% in 2019 [11,12].
Few studies have reported CPEs in the clinical environment of a private hospital in the country, notably carbapenemase-producing E. coli (18.87%) [13]. Another study (Markkanen et al.) reported carbapenem resistance genes in the hospital’s wastewater [14]. These findings highlight the need to further screen the environment, and the paucity of data available on the presence and characteristics of CPEs in the environment, particularly in hospital effluents in Burkina Faso, hence the interest of our study. The aim of the study was to identify the presence and characteristics of carbapenemase-producing E. coli and K. pneumoniae in Ouagadougou hospital effluents.
2. Results
2.1. Carbapenemase-Producing E. coli and K. pneumoniae Detection
A total of 209 (77.40%) third-generation cephalosporin-resistant isolates (103 E. coli and 106 K. pneumoniae) were identified from the 270 samples collected during the course of this study by Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight (MALDI-TOF. At the Yalgado Ouedraogo University Hospital (CHU-YO), 28 (27.18%) E. coli and 31 (29.24%) K. pneumoniae were isolated from raw wastewater. At the Bogodogo University Hospital (CHU-B), 37 (35.92%) E. coli and 29 (27.36%) K. pneumoniae were isolated from raw wastewater versus 38 (36.90%) E. coli and 46 (43.40%) K. pneumoniae from treated wastewater (Table 1). Among these isolates, 33 (15.79%) were carbapenem-resistant: 14 (13.59%, 95% CI: 7.63–21.75) E. coli and 19 (17.92%, 95% CI: 11.15–26.57) K. pneumoniae (Table 1).

Table 1.
The rate of detection of carbapenemase producing E. coli and K. pneumoniae.
2.2. Antibiotic Resistance Profile
As expected, a high rate of β-lactam resistance was observed in the isolates. The resistance rate of carbapenemase-producing E. coli and K. pneumoniae was higher compared to the resistance rate of non-carbapenemase-producing E. coli and K. pneumoniae (Table 2). This is particularly true for cefoxitin: 85.71% in carbapenemase-producing E. coli versus 12.36% in non-carbapenemase-producing E. coli; 78.95% in carbapenemase-producing K. pneumoniae versus 16.09% in non-carbapenemase-producing K. pneumoniae. This resistance was highly plausible, as our selection method targeted third-generation cephalosporine-resistant isolates. However, a high rate of resistance to non-β-lactam antimicrobials was noted: 100% and 85.71% to ciprofloxacin in carbapenemases-producing K. pneumoniae and E. coli, respectively. For non-carbapenemase producers, ciprofloxacin resistance in K. pneumoniae and E. coli was 63.2% and 51.7%, respectively. In addition, a very low rate of resistance was observed in amikacin. Interestingly, all carbapenemase-producing E. coli and K. pneumoniae isolates were resistant to ertapenem, but not to imipenem.

Table 2.
Resistance rate of non-carbapenemases-producing and carbapenemase-producing isolates.
Carbapenemase-producing E. coli and K. pneumoniae of the effluents from CHU-YO and CHU-B showed 100% resistance to antibiotics such as ampicillin, piperacillin, cefepime, aztreonam, amoxicillin-clavulanic acid, ceftazidime, and ceftriaxone (Table 3).

Table 3.
Carbapenemase-producing isolates according to the origin and nature of the effluents.
As shown in Table 4, the proportion of MDR isolates was very similar between raw and treated effluents from CHU-B, except that three isolates from the treated wastewater were resistant to 14 antibiotics compared to only one isolate from the raw wastewater.

Table 4.
Antibiotic resistance phenotype and carbapenemase genes harbored by E. coli and K. pneumoniae.
The Multiple Antibiotic Resistance (MAR) indices of the effluents from CHU-YO (0.77) and the raw and treated wastewater from CHU-B (0.84 and 0.83, respectively) are above the threshold of 0.2 (Table 5).

Table 5.
Multiple Antibiotic Resistance (MAR) index of wastewater samples.
2.3. Carbapenemase Genes in E. coli and K. pneumoniae
The carbapenemase genes recovered in E. coli were 10 blaNDM (71.43%), 6 blaVIM (42.86%), 4 blaIMP (28.57%), 2 blaKPC (14.29%), and 2 blaOXA-48 (14.29%). In K. pneumoniae, these genes were 13 blaKPC (68.42%), 13 blaNDM (68.42%), 2 blaIMP (10.53%), 2 blaVIM (10.53%), and 1 blaOXA-48 (5.26%). The different genes detected in E. coli and K. pneumoniae are presented in supplementary files as follow: blaKPC, blaVIM and blaKPC (Supplementary Figure S1), blaNDM (Supplementary Figure S2) and blaOXA-48 (Supplementary Figure S3).
A high proportion of NDM-producing isolates was observed in each type of effluent. This proportion was 66.67% (95% CI: 29.93–92.51) for raw effluents from CHU-YO, 100% (95% CI: 15.81–100.00) for raw effluents from CHU-B, and 66.67% (95% CI: 9.43–99.16) for treated effluents from CHU-B (Figure 1). IMP- and VIM-producing isolates were detected at 44.44% in CHU-YO effluents, but were not detected in CHU-B effluents untreated for E. coli isolates. The proportion of KPC-producing isolates was very high (100%) in CHU-YO wastewater, followed by NDM-producing isolates, whose proportion was higher in CHU-B effluents (100%) (Figure 1).
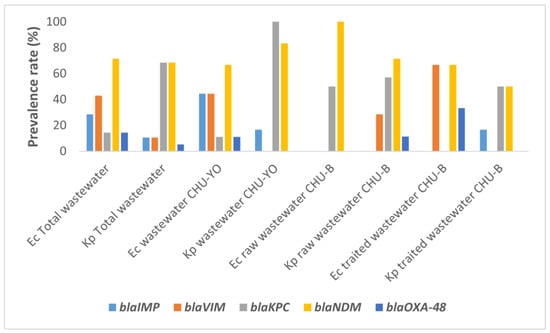
Figure 1.
Prevalence of blaIMP, blaVIM, blaKPC, blaNDM and blaOXA-48 in carbapenemase-producing E. coli (Ec) and K. pneumoniae (Kp).
Interestingly, eight (57.14%) E. coli and eleven (57.89%) K. pneumoniae isolates (Figure 2) co-produced carbapenemases. Two or more had a high proportion of KPC + NDM (Table 4). However, whether the isolates produced only one or several enzymes at a time, they were resistant to more than nine antibiotics. Moreover, all blaNDM-producing E. coli and K. pneumoniae were resistant to aztreonam (Table 4).
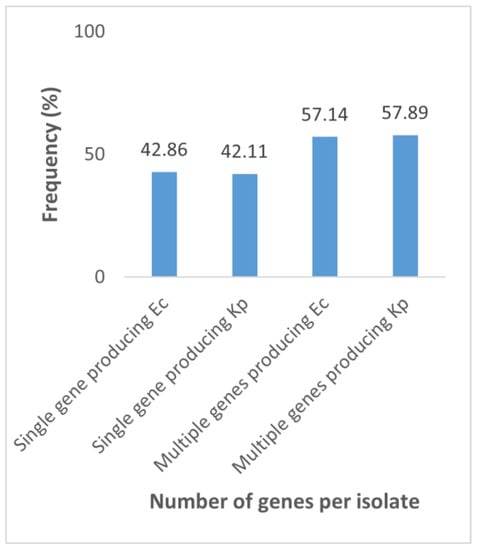
Figure 2.
Proportion of the number of genes produced by carbapenemase-producing E. coli (Ec) and K. pneumoniae (Kp). Single gene producing Ec: E. coli producing a single gene; Single gene producing Kp: K. pneumoniae producing a single gene; Multiple genes producing Ec: E. coli producing multiple genes; Multiple genes producing Kp: K. pneumoniae producing multiple genes.
3. Discussion
We present here a comprehensive study on the occurrence and characteristisation of carbapenemase-producing E. coli and K. pneumoniae isolates in wastewater samples over a twelve-month period from two hospitals of Ouagadougou, the capital of Burkina Faso. The study revealed the presence of third-generation cephalosporin-resistant E. coli (103/209) and K. pneumoniae (106/209) in hospital wastewater samples. The presence of E. coli and K. pneumoniae could be explained, as these bacterial species are part of the microbiota of the digestive tract of healthy and sick people and are widely distributed in the environment. Hospital effluents are considered a reservoir for multi-drug-resistant pathogens [15,16,17], playing a key role in the spread of antimicrobial-resistant bacteria in the environment [18,19]. Indeed, the present study reports for the first time the presence of carbapenemase-producing E. coli and K. pneumoniae in hospital wastewater in Ouagadougou with respective prevalence of 13.59% and 17.92%. Similar results were reported in several works including those of Cahill and collaborators who, in 2019, collected 142 isolates of Enterobacterales resistant to ertapenem and/or meropenem in hospital effluents [20] and those reported by Zagui and collaborators who, in 2020, isolated strains of K. pneumoniae with resistance patterns to broad-spectrum cephalosporins and carbapenems [21]. This proportion of carbapenemase-producing E. coli and K. pneumoniae in hospital effluents may be explained by a higher frequency of carriage in hospitalized patients than in healthy community carriers [22]. It was shown that the constant load of E. coli isolates and the enrichment of K. pneumoniae during wastewater treatment may be due to either a shorter generation time of these bacteria or to horizontal transfer of resistance genes [23]. These findings are corroborated by a previous study in Burkina Faso, which reported 18.87% of carbapenemase-producing E. coli in clinical settings [13]. The presence of carbapenemase-producing bacteria in the CHU-B raw and treated wastewater highlights the inefficiency of the treatment, which normally should reduce the load of antibiotic-resistant pathogenic bacteria. This situation is worrisome with Enterobacterales, especially E. coli and K. pneumoniae, which are now emerging as difficult-to-treat bacteria [24]. Multidrug resistance of carbapenemase-producing E. coli and K. pneumoniae was confirmed, and even pan resistance of the latter to almost all antibiotics tested. The resistance patterns of E. coli and K. pneumoniae were relatively similar between the two hospitals studied. The MAR indices of the raw wastewater from CHU-YO (0.77) and the raw and treated wastewater from CHU-B (0.84 and 0.83, respectively) are higher than the threshold of 0.2, indicating a high risk of environmental contamination and, consequently, public health concerns. The presence of MDR bacteria in hospital effluents could be explained by the release of MDR clinical isolates and the transfer of resistance genes from MDR clinical isolates found in hospital effluents to other environmental bacteria [25]. Moreover, some of the consumed antimicrobials may be excreted unchanged and end up in the environment, contributing to the selection pressure that may lead to the emergence of novel antimicrobial-resistant genes in the environment [26]. Indeed, large amounts of antibiotics are discharged in sewer systems due to incomplete metabolism in humans and sometimes to the disposal of unused antibiotics [27,28]. The presence of disinfectants and antiseptics in sewage can also promote the emergence of antibiotic-resistant bacteria in hospital effluents [29].
The predominance of blaNDM recovered in our study in E. coli (71.43%) and K. pneumoniae (68.42%) is consistent with existing reports [20,30,31,32]. NDM production is a mechanism of carbapenem resistance and is mediated by a ubiquitous plasmid not associated with dominant clonal strains [33]. E. coli- and K. pneumoniae-producing blaOXA-48 were also found in hospital wastewater from Algeria [34,35], Tunisia [36], Spain [37], Germany [38], India [39,40], and Brazil [41,42]. This could support the idea that E. coli- and K. pneumoniae-producing carbapenemases of blaOXA-48 types (weakly represented in our study) originate from the hospital environment. The presence of carbapenemases producing E. coli and K. pneumoniae in aqueous environments is a phantom threat [43,44], especially when it is reported from hospital-treated wastewater, as in the case of this study. Recently, inadequate wastewater management by bulk drug manufactures facilities in India led to the contamination of water resources with antimicrobial agents, associated with the selection and dissemination of blaOXA-48 producers [40]. This means that even after wastewater treatment, significant numbers of carbapenemase-producing bacteria could still survive and be subsequently released into the aquatic environment.
Other genes such as blaVIM and blaIMP were also recovered from Irish hospitals [20,45]. blaVIM and blaIMP are in higher proportions in E. coli (42.86% and 28.57%, respectively) than in K. pneumoniae (10.53% and 10.53%, respectively). The high proportion of VIM and, to a lesser extent, IMP in E. coli compared with K. pneumoniae could be explained by a more rapid diffusion of these genes within E. coli. blaKPC was also detected in isolates of hospital wastewater in Tunisia [36], Spain [37], Germany [38], and India [39]. However, the proportion of blaKPC was higher in K. pneumoniae (68.42%) than in E. coli (14.29%). KPC is the most common transmissible class A carbapenemase circulating in Enterobacterales, especially in K. pneumoniae worldwide, mainly due to the clonal expansion of K. pneumoniae strains [46]. These results are somewhat surprising since, according to the first study of hospital wastewater in Burkina Faso using a shotgun metagenomic approach [14], no blaKPC was found.
Analysis of the number of genes expressed by bacterial isolates showed that the majority of our isolates expressed more than one gene, suggesting that isolates harbored multiple plasmid-mediated drug resistant determinants.
Comparison of the antibiotic resistance phenotype with the genotypic profile showed that E. coli and K. pneumoniae producing only one or several carbapenemases were resistant to more than nine antibiotics, even in wastewater treated before discharge. The emergence and spread of carbapenem resistance in Gram-negative bacilli such as K. pneumoniae and E. coli via the production of carbapenemases is a global phenomenon. It threatens patient care and could lead to therapeutic failure. This high rate of multi-resistance is of risk as the resistance could spread wider in the environment and attain for instance the food chain. Indeed, a recent study highlighted the presence of resistance genes NDM and VIM among the clinical strains of E. coli in a hospital in Ouagadougou [13]. Furthermore, these results constitute scientific evidence that could help public authorities, health ministry officials, and the Ouagadougou municipal hygiene department to take appropriate measures to protect and preserve the health of the population. In fact, in the vicinity of the study sites, particularly the CHU-YO, salads and other foodstuffs are produced and consumed by the population.
The potential global spread of critical-priority antimicrobial-resistant Enterobacterale is a public health problem. Of course, it was shown that the release of raw and improperly treated wastewater onto water courses has both short- and long-term effects on the environment and human health [47]. Hence, there should be proper enforcement of water and environmental laws to protect the health of inhabitants of both rural and urban communities.
4. Materials and Methods
4.1. Study Design, Sites and Sampling
This was a cross-sectional study from July 2021 to April 2022 in the CHU-YO and the CHU-B, which are the two most frequented university hospitals in Ouagadougou. Raw wastewater discharged by the CHU-YO flows directly into the city’s sewer system. This is because the hospital has no treatment system. On the other hand, raw wastewater and treated wastewater were collected from CHU-B, which has a functional treatment system. A total of 270 wastewater samples were collected at daily intervals via randomized technique in sterile 250 mL containers: eighty raw wastewater samples from the CHU-YO, ninety-five raw, and ninety-five treated wastewater samples from the CHU-B. The number of samples from CHU-B exceeds that of CHU-YO due to the categorization of its effluents, to lend assiduous credibility to the results that would emanate from the analysis of said effluents. All the samples were transported at <4 °C to the Laboratory of Molecular Biology, Epidemiology and Surveillance of Foodborne Bacteria and Viruses (LaBESTA), within two hours of collection for processing.
4.2. Microbial Culturing and Identification
The stock solution was obtained by diluting 10 mL of hospital wastewater in 90 mL of sterile peptone water. A series of decimal dilutions was performed from the stock solution to 1/1000. The following agar plates were inoculated from 1/1000 dilution. First, Chromocult® Coliform Agar (CCA) (Laboratorios Conda S.A), and then Plate Count Agar (PCA) + Tri-phenyl Tetrazolium Chloride (TTC) agar supplemented with ceftriaxone to maximize isolation of extended-spectrum beta-lactamase-producing strains (after CCA exhaustion). Plates were incubated at 37 °C for 24 h. Presumptive colonies were then transferred to Eosin Methylene Blue (EMB) agar (Laboratorios Conda S.A) at 37 °C for 24 h for purification and conservation. Muller Hinton (MH) agar (Laboratorios Conda S.A r) was used to subculture the isolates at 37 ± 0.5 °C for 24 h and confirm the identification via MALDI-TOF mass spectrometry using the Microflex MALDI-TOF MS® (Bruker Daltonics, Bremen, Germany) equipment. As the identification of E. coli with MALDI-TOF was not accurate (confusion between E. coli and Shigella), all E. coli identified by MALDI-TOF were confirmed on Bromocresol Purple (BCP) Agar plate and subsequent API 20E gallery (bioMerieux, Marcy-l’Etoile France) for lactose negative isolates.
4.3. Antibiotic Susceptibility Testing
Antimicrobial susceptibility testing to 16 antibiotics was determined via the agar disk diffusion method on MH agar plates according to the Antimicrobial Susceptibility Testing Committee of the French Microbiology Society/European Committee on Antimicrobial Susceptibility Testing (CA-SFM/EUCAST), 2021 guidelines [48]. The following antibiotics (MAST Amiens France) were used: Ampicillin (AMP) 10 μg, Piperacillin (PIP) 30 μg, Piperacillin-Tazobatam (PTZ) 36 μg, Amoxicillin-Clavulanic acid (AUG) 30 μg, Aztreonam (ATM) 30 μg, Imipenem (IMP) 10 μg, Ertapenem (ETP) 10 μg, Ceftriaxone (CRO) 30 μg, Cefoxitin (FOX) 30 μg, Cefepime (FEP) 30 μg, Ceftazidime (CAZ) 30 μg, Gentamicin (GM) 10 μg, Amikacin (AK) 30 μg, Nalidixic acid (NA) 30 μg, Ciprofloxacin (CIP) 5 μg, and Trimethoprim-Sulfamethoxazole (SXT) 25 μg). Antibiotic susceptibility was interpreted according to the CA-SFM/EUCAST, 2021 guidelines recommendations [48]. For isolates with reduced susceptibility to carbapenems (ertapenem), DNA was extracted for carbapenemase genes. For quality control, we used E. coli ATCC 25922 and K. pneumoniae ATCC 700603, which produces an ESBL (SHV-18).
The Multiple Antibiotic Resistance (MAR) Index per sampling site was calculated using the Krumperman method [49] to compare the contribution of each human activity to the risk to the environment and public health. Fifteen of the antibiotics tested were included in the index calculation, with CRO excluded because the medium used to select isolates was CRO-supplemented.
Sites with an index of 0.2 or less were considered to present a low risk of contamination because antibiotics are rarely or never used, whereas sites with an index greater than 0.2 were considered to present a high risk of contamination because of constant and significant exposure to antibiotics.
4.4. Detection of Carbapenemase Genes
DNA was extracted from E. coli or K. pneumoniae isolates using organic extraction (phenol–chloroform method) as previously described [50]. The quantity, purity, and integrity of the extracted DNA were verified via 1% agarose gel electrophoresis and nanodrop measurement.
Multiplex PCR was performed for the detection of carbapenemase genes (blaIMP, blaVIM, blaKPC, blaNDM, and blaOXA-48) using primers previously described [51,52]. Five μL of sample DNA [C < 250 ng, https://www.promega.com.au/-/media/files/resources/protocols/product-information-sheets/g/gotaq-hot-start-green-master-mix-protocol.pdf, accessed on 28 August 2023] was subjected to each multiplex PCR in a 25 μL reaction mixture containing PCR buffer (5X, primers (10 µM), and PCR water using the GoTaq® Master mixes (Promega, Charbonnières-les-Bains, France). Amplification was performed as follows: initial denaturation at 95 °C for 3 min; 34 cycles of 95 °C for 40 s, 55 °C for 40 s, and 72 °C for 1 min; and a final elongation step at 72 °C for 5 min. For amplification of blaOXA-48 genes, the annealing temperature was optimal at 60 °C. Amplicons were visualized on a 1% agarose gel containing ethidium bromide. For quality control, we used clinical strains obtained from the Pelegrin Hospital of Bordeaux, for which the genes were sequenced [50].
5. Conclusions
This study constitutes the first report of enterobacterial carbapenemase producers in hospital wastewater in Burkina Faso. One limitation of our study is that we did not have the opportunity to confirm the identity of carbapenemase genes via the DNA sequencing technique. In the future, it would be interesting to extract the potential plasmids carrying these genes and pass them into reference strains of E. coli by transformation (TOP10 for example) or by conjugation (E. coli K12). The high level of resistance in these strains, particularly to carbapenems, shows that at least one of the two genes is expressed. Indeed, it is well known that the rapid and global dissemination of critical-priority antimicrobial-resistant Enterobacterales is a public health problem that demands mitigation strategies and strengthening via epidemiological surveillance investigations. However, the results of the study highlight the danger to public health posed by hospital effluents to the population of Ouagadougou and show that measures need to be taken to protect public health. A significant occurrence of carbapenemase E. coli and K. pneumoniae was detected in this study, suggesting that hospital effluents may act as a potential reservoir of MDR bacterial pathogens and play a key role in their dissemination in the environment that can negatively impact human health. Indeed, our results show very high levels of carbapenemase genes including blaNDM, blaVIM, blaIMP, blaKPC, and blaOXA-48 in both E. coli and K. pneumoniae strains. This spread of carbapenemase-producing Enterobacterales in hospital wastewater warrants the need for intensive surveillance of AMR and the implementation of an efficient infection control program in Burkina Faso for the management of such infections.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12101494/s1, Figure S1: blaKPC, blaVIM and blaKPC genes detected in E. coli and K. pneumoniae; Figure S2: blaNDM gene detected in E. coli and K. pneumoniae; Figure S3: blaOXA-48 gene detected in E. coli and K. pneumoniae.
Author Contributions
A.B.K. is responsible for the initiation of the study and the analysis of the data. Examinations. Laboratory examinations were performed by A.B.K. under the direction of R.D., L.M., V.D., N.B., A.B.K., R.D., L.B., A.H.M., H.C., F.M., V.D. and N.B. participated in data analysis and preparation of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
A.B.K. is a doctoral student with a SCAC grant from the French government. The Cooperation and Cultural Action Department of the French Embassy in Burkina Faso financed the study under the following funding number: 130446Z.
Institutional Review Board Statement
Authorization to conduct the study was obtained from the hospital authorities of the two hospitals (CHU-YO and CHU-B) in the city of Ouagadougou, Burkina Faso. The Ethical Approval code is 2022-05-097.
Informed Consent Statement
The study did not involve humans.
Data Availability Statement
No new data were created.
Acknowledgments
We thank the hospital authorities for their agreement and the staff of the hygienic department of both the CHU-YO and CHU-B for their support in obtaining the samples.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- World Health Organization Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 5 May 2023).
- Beattie, R.E.; Bakke, E.; Konopek, N.; Thill, R.; Munson, E.; Hristova, K.R. Antimicrobial Resistance Traits of Escherichia coli Isolated from Dairy Manure and Freshwater Ecosystems Are Similar to One Another but Differ from Associated Clinical Isolates. Microorganisms 2020, 8, 747. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR) wholist of Critically Important Antimicrobials (CIA). In Report of the 7th Meeting; WHO: Geneva, Switzerland, 2018; ISBN 978-92-4-151552-8. [Google Scholar]
- Dembélé, R.; Soulama, I.; Kaboré, W.A.D.; Konaté, A.; Kagambèga, A.; Coulibaly, D.N.; Traoré, O.; Seck, A.; Traoré, A.S.; Guessennd, N.; et al. Molecular characterization of carbapenemase-producing Enterobacterales in children with diarrhea in rural Burkina Faso. J. Drug Deliv. Therapeutics 2021, 1, 84–92. [Google Scholar] [CrossRef]
- Mariappan, S.; Sekar, U.; Kamalanathan, A. Carbapenemase-producing Enterobacteriaceae: Risk factors for infection and impact of resistance on outcomes. Int. J. Appl. Basic Med. Res. 2018, 7, 32–39. [Google Scholar] [CrossRef]
- Halat, D.H.; Moubareck, C.A. Thecurrent burden of carbapenemases: Review of significant properties and dissemination among gram-negative bacteria. Antibiotica 2020, 9, 186. [Google Scholar]
- Chia, P.Y.; Sengupta, S.; Kukreja, A.; Ponnampalavanar, S.S.; Ng, O.T.; Marimuthu, K. The role of hospital environment in transmissions of multidrug-resistant Gram-negative organisms. Antimicrob. Resist. Infect. Control 2020, 9, 29. [Google Scholar] [CrossRef]
- Suay-García, B.; Pérez-Gracia, M.T. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics 2019, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Vegesana, K.; German-Mesner, I.; Ainsworth, J.; Pannone, A.; Crook, D.W.; Sifri, C.D.; Sheppard, A.; Stoesser, N.; Peto, T.; et al. Risk factors for Klebsiella pneumoniae carbapenemase (KPC) gene acquisition and clinical outcomes across multiple bacterial species. J. Hosp. Infect. 2020, 104, 456–468. [Google Scholar] [CrossRef]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae–clinical and molecular perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef]
- Secrétariat général, Direction des laboratoires de biologie médicale, Direction générale de l’accès aux produits de santé. Rapport synthèse de la surveillance de la résistance aux antimicrobiens au laboratoire. In Rapport Année 2018; Ministère de la Santé de Burkina Faso: Ouagadougou, Burkina Faso, 2018; Available online: https://drive.google.com/file/d/1qwqwikowd7frnj5pegtbzue-7_6fz84i/view (accessed on 5 November 2021).
- Secrétariat général, Direction des laboratoires de biologie médicale, Direction générale de l’accès aux produits de santé. Rapport synthèse de la surveillance de la résistance aux antimicrobiens au laboratoire. In Rapport Année 2019; Ministère de la santé de Burkina Faso: Ouagadougou, Burkina Faso, 2019; Available online: https://drive.google.com/file/d/1UT6u9kSDGs1W3cl_td1imlazaq5yuva_/view (accessed on 5 November 2021).
- Kaboré, B.; Ouédraogo, H.S.; Zongo, O.; Ouédraogo, G.A.; Tapsoba, F.; Bougma, S.; Zongo, K.J.; Zeba, B.; Traoré, Y.; Sanou, I.; et al. Emergence of New Delhi Metallo-β-Lactamase (NDM) Genes Detected from Clinical Strains of Escherichia coli Isolated in Ouagadougou, Burkina Faso. Int. J. Microbiol. 2023, 2023, 4813225. [Google Scholar] [CrossRef]
- Markkanen, M.A.; Haukka, K.; Pärnänen, K.M.M.; Dougnon, V.T.; Bonkoungou, I.J.O.; Garba, Z.; Tinto, H.; Sarekoski, A.; Karkman, A.; Kantele, A.; et al. Metagenomic Analysis of the Abundance and Composition of Antibiotic Resistance Genes in Hospital Wastewater in Benin, Burkina Faso, and Finland. mSphere 2023, 8, e0053822. [Google Scholar] [CrossRef]
- Kanamori, H.; Weber, D.J.; Rutala, W.A. Healthcare Outbreaks Associated with a Water Reservoir and Infection Prevention Strategies. Clin. Infect. Dis. 2016, 62, 1423–1435. [Google Scholar] [CrossRef]
- Kizny Gordon, A.E.; Mathers, A.J.; Cheong, E.Y.; Gottlieb, T.; Kotay, S.; Walker, A.S.; Peto, T.E.; Crook, D.W.; Stoesser, N. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections-A systematic review of the literature. Clin. Infect. Diseases 2017, 64, 1435–1444. [Google Scholar] [CrossRef]
- Park, S.C.; Parikh, H.; Vegesana, K.; Stoesser, N.; Barry, K.E.; Kotay, S.M.; Dudley, S.; Peto, T.E.; Crook, D.W.; Walker, A.S.; et al. Risk factors associated with carbapenemase-producing Enterobacterales (CPE) positivity in the hospital wastewater environment. Appl. Environ. Microbiol. 2020, 86, e01715-20. [Google Scholar] [CrossRef]
- Obasi, A.I.; Ugoji, E.O.; Nwachukwu, S.U. Incidence and molecular characterization of multidrug resistance in Gram-negative bacteria of clinical importance from pharmaceutical wastewaters in South-western Nigeria. Environ. DNA 2019, 1, 268–280. [Google Scholar] [CrossRef]
- Galarde-López, M.; Velazquez-Meza, M.E.; Bobadilla-del-Valle, M.; Cornejo-Juárez, P.; Carrillo-Quiroz, B.A.; Ponce-de-León, A.; Sassoé-González, A.; Saturno-Hernández, P.; Alpuche-Aranda, C.M. Antimicrobial Resistance Patterns and Clonal Distribution of E. coli, Enterobacter spp. and Acinetobacter spp. Strains Isolated from Two Hospital Wastewater Plants. Antibiotics 2022, 11, 601. [Google Scholar] [CrossRef]
- Cahill, N.; O’Connor, L.; Mahon, B.; Varley, A.; McGrath, E.; Ryan, P.; Cormican, M.; Brehony, C.; Jolley, K.A.; Maiden, M.C. Hospital effluent: A reservoir for carbapenemase-producing Enterobacterales? Sci. Total Environ. 2019, 672, 618–624. [Google Scholar] [CrossRef]
- Zagui, G.S.; De Andrade, L.N.; Moreira, N.C.; Silva, T.V.; Machado, G.P.; da Costa Darini, A.L.; Segura-Muñoz, S.I. Gram-negative bacteria carrying β-lactamase encoding genes in hospital and urban wastewater in Brazil. Environ. Monit. Assess. 2020, 192, 376. [Google Scholar] [CrossRef] [PubMed]
- Aldali, H.J.; Khan, A.; Alshehri, A.A.; Aldali, J.A.; Meo, S.A.; Hindi, A.; Elsokkary, E.M. Hospital-Acquired Infections Caused by Carbapenem-Resistant Enterobacteriaceae: An Observational Study. Microorganisms 2023, 11, 1595. [Google Scholar] [CrossRef] [PubMed]
- Martak, D. Epidémiologie des Bacilles à Gram Négatif dans la Communauté, L’environnement et la Nourriture. Microbiologie et Parasitologie. Ph.D. Thesis, Université Bourgogne Franche-Comité, Besançon, France, 2021; 234p. [Google Scholar]
- Aggarwal, A.; Bhalla, M.; Fatima, K.H. Detection of New Delhi metallo-beta-lactamase enzyme gene blaNDM-1 associated with the Int-1 gene in Gram-negative bacteria collected from the effluent treatment plant of a tuberculosis care hospital in Delhi, India. Access Microbiol. 2020, 2, acmi000125. [Google Scholar] [CrossRef]
- Petit, F. Antibiotic resistance in aquatic environments: A microbial ecology and public health issue. Environ. Risks Health 2018, 17, 40–46. [Google Scholar]
- Nwafia, I.N.; Ike, A.C.; Orabueze, I.N.; Nwafia, W.C. Carbapenemase producing Enterobacteriaceae: Environmental reservoirs as primary targets for control and prevention strategies. Niger. Postgrad. Med. J. 2022, 29, 183–191. [Google Scholar]
- Nagulapally, S.R.; Ahmad, A.; Henry, A.; Marchin, G.L.; Zurek, L.; Bhandari, A. Occurrence of ciprofloxacin-, trimethoprim-sulfamethoxazole-, and vancomycin-resistant bacteria in a municipal wastewater treatment plant. Water Environ. Res. 2009, 81, 82–90. [Google Scholar] [CrossRef]
- Pereira, A.L.; de Oliveira, P.M.; Faria-Junior, C.; Alves, E.G.; de Castro e Caldo Lima, G.R.; da Costa Lamounier, T.A.; Haddad, R.; de Araujo, W.N. Environmental spreading of clinically relevant carbapenem-resistant gram-negative bacilli: The occurrence of blaKPC-or-NDM strains relates to local hospital activities. BMC Microbiol. 2022, 22, 6. [Google Scholar] [CrossRef] [PubMed]
- Carenco, P. Antibiorésistance et biocides. Bull. Clin. Arlin. 2017, 7, 1–9. [Google Scholar]
- Sakkas, H.; Bozidis, P.; Ilia, A.; Mpekoulis, G.; Papadopoulou, C. Antimicrobial Resistance in Bacterial Pathogens and Detection of Carbapenemases in Klebsiella pneumoniae Isolates from Hospital Wastewater. Antibiotics 2019, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Parvez, S.; Khan, A.U. Hospital sewage water: A reservoir for variants of New Delhi metallo-β-lactamase (NDM)- and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2018, 51, 82–88. [Google Scholar] [CrossRef]
- Müller, H.; Sib, E.; Gajdiss, M.; Klanke, U.; Lenz-Plet, F.; Barabasch, V.; Albert, C.; Schallenberg, A.; Timm, C.; Zacharias, N.; et al. Dissemination of multi-resistant Gram-negative bacteria into German wastewater and surface waters. FEMS Microbiol. Ecol. 2018, 94, fiy057. [Google Scholar] [CrossRef] [PubMed]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef]
- Tafoukt, R.; Leangapichart, T.; Hadjadj, L.; Bakour, S.; Diene, S.M.; Rolain, J.M.; Touati, A. Characterisation of blaoxa-538, a new variant of blaoxa-48, in Shewanella xiamenensis isolated from river water in Algeria. J. Glob. Antimicrob. Resist. 2018, 13, 70–73. [Google Scholar] [CrossRef]
- Yousfi, K.; Touati, A.; Lefebvre, B.; Philippe, G.; Brahim, S.; Gharout-Sait, A.; Harel, J.; Bekal, S. Characterization of multidrug-resistant Gram-negative bacilli isolated from hospitals effluents: First report of a blaoxa-48-like in Klebsiella oxytoca, Algeria. Braz. J. Microbiol. 2019, 50, 175–183. [Google Scholar] [CrossRef]
- Nasri, E.; Subirats, J.; Sànchez-Melsió, A.; Mansour, H.B.; Borrego, C.M.; Balcázar, J.L. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ. Pollut. 2017, 229, 371–374. [Google Scholar] [CrossRef]
- Subirats, J.; Royo, E.; Balcázar, J.L.; Borrego, C.M. Real-time PCR assays for the detection and quantification of carbapenemase genes (bla KPC, bla NDM, and bla OXA-48) in environmental samples. Environ. Sci. Pollut. Res. Int. 2017, 24, 6710–6714. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.M.; Zacharias, N.; Timm, C.; Wasser, F.; Sib, E.; Skutlarek, D.; Parcina, M.; Schmithausen, R.M.; Schwartz, T.; Hembach, N.; et al. Association between antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in anthropogenic wastewater—An evaluation of clinical influences. Chemosphere 2020, 241, 125032. [Google Scholar] [CrossRef] [PubMed]
- Marathe, N.P.; Berglund, F.; Razavi, M.; Pal, C.; Dröge, J.; Samant, S.; Kristiansson, E.; Larsson, D.G.J. Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 2019, 7, 97. [Google Scholar] [CrossRef]
- Lübbert, C.; Baars, C.; Dayakar, A.; Lippmann, N.; Rodloff, A.C.; Kinzig, M.; Sörgel, F. Environmental pollution with antimicrobial agents from bulk drug manufacturing industries in Hyderabad, South India, is associated with dissemination of extended-spectrum beta-lactamase and carbapenemase-producing pathogens. Infection 2017, 45, 479–491. [Google Scholar] [CrossRef]
- Sanchez, D.G.; de Melo, F.M.; Savazzi, E.A.; Stehling, E.G. Detection of different β-lactamases encoding genes, including blaNDM, and plasmid-mediated quinolone resistance genes in different water sources from Brazil. Environ. Monit. Assess. 2018, 190, 407. [Google Scholar] [CrossRef] [PubMed]
- Bartley, P.S.; Domitrovic, T.N.; Moretto, V.T.; Santos, C.S.; Ponce-Terashima, R.; Reis, M.G.; Barbosa, L.M.; Blanton, R.E.; Bonomo, R.A.; Perez, F. Antibiotic Resistance in Enterobacteriaceae from Surface Waters in Urban Brazil Highlights the Risks of Poor Sanitation. Am. J. Trop. Med. Hyg. 2019, 100, 1369–1377. [Google Scholar] [CrossRef]
- Tanner, W.D.; VanDerslice, J.A.; Goel, R.K.; Leecaster, M.K.; Fisher, M.A.; Olstadt, J.; Gurley, C.M.; Morris, A.G.; Seely, K.A.; Chapman, L.; et al. Multi-state study of Enterobacteriaceae harboring extended-spectrum beta-lactamase and carbapenemase genes in U.S. drinking water. Sci. Rep. 2019, 9, 3938. [Google Scholar] [CrossRef]
- Schages, L.; Wichern, F.; Kalscheuer, R.; Bockmühl, D. Winter is coming—Impact of temperature on the variation of beta-lactamase and mcr genes in a wastewater treatment plant. Sci. Total Environ. 2020, 712, 136499. [Google Scholar] [CrossRef]
- Hoelle, J.; Johnson, J.R.; Johnsto, B.D.; Kinkle, B.; Boczek, L.; Ryu, H.; Hayes, S. Survey of US wastewater for carbapenem-resistant Enterobacteriaceae. J. Water Health 2019, 17, 219–226. [Google Scholar] [CrossRef]
- Awoke, T.; Teka, B.; Aseffa, A.; Sebre, S.; Seman, A.; Yeshitela, B.; Abebe, T.; Mihret, A. Detection of blaKPC and blaNDM carbapenemase genes among Klebsiella pneumoniae isolates in Addis Ababa, Ethiopia: Dominance of blaNDM. PLoS ONE 2022, 17, e0267657. [Google Scholar] [CrossRef]
- Edokpayi, J.N.; Odiyo, J.O.; Durowoju, O.S. Impact of Wastewater on Surface Water Quality in Developing Countries: A Case Study of South Africa. In Water Quality; Tutu, H., Ed.; Intech Open Science: London, UK, 2017; pp. 401–416. ISBN 978-953-51-5466-2. [Google Scholar] [CrossRef]
- Aubin, G.; Caron, F.; Cattoir, V.; Dubreuil, L.; Goutelle, S.; Jeannot, K.; Lepeule, R.; Lina, G.; Marchandin, H.; Merens, A.; et al. Comite de l’antibiogramme de la Societe Francaise de Microbiologie. Soc. Fr. Microbiol. 2021, 1, 188. [Google Scholar]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Dubois, V.; De Barbeyrac, B.; Rogues, A.M.; Arpin, C.; Coulange, L.; Andre, C.; M’zali, F.; Megraud, F.; Quentin, C. CTX-M-producing Escherichia coli in a maternity ward: A likely community importation and evidence of mother-to-neonate transmission. J. Antimicrob. Chemother. 2010, 65, 1368–1371. [Google Scholar] [CrossRef] [PubMed]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Belotti, P.T.; Thabet, L.; Laffargue, A.; André, C.; Coulange-Mayonnove, L.; Arpin, C.; Messadi, A.; M’Zali, F.; Quentin, C.; Dubois, V. Description of an original integron encompassing blaVIM-2, qnrVC1 and genes encoding bacterial group II intron. J. Antimicrob. Chemother. 2015, 70, 2237–2240. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).