Abstract
In recent years, bacterial pathogens have developed resistance to antimicrobial agents that have created a global threat to human health and environment. As a novel approach to combating antimicrobial resistance (AMR), targeting bacteria’s virulent traits that can be explained by quorum sensing (QS) is considered to be one of the most promising approaches. In the present study, biologically synthesized silver nanoparticles derived from Lactobacillus rhamnosus (AgNPs-LR) were tested against three Gram-negative bacteria to determine whether they inhibited the formation of biofilms and triggered the virulence factors controlled by QS. In C. violaceum and S. marcescens, a remarkable inhibition (>70%) of QS-mediated violacein and prodigiosin production was recorded, respectively. A dose-dependent decrease in virulence factors of P. aeruginosa (pyocyanin, pyoverdine, LasA protease, LasB elastase and rhamnolipid production) was also observed with AgNPs-LR. The biofilm development was reduced by 72.56%, 61.70%, and 64.66% at highest sub-MIC for C. violaceum, S. marcescens and P. aeruginosa, respectively. Observations on glass surfaces have shown remarkable reductions in biofilm formation, with less aggregation of bacteria and a reduced amount of extra polymeric materials being formed from the bacteria. Moreover, swimming motility and exopolysaccharides (EPS) was also found to reduce in the presence of AgNPs-LR. Therefore, these results clearly demonstrate that AgNPs-LR is highly effective in inhibiting the development of biofilms and the QS-mediated virulent traits of Gram-negative bacteria. In the future, AgNPs-LR may be used as an alternative to conventional antibiotics for the treatment of bacterial infections after careful evaluation in animal models, especially for the development of topical antimicrobial agents.
1. Introduction
As a global public health issue, antimicrobial resistance (AMR) is a growing problem that occurs when microorganisms such as bacteria, viruses, fungi, and parasites become resistant to antimicrobial drugs that have previously served as effective treatments for infections [1,2]. When bacteria become resistant to antibiotics, they can spread infections which are difficult to treat, which can result in prolonged illness, disability and even death as a result [3,4]. Multi-drug resistance (MDR) is a form of AMR in which microorganisms become resistant to multiple drugs, making it more difficult to treat infections [5]. This can happen when antibiotics are overused or misused, as well as when there is poor infection prevention and control in healthcare settings [6]. The current problem of AMR and MDR poses a serious problem to global health [7]. According to the World Health Organization (WHO), by 2050 if no action will be taken against drug-resistant infection, the number of deaths will rise to 10 million annually [8]. AMR and MDR also have significant economic impacts. A drug-resistant infection costs more to treat than a non-resistant infection due to the need for more expensive drugs and longer hospital stays [5]. AMR also affects agricultural productivity as it can lead to the loss of livestock and crops [9]. Efforts to address AMR and MDR require a multi-sectoral approach, including reducing unnecessary antibiotic use, improving prevention of infection and measures of prevention, developing new antimicrobial drugs and diagnostic tools, and promoting global cooperation and coordination [10,11,12].
As a strategy to combat the AMR epidemic, anti-infective drugs should be designed, which can target quorum sensing (QS)-regulated virulence factors and biofilm formation [13]. Quorum sensing is a process used by bacteria to communicate with one another through the release and detection of chemical signals called autoinducers. This process permits bacteria to coordinate their behaviour and synchronize their gene expression against the changed environmental conditions [14]. Biofilms are communities of microorganisms embedded in a protective extracellular matrix that adhere to surfaces. QS has a key role in the formation and maintenance of these communities. The eradication of biofilms is extremely difficult and they contribute to the development of antimicrobial resistance by providing a protective environment for bacteria to grow and exchange genetic material, including antibiotic resistance genes [15]. Quorum sensing inhibitors (QSIs) are compounds that can disrupt the communication process between bacteria and prevent the formation of biofilms and reducing the spread of bacterial infections. QSIs have shown promise as a potential strategy to inhibit AMR by reducing resistance to antibiotics in biofilms. Additionally, by preventing the formation of biofilms, QSIs can make bacteria more susceptible to antibiotic treatment, lowering the likelihood of the development of resistance. Several natural and synthetic QSIs have been identified and research is ongoing to develop more effective QSIs and explore their potential clinical applications [16,17]. However, in this regard it is imperative to note that QSIs alone are unlikely to be a promising one for addressing AMR and that a comprehensive approach that includes reducing unnecessary antibiotic use and improving infection prevention and control measures is necessary to combat this global public health challenge [18,19].
Recently, a lot of attention has been paid in finding ways to produce and use the nanomaterials, and the interest is growing every day. In terms of manufacturing nanoparticles, one of the methods to be considered is the bio-approach (green) [20]. Using microorganisms for synthesis of nanoparticles, for example, is referred to as nanoparticles synthesis by biological means. Nanoparticles are produced both by living and dead microorganisms, contribute greatly to nanoparticle production [21]. Material can be controlled at the molecular level using nanotechnology. Silver nanoparticles resulted in significant attention because of their antimicrobial properties, and these metal nanoparticles are being used in different fields such as industrial packaging, agriculture, medicine, cosmetics, and in the military [22]. An array of microorganisms such as E. coli, S. aureus, P. aeruginosa, B. subtilis, V. cholera and S. typhus may be susceptible to AgNPs as antimicrobial agents [22,23,24]. Hence, as an emerging method for discovering antibacterial involves using green synthesized nanoparticles to target bacterial biofilm and QS. Utilization of silver nanoparticles as an alternative antimicrobial agent has been suggested in previous studies and it may prove useful as an alternative [17]. Furthermore, it has become apparent in recent years that nanotechnology has grasped a lot of attention from scientists because of the possibility of its application to medicine, diagnostics, agriculture, bioremediation and many other fields [18]. Nano-scaled materials appear to exhibit better biological effects than their bulk counterparts because their chemical and physical properties are different at this scale, which is mainly why the nano-scaled materials shown improved biological activity [23]. In the future, nanotechnology is expected to have potential applications in different fields of health care, such as for the purposes of new drugs, for drug delivery and diagnostics, and for the creation of improved biomaterials [24].
Lactobacillus rhamnosus (L. rhamnosus) is a probiotic bacterium that has gained considerable attention in the field of medicine due to its potential health benefits. As a naturally occurring bacterium in the human gastrointestinal tract, L. rhamnosus has been extensively studied for its various applications in promoting and supporting human health. With its ability to positively influence the gut microbiota and modulate the immune system, L. rhamnosus has shown promising potential in the prevention and management of several medical conditions [25]. This strain is therefore given more importance nowadays for its health benefits, ability to defeat intestinal pathogens, maintain intestinal flora balance, and maintain intestinal barriers [26,27]. The strain also produced antimicrobial metabolites that had an antagonistic effect on harmful bacteria, including E. coli [28], S. enterica [29] and S. aureus [28]. In addition to this, several studies have also shown that L. rhamnosus has the ability to reduce the bioavailability of mycotoxins in the gastrointestinal tract as well [30]. Therefore, in the present study, L. rhamnosus was used in order to synthesize silver nanoparticles (AgNPs-LR). The synthesized AgNPs-LR were investigated for their broad-spectrum effect on inhibiting the virulence factors controlled by QS in bacterial pathogens namely, C. violaceum, P. aeruginosa and S. marcescens in conjunction with the suppression of biofilm development.
2. Materials and Methods
2.1. Strains of Bacteria and Growth Conditions
The strain of lactic acid bacteria (LAB), Lactobacillus rhamnosus MTCC-1423 (L. rhamnosus) and pathogenic Gram-negative bacterial strain C. violaceum MTCC-2656 (C. violaceum) P. aeruginosa MTCC-741 (P. aeruginosa) and Serratia marcescens MTCC-97 (S. marcescens) were collected from the Microbial Type Culture Collection (IMTECH, Chandigarh, India). The De Man, Rogosa and Sharpe (MRS) agar plate (HiMedia®, Mumbai, India) was used for the growth and maintenance of L. rhamnosus, whereas, Luria-Bertani agar (LB) (HiMedia®, Mumbai, India) was used for the bacterial pathogens. All the bacterial strains were stored at 4 °C for further use.
2.2. Biosynthesis of Silver Nanoparticles (AgNPs) Using L. rhamnosus (AgNPs-LR)
The active culture of L. rhamnosus was added into a fresh MRS media and incubated at 37 °C for overnight. Following incubation, the grown culture was centrifuged for 10 min. at 10,000 rpm and 4 °C to collect the culture supernatant. Then, culture supernatant (10 mL) was mixed with 0.1 mM silver nitrate solution (90 mL) and incubated at 30 °C for 24 h in dark condition. Observations were made of the colour change of AgNPs after 24 h of synthesis. As part of the characterization of the AgNPs-LR, UV-Vis, FTIR and TEM analysis were performed [31].
2.3. Characterization of AgNPs-LR
2.3.1. Ultraviolet-Vis Analysis
In order to characterize AgNPs-LR, a spectrophotometric analysis was performed as a first step. With a resolution of 1 nm, AgNPs-LR were scanned in the range of 300 to 700 nm [31]. The UV-Vis analysis was further used to determine the size of AgNPs-LR using Haiss equation, d = ln((λSPR − λ0)/L1)/L2. Where λSPR is the wavelength at which maximum absorption occurs, λ0 is the wavelength at which minimum absorption occurs at the start of SPR, L1 and L2 are the values taken from the data fit of TEM vs. UV-Vis, whose values are L1 = 6.53 and L2 = 0.0216 [32].
2.3.2. FTIR Analysis
The potential interaction between the culture supernatant of L. rhamnosus and AgNO3 was examined using Fourier Transform Infrared spectroscopy (FT-IR) (Bruker®, Billerica, MA, USA). The spectra were recorded from 500 to 4000 cm−1 [33].
2.3.3. Transmission Electron Microscopy (TEM)
Additionally, TEM measurements were performed on the AgNPs-LR for determining the size and shape. For TEM analysis, a JEM-1400 Plus, Jeol, India was used. By applying the AgNPs-LR sample on a grid made of carbon-coated copper and water content was then evaporated within a vacuum dryer for 1 h, TEM analysis was performed [34].
2.4. Antibacterial Activity of AgNPs-LA
The antibacterial activity of AgNPs-LR was tested using the agar well diffusion method against C. violaceum, S. marcescens, and P. aeruginosa [35]. A sterilized swab was used to streak the inoculum of the bacterial culture onto a MHB agar plate. With the help of a sterile Cork Borer, wells were punched and each well was filled with AgNPs-LR. Afterwards, zone of inhibition was determined after 24 h of incubation at 37 °C.
2.5. Determination of Minimum Inhibitory Concentration (MIC)
In order to evaluate the antibacterial efficacy of AgNPs-LR, the standard broth dilution assay was performed [36]. A series of two-fold dilutions of AgNPs-LR from 1698.7 µg/mL to 0.10 µg/mL concentrations were used to determine MICs in LB broth with active bacterial culture (108 CFU/mL, 0.5 McFarland standard). Only inoculated broth was used for the control, which was incubated at 37 °C for 24 h. MIC is defined as the lowest concentration of AgNPs-LR at which no visible growth can be seen in the tubes at the end of the experiment. In order to confirm the MIC value, a visual examination of the tubes was performed before and after incubation to determine the turbidity.
2.6. Assessment of the Quantity of Violacein Pigment in C. violaceum
Violacein production was quantitatively assessed according to standard procedure [37]. In brief, C. violaceum was grown for 18 h without and with varying sub-MIC concentrations of AgNPs-LR at 30 °C. For the separation of the insoluble pigment (violacein) from the bacterial cells, centrifugation of 1 mL of culture was carried out at 10,000 rpm for 5 min. To dissolve the pigment, the cell pellet was resuspended in 1 mL of DMSO and vortexed vigorously for 5 min. To spin down the bacteria debris, the suspension was again centrifuged. The UV-spectrophotometer (UV-2600, Shimadzu, Japan) at 585 nm was used for measuring the absorbance of the supernatant.
2.7. Assessment of the Quantity of Prodigiosin Pigment in S. marcescens
The production of prodigiosin pigment was assessed using LB medium according to the standard method [38]. The active culture of S. marcescens was added into sterile LB medium with and without AgNPs-LR and grown at 30 °C for overnight. After incubation, centrifugation of 2 mL of grown culture was performed for 10 min. at 10,000 rpm to collect the cell pellet. Acidified ethanol was used to dissolve the obtained cell pellet (96 mL ethanol + 4 mL 1 M HCl) by vigorous shaking at room temperature. After centrifuging the sample once again to remove debris, the absorbance of the supernatant was measured at 534 nm using a spectrophotometer (UV-2600, Shimadzu, Japan).
2.8. Assessment of QS-Mediated Virulence Factors of P. aeruginosa
2.8.1. Estimation of Pyocyanin
The synthesis of pyocyanin by P. aeruginosa was evaluated in LB broth with (sub-MICs) and without AgNPs-LR [39]. The active culture of P. aeruginosa was inoculated into a sterile LB medium with and without AgNPs-LR and incubated for overnight at 30 °C. To collect the culture supernatant, 5 mL of grown culture was centrifuged for 10 min. at 10,000 rpm after incubation. Chloroform (3 mL) was then used to extract the pyocyanin from the culture supernatant. The organic phase was collected and further extracted with 1.2 mL of 0.2 N HCl. At last, the absorbance of aqueous phase was taken at 520 nm via spectrophotometer (UV-2600, Shimadzu, Japan).
2.8.2. Assessment of Pyoverdine
As per the standard procedure, the levels of pyoverdine were analysed via performing the standard method [40]. The P. aeruginosa was grown overnight at 37 °C without and with sub-MIC amounts of AgNPs-LR. A centrifugation process was performed to obtain a supernatant that was free of cells. Then, 900 µL of 50 mM Tris-HCl (pH 7.4) was mixed with 100 µL of culture supernatant. A multi-mode microplate reader was used to measure the fluorescence emission signal (460 nm) of the sample.
2.8.3. Assessment of LasA Staphylolytic Assay
The P. aeruginosa culture supernatant was applied to boiled S. aureus cells to determine LasA protease activity [41]. To collect the cell pellets from S. aureus, centrifugation of culture was carried out at 8000 rpm for 5 min. After collecting the cell pellet, 0.02 M Tris-HCl (pH-8.5) was added and boiled for 10 min. After that, to adjust the absorbance of 0.8 at 595 nm, it was diluted with 0.02 M Tris-HCl. In the following step, a diluted suspension of S. aureus was added to the culture supernatants of P. aeruginosa grown with and without AgNPs-LR (sub-MICs) (9:1). Afterwards, the absorbance was measured at 595 nm.
2.8.4. Assessment of LasB Elastinolytic Activity
The measurements of elastolytic activity were carried out using the procedure mentioned by Adonizio et al., (2008) [42]. The method was based on the treatment of a grown P. aeruginosa culture with or without AgNPs-LR (sub-MICs). The supernatant was collected and added to 900 µL of a buffer containing 20 mg of elastin congo red (100 mM Tris, 1 mM CaCl2, pH-7.5), containing 20 mg of elastin (Sigma®, Bengaluru, India). To remove the insoluble components (elastin Congo red), centrifugation was performed after 3 h of incubation at 37 °C. The absorbance of supernatant was measured at 495 nm.
2.8.5. Assessment of Rhamnolipid
The active culture of P. aeruginosa was added into sterile LB broth in the presence or absence of AgNPs-LR and incubated at 37 °C for 24 h. Ethyl acetate evaporation was used to extract rhamnolipids from culture supernatant. Using the modified method of Pinzon and Ju (2009) [43], an estimate of P. aeruginosa rhamnolipid production was made by dissolving the extracted rhamnolipid in chloroform. A suspension of rhamnolipids was prepared by adding 200 µL of methylene blue solution (0.025%) to 2 mL of solution. A vortexing step was performed for 5 min. and the mixture was incubated at room temperature for 15 min. For phase separation, 0.2 N HCl was added to new tubes containing the chloroform layer after incubation. The mixture was mixed well and left at room temperature for 10 min. By using 0.2 N HCl as blank, 200 µL of the acidic phase holding methylene blue was spectrophotometrically measured at 638 nm.
2.9. Determination of the Swimming Motility of P. aeruginosa and S. marcescens
The LB plate containing sub-MIC (1/2 MIC) of AgNPs-LR were spotted with 5 µL of the active culture of P. aeruginosa and S. marcescens. The control plate was not amended. Incubation was carried out overnight at 30 °C. At the end of incubation, swimming zone was observed in the control and treatment plates [44].
2.10. Assessment of Antibiofilm Activity
The glass test tubes were used to determine the antibiofilm effect of AgNPs-LR as the hydrophilic surface [45]. Briefly, sterilized LB medium (3 mL) was transferred to tubes containing 1 mL of active bacterial culture and 500 µL of AgNPs-LR (sub-MICs). In the following step, the contents of the tube were mixed thoroughly and then tubes were incubated at room temperature in a shaker for 72 h. After incubation, the planktonic cells were removed from the tubes and washed with PBS. Then, the formed biofilm inside the tubes was stained with crystal violet. After removing the excess dye via washing with PBS, the stained biofilm was dissolved in acetic acid and absorbance was determined using a spectrophotometer at 595 nm. As a control for the growth of biofilms, LB medium containing individual bacterial strains was used. Estimates of the percentage inhibition of biofilm was made as follows:
O.D. control − O.D. test/O.D. control × 100
2.11. Light Microscopic Assessment of Biofilm Inhibition
In brief, 60 µL of active culture of bacteria was added in 6-well plates consisting 3 mL of sterile culture medium and allow to grown overnight. Sterile glass coverslips (1 × 1 cm) were placed in the wells along with the respective sub-MIC values of AgNPs-LR. No treatment was administered to the control group. Following incubation of 24 h, the planktonic cells were removed via washing with PBS and then air dried at room temperature for 20 min. The biofilm was stained with crystal violet for 15 min. Afterwards, the slides were washed and air dried for 30 min. to remove any excess dye. In order to visualize the biofilms, a light microscope was used (Axioscope A1, Zeiss, Jena, Germany). Magnification was set at 40× for the images [46].
2.12. Assessment of Exopolysaccharide (EPS) Production
A staining assay using ruthenium red was conducted to determine AgNPs-LR activity in reducing EPS content in biofilms [47]. Each of the bacterial test strains (100 µL) and AgNPs-LR were incubated for 24 h at 37 °C. After incubation, planktonic cells were removed and the cell pellet was washed with sterile PBS (200 µL). In order to stain the biofilms formed by the adherent cells, 200 µL of ruthenium red (0.01%) (Sigma-Aldrich®, Bangalore, India). A well that was free of biofilm and a well containing ruthenium red served as a blank. For the next step, the plate was incubated for 1 h at 37 °C. The residual stain was then re-dispersed into a new microtiter plate and the absorbance measured at 450 nm. Amount of dye fixed to biofilm matrix measured as follows:
where, AbsB = absorbance of the blanks, and AbsS = absorbance of the residual stain collected from the sample wells.
AbsBF = AbsB − AbsS
3. Results
3.1. Synthesis and Characterization of AgNPs-LR
As a result of adding 1 mL of culture supernatant into 10 mL of 0.1 mM silver nitrate solution (1:10), the biosynthesis of AgNPs-LR has been achieved. It was observed during the period of incubation that the colour changed from yellow to dark brown, while the intensity increased over the course of the period of incubation, as a result of a successful biosynthesis of AgNPs-LR. As a first step, UV-Vis analysis of AgNPs-LR was conducted in order to confirm their biosynthesis. According to spectroscopic measurements made after 24 h of synthesis of AgNPs-LR, absorption spectrum with a clear symmetry was observed with a highest absorption at 464 nm (Figure 1A). Bu using information available from the UV-Vis analysis and Haiss equation, we calculated the size of AgNPs-LR as 6.26 nm. By using FTIR spectrum, different functional groups can be identified, as a result of which silver ions are reduced and stabilizing silver nanoparticles. A spectrum of AgNPs-LR is shown in Figure 1B. As shown in the figure, there are several vibrational bands that can be observed in the spectrum, which indicates the existence of several functional groups. The vibrational bands at 3285.38 cm−1, 2934.78 cm−1, 2125.44 cm−1, 1639.12 cm−1, 1401.45 cm−1, 1243.24 cm−1 and 551.64 cm−1 are hydroxyl, C-H/methylene, thiocyanate, alkanyl, carboxylate, aromatic ethers, respectively. Study of infrared spectroscopy suggested the predicted factor groups bind to metals with the most strength, and by coating particles, they can be prevented from agglomerating and maintained for a prolonged period of time. Furthermore, the TEM studies of the AgNPs-LR revealed that most of the nanoparticles were spherical and polyhedral in shape, as well as poly dispersed nanoparticles (Figure 1C–E). It was found that the diameter of the AgNPs-LR ranged from 5 to 70 nm.
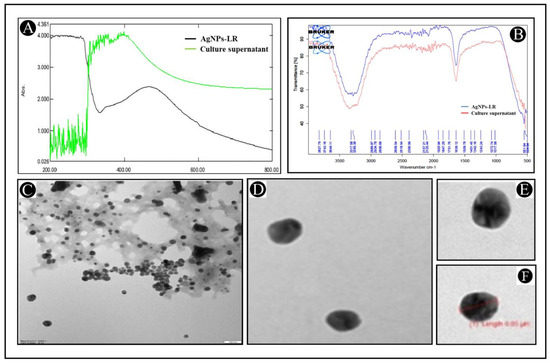
Figure 1.
Characterization of AgNPs-LR. (A) UV-visible absorption spectra of AgNPs-LR. (B) FT-IR analysis of AgNPs-LR. (C–F) Morphological analysis of AgNPs-LR via TEM analysis.
3.2. Antibacterial Potential of Biosynthesized AgNPs-LR
Biosynthesized AgNPs-LR shown to possess antibacterial properties against all of the tested bacterial strains in well diffusion assays. Among the tested bacteria, C. violaceum showed the highest zone of inhibition, followed by P. aeruginosa and S. marcescens Figure 2. Additionally, a broth microdilution was employed in order to determine the MIC of AgNPs-LR. The AgNPs-LR had MIC values of 13.27 μg/mL against C. violaceum, 26.54 μg/mL against P. aeruginosa and 53.08 μg/mL against S. marcescens, respectively. In order to test the efficacy of biosynthesized AgNPs against formation of biofilm as well as QS-regulated virulence factors, the concentrations of AgNPs were below the inhibitory concentrations (sub-MICs).
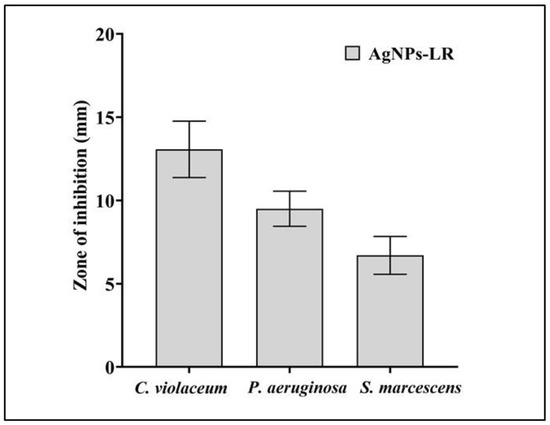
Figure 2.
Antibacterial activity of AgNPs-LR against different Gram-negative bacterial pathogens. Values are represented as the mean ± SD of three independent experiments.
3.3. Inhibition of Inhibition of Virulence Factors of C. violaceum
The AgNPs-LR has been checked for their preliminary anti-QS activity by determining their impact on C. violaceum pigment production. The pigment production in this strain is controlled by QS. Reduced pigment production can be considered as an indication of the presence of anti-QS activity. A treatment with 1/2, 1/4 and 1/8 MIC of AgNPs-LR in C. violaceum resulted in a 75.49%, 59.22% and 47.23% reduction in the synthesis of violacein, respectively (Figure 3A). This clearly indicates that green synthesized AgNPs-LR are capable of exhibiting anti-QS activity.
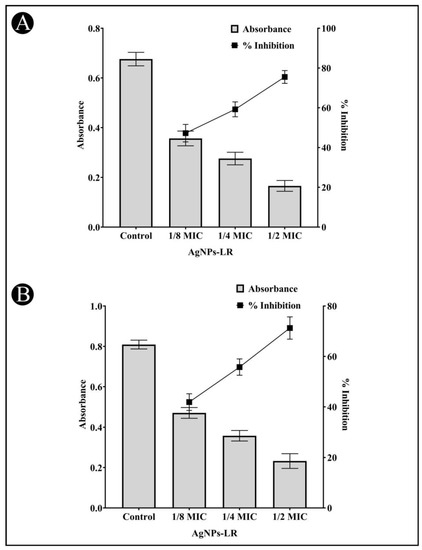
Figure 3.
Anti-QS activity of AgNPs-LR against C. violaceum and S. marcescens. (A) An analysis of the quantitative inhibition of violacein in C. violaceum using AgNPs-LR. (B) An analysis of the quantitative inhibition of prodigiosin in S. marcescens using AgNPs-LR. Values are represented as the mean ± SD of three independent experiments. A secondary y-axis shows the percentage inhibition.
3.4. Inhibition of Virulence Factors of S. marcescens
AgNPs-LR were also tested for anti-QS activity against S. marcescens in an effort to determine whether they inhibit the broad spectrum of QS. S. marcescens produces a pink-red pigment called prodigiosin that is regulated via QS. As per Figure 3B, a range of sub-MICs of AgNPs-LR were found to reduced production of prodigiosin in S. marcescens. At the concentration of 1/2, 1/4 and 1/8 MIC, AgNPs-LR led to a 71.28%, 55.78% and 41.90% inhibition of prodigiosin, respectively.
3.5. Inhibition of Virulence Factors of P. aeruginosa
The virulence factor of P. aeruginosa mediated by QS was examined against AgNPs-LR. A blue-green pigment pyocyanin is produced by P. aeruginosa and is controlled by the communication between bacterial cells. The pigment production of the cells was gradually decreased following treatment with AgNPs-LR. A concentration of 1/2, 1/4 and 1/8 MIC reduced the pigment production of P. aeruginosa by 72.60%, 50.07% and 38.65% (Figure 4A). The pyocyanin contained in P. aeruginosa has been shown to be a significant contributor to its pathogenic potential through the interference with cellular functions of the host.
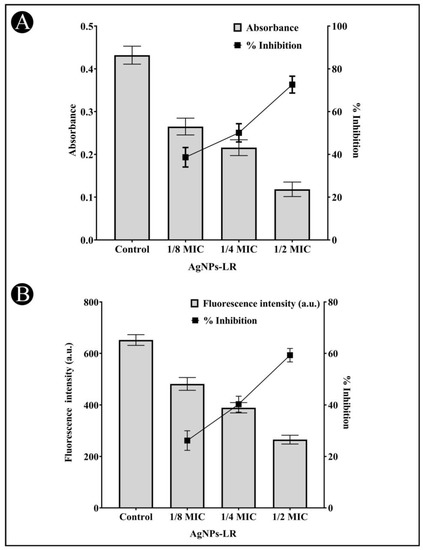
Figure 4.
Anti-QS activity of AgNPs-LR against P. aeruginosa. (A) An analysis of the quantitative inhibition of pyocyanin production in P. aeruginosa using AgNPs-LR. (B) An analysis of the quantitative inhibition of pyoverdine production in P. aeruginosa using AgNPs-LR. Values are represented as the mean ± SD of three independent experiments. A secondary y-axis shows the percentage inhibition.
A pigment known as pyoverdine can also be produced by several strains of P. aeruginosa which are virulent. There was an inhibition of pyoverdine production in the supernatant by 59.30%, 40.28%, and 26.17%, respectively, when sub-MICs of AgNPs-LR is present (Figure 4B).
A virulent strain of bacteria produces proteolytic enzymes that cause damage to the tissues of the host upon successful infection. A staphylolytic assay was used to determine whether AgNPs-LR inhibit LasA protease activity. After treatment with sub-MICs of AgNPs-LR, LasA protease activity decreased by 62.41%, 41.55%, and 18.36%, respectively (Figure 5A).
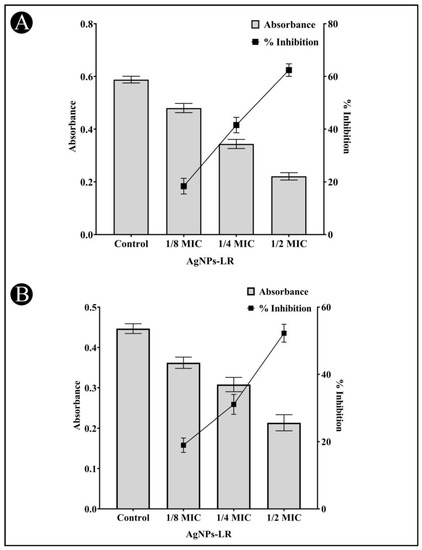
Figure 5.
Anti-QS activity of AgNPs-LR against P. aeruginosa. (A) An analysis of the quantitative inhibition of LasA protease production in P. aeruginosa using AgNPs-LR. (B) An analysis of the quantitative inhibition of LasB elastase production in P. aeruginosa using AgNPs-LR. Values are represented as the mean ± SD of three independent experiments. A secondary y-axis shows the percentage inhibition.
An eleastase is a hydrolytic enzyme produced by bacteria during an infection that destroys and inhibit the host immune system. In the presence of AgNPs-LR, P. aeruginosa showed a concentration-dependent inhibition of its elastinolytic activity (Figure 5B).
In addition to maintaining the structure of biofilms, rhamnolipids play an important role in adhering bacterial cells to solid surfaces. Rhamnolipid production by P. aeruginosa is regulated by RhlR-RhlI QS. Following treatment with AgNPs-LR, rhamnolipid production was reduced (Figure 6). Rhamnolipid production was decreased by 55.86%, 42.76% and 31.82%, respectively, in the presence of sub-MICs of AgNPs-LR.
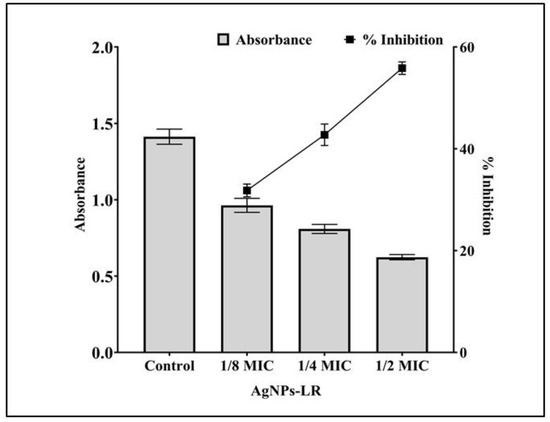
Figure 6.
An analysis of the quantitative inhibition of rhamnolipid production in P. aeruginosa using AgNPs-LR. Values are represented as the mean ± SD of three independent experiments.
3.6. Quantitative Inhibition of the Formation of Biofilms
An AI-mediated QS phenomenon is often responsible for regulating the formation of a biofilm by regulating its mechanisms. The results obtained from the study on the effects of the AgNPs-LR on the formation of biofilms is shown in Figure 7A for all three bacteria tested. The development of bio-films of C. violaceum was inhibited by 72.56%, 55.37% and 42.70%, when 1/2, 1/4, and 1/8 MIC concentrations were used as treatment. The biofilm of P. aeruginosa was inhibited by 64.66%, 46.58% and 38.93% at sub-MICs. Similarly, when sub-MICs are present, the biofilms of S. marcescens decreased by 61.70%, 41.72%, and 36.50%, respectively.
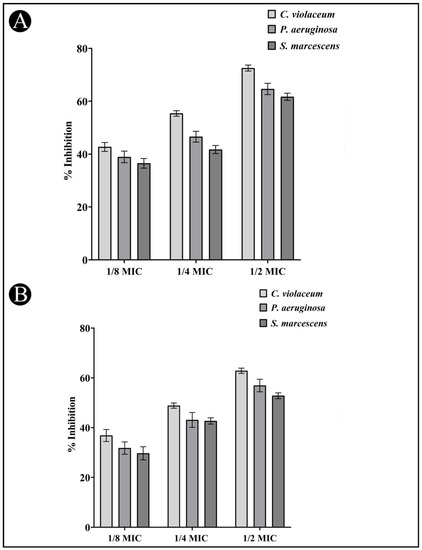
Figure 7.
Anti-biofilm and EPS inhibition activity of AgNPs-LR against different Gram-negative bacterial pathogens. (A) An analysis of the quantitative inhibition of biofilm production using AgNPs-LR. (B) An analysis of the quantitative inhibition of EPS production using AgNPs-LR. Values are represented as the mean ± SD of three independent experiments.
3.7. Inhibition of EPS Production
The EPS matrix provides protection and support for the biofilm and is critical for its formation, stability, and function. EPS can help bacteria adhere to surfaces, create channels for nutrient flow, and provide protection against antibiotics and other stressors. The present study found that EPS production decreased upon the treatment of AgNPs-LR as 62.84%, 48.84% and 36.83% in C. violaceum, 56.91%, 43.12% and 31.77% in P. aeruginosa and 52.80%, 42.68% and 29.67% in S. marcescens at sub-MICs, respectively (Figure 7B).
3.8. Analysis of Biofilm Inhibition on Glass Surfaces Using Microscope
Using a microscopy technique, it was possible to perform a further evaluation of the inhibition of biofilms. For the purpose of examining a change in the biofilm architecture, the test bacteria were cultured without and with the maximum sub-MIC of AgNPs-LR in order to visualize biofilm architecture changes. All the tested bacteria displayed a dense cluster of cells on the glass coverslips, as can be seen from the light microscopy images (Figure 8A–E). With AgNPs-LR treatment, cells were seen in a scattered form on the glass surface and were significantly reduced in clustering.
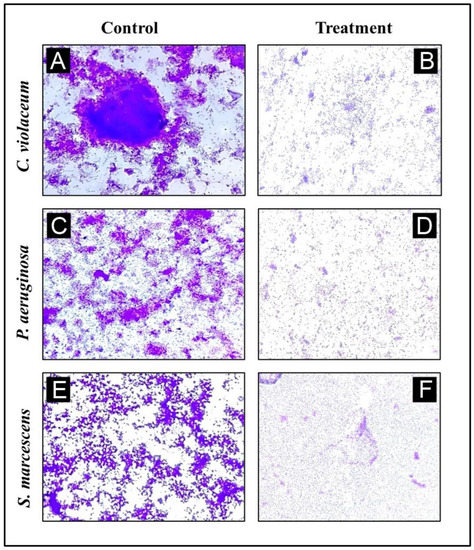
Figure 8.
A representative light micrograph of a biofilm showing the effects of AgNPs-LR at their highest sub-MICs. (A) Control of C. violaceum, (B) Treatment of C. violaceum with ½ MIC, (C) Control of P. aeruginosa, (D) Treatment of P. aeruginosa with ½ MIC, (E) Control of S. marcescens, (F) Treatment of S. marcescens with ½ MIC.
3.9. Inhibition of Swimming Motility
QS has a very important role in controlling the movement of P. aeruginosa and S. marcescens, a crucial factor in determining the spread of infection in a host. It is also considered as a crucial factor in the pathogenicity of both bacteria, so it is an important factor in virulence. In Figure 9A–D, it is shown that the control P. aeruginosa and S. marcescens swims across the entire Petriplate after overnight incubation. Whereas, there was a decrease in the zone of swimming in the treatment plates.
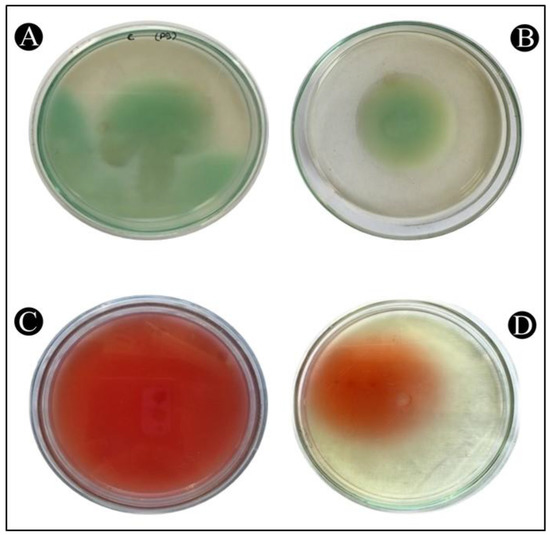
Figure 9.
The inhibition of the swimming motility of P. aeruginosa and S. marcescens by AgNPs-MK. (A) Control of P. aeruginosa, (B) Treatment of P. aeruginosa with ½ MIC, (C) Control of S. marcescens, (D) Treatment of S. marcescens with ½ MIC.
4. Discussion
In order to synthesize metal nanoparticles, there are a variety of approaches such as chemical, physical and biological methods that can be utilized. There are significant drawbacks associated with chemical synthesis, primarily the fact that dangerous and non-biodegradable by-products are formed, making them harmful for the environment. The concept of green synthesis is gaining increasing popularity because of its environmental benefits, such as minimizing waste production, using non-hazardous materials and enhancing its environmental friendliness [19]. Besides not causing disease, probiotic bacteria also prevent pathogenic bacteria from multiplying in animals’ digestive system and increase intestinal microflora that is beneficial for the animal. There has been a growing market for probiotics in the majority of countries [48].
Using L. rhamnosus as a basis for the production of AgNPs, the purpose of the present study was to establish a simple, green, and inexpensive approach that could be utilized to synthesize AgNPs. A prepared sample of L. rhamnosus was transferred to a final concentration of 1 mM of silver nitrate in order to observe the formation of AgNPs-LR. The colour change in the sample indicated AgNPs-LR formation, which were then characterized using UV-Vis, FT-IR and TEM analysis. Analyzing AgNPs with UV-Vis is a common technique used to characterize the optical properties of these nanoparticles. The technique is based on the principle of interaction of light with the localized surface plasmon resonance of AgNPs [49]. Therefore, the electron transition that occurs at 464 nm during AgNPs synthesis can be interpreted as a result of the interaction between incident light and the localized surface plasmon resonance of the AgNPs. At 464 nm, the absorption peak suggests that the electron transition involved is primarily a dipole transition. In a dipole transition, the conduction electrons of the AgNPs are excited from the ground state to a higher energy state by absorbing a photon with a specific energy corresponding to the wavelength of 464 nm [50,51,52]. FT-IR analysis of AgNPs is a technique utilized to determine the chemical composition and bonding properties of these nanoparticles. This technique is based on principle of the interaction of infrared radiation with the molecular vibrations of the nanoparticles [53]. Whereas, TEM analysis was performed to characterize the size, shape and distribution of synthesized AgNPs.
Previously, an iron oxide nanoparticle has been synthesized by Torabian et al. [54] using the green synthesis approach. Upon synthesis, iron oxide nanoparticles were found to be around 15 nm in size. Nanoparticles had round to spherical shape and they found its application in drug delivery or therapy as safe, effective and inexpensive [54]. By synthesizing AgNPs from the supernatant filtrate of L. acidophilus, Rajesh et al. [55] designed eco-friendly antibacterial components. Using electron microscopy, they determined that the particles had spherical shapes with sizes ranging from 4–50 nm. According to their report, AgNPs showed antibacterial properties when used against Klebsiella pneumoniae by causing cytolysis and destroying the membrane of the bacterial cell [55]. A further study conducted by Nithya et al. [56] examined the antimicrobial efficacy of AgNPs synthesized from Brevibacterium linens against multidrug-resistant clinical isolates and demonstrated that these nanoparticles were highly effective. As a result of the AgNPs incubated with Escherichia coli colonies for 3 h, it was observed that viable cells had decreased. In contrast, it can cause Staphylococcus aureus inhibition zones similar to those of Amikacin. Selenium nanoparticles (SeNP) with 50–80 nm size was prepared by Xu et al. [57] using L. casei. Furthermore, in mice infected with Enterotoxigenic E. coli K88, SeNP treatment resulted in lowering inflammatory cytokines and oxidative stress in the mice [57]. Using the probiotic L. kimchicus DCY51 isolated from traditional Korean kimchi, Markus et al. [58] synthesized gold nanoparticles (AuNPs). Synthesized AuNPs were found to be spherical structure with a size between 5–30 nm. The antioxidant properties of AuNP were demonstrated by its ability to remove free radicals [58]. The antimicrobial efficacy of AgNPs synthesized from one more probiotic L. amylophilus GV6 has been tested by Kumar et al. [59] using agar well plate assay against several bacterial pathogens such as, B. subtilis MTCC 121, S. aureus MTCC 96, P. aeruginosa MTCC 424, K. pneumoniae MTCC 109 and E. coli MTCC 43, which reported the 1.5 cm inhibition zone against S. aureus MTCC 96. Using two probiotic bacteria, L. acidophilus 58p and L. plantarum 92T, Garmasheva et al. [60] synthesized AgNPs and studied their antibacterial activity. The AgNPs obtained from L. acidophilus 58p were found more active against E. coli, S. epidermidis, S. flexneri, K. pneumonia, and S. sonnei than L. plantarum 92T AgNPs. In a similar study, Naseer et al. [61] synthesized AgNPs from L. bulgaricus and evaluated their antibacterial effectiveness against S. aureus, S. epidermidis and S. typhi. Gram-negative bacteria were found to be more susceptible than Gram-positive bacteria. A recent study by Sharma et al. [62] also synthesized safe, inexpensive and biocompatible AgNPs using four different probiotic isolates such as, L. plantarum F22, L. paraplantarum KM1, L. pentosus S6 and L. crustorum F11. Various bacterial and fungi pathogens viz., B. cereus, L. monocytogenes, antibiotic-resistant S. aureus, P. aphanidermatum, P. parasitica and F. oxysporum were also found to be susceptible to these AgNPs. Among them, AgNP synthesized by L. crustorum F11 showed maximum inhibition against all pathogens, with maximum activity against S. aureus (20 ± 0.61 mm) and F. oxysporum (23 ± 0.37).
The current study further explores the antibacterial, antibiofilm and anti-QS activities of biosynthesized AgNPs-LR. As a first step, biosynthesized AgNPs were tested against different bacteria pathogens using well diffusion assays. According to our results, the biosynthesized AgNPs-LR display the highest antibacterial activity against C. violaceum, S. marcescens and P. aeruginosa, respectively. As reported earlier, the high antibacterial activity of biosynthesized AgNPs resulting from reactive oxygen species production and the damage to membranes [63,64]. By virtue of the low MIC of biosynthesized AgNPs against bacterial pathogens, it is not necessary to use conventional antibiotics in conjunction with the biosynthesized AgNPs. Additionally, biosynthesized AgNPs showed higher susceptibility to C. violaceum and P. aeruginosa than S. marcescens. In general, each of these bacteria is capable of forming biofilms, which are difficult to eradicate even with conventional antibiotics available on the market, since their extracellular matrix also reveals resistance to both environmental factors and antibiotics [65,66]. The formation of biofilms contributes to pathogenesis, with almost 80% of infections attributed to biofilm formation [17]. Biofilms are responsible for reducing the efficiency of antibiotics by up to a thousand-fold, which places a burden on the health care system to treat infections. The AI often contributes to QS that controls the formation of a biofilm in a given environment. The synthesized AgNPs-LR were tested for their effect on the development of biofilms in all three bacterial pathogens and the results showed that they effectively inhibited the formation of biofilms at sub-MIC levels. It has been discovered that bacteria that form biofilms resist both chemical and physical therapy and their virulence genes are coordinated in their expression [67]. Previous studies found tobramycin resistance in biofilms of P. aeruginosa to be 1000 times higher than in planktonic bacteria [68]. A study carried out using AgNPs derived from the bark extract of Holarrhena pubescens reported that the AgNPs inhibited the development of biofilms of imipenem-resistant P. aeruginosa [69]. Additionally, AgNPs also inhibited the biofilm development of methicillin-resistant S. aureus and P. aeruginosa producing extended-spectrum beta-lactamases (EsbL) [70]. Histidine-capped AgNPs were also known to eradicate mature biofilm of K. pneumoniae by Chibber et al. [71].
The bacterial QS process is a unique way in which bacteria communicate between themselves, through cell density can be sensed by bacteria in their adjacent atmosphere and this in turn results in the activation or suppression of specific genes in the bacteria [72,73]. QS-dependent gene expression is responsible for many important bacterial characteristics such as physiology, virulence, and the formation of biofilms in bacteria. A huge attention has been given to research on QS because of its potential for human medicine over the past 15–20 years [74]. Thus, the current study also examined an inhibitory effect on QS of AgNPs-LR against C. violaceum, P. aeruginosa and S. marcescens.
Violacein production by C. violaceum is regulated by the QS system based on the density of bacteria in the population. Although, violacein itself is not usually considered a pathogenic factor, infections caused by C. violaceum can be serious and even life-threatening in individuals with compromised immune systems [75]. In these instances, violacein may help bacteria evade the immune system and establish infection by contributing to their virulence. In spite of the fact that cell-to-cell communication is critical to bacterial physiology and virulence, QS inhibitors have been shown to inhibit the production of violacein by C. violaceum, thus providing insights into the potential of QS inhibitors as therapeutic agents against bacterial infections [76].
Pyocyanin production in P. aeruginosa is also regulated by QS, just as violacein production. Pyocyanin is a blue-green pigment that is crucial for the pathogenesis of P. aeruginosa infections [77]. Pyocyanin plays a key role in P. aeruginosa infections by causing oxidative stress in the host. The pyocyanin pigment damages host cell membranes and contributes to the destruction of host tissues by generating reactive oxygen species (ROS). Aside from causing tissue damage and exacerbated infection, oxidative stress triggers inflammation [78]. The immune response to infection has also been interfered with by pyocyanin. Immune cells such as neutrophils and macrophages, which fight bacterial infections, are inhibited by it. As a result, P. aeruginosa infection that evades the immune system can become persistent [79,80]. Additionally, P. aeruginosa produces pyocyanin, a fluorescent green-yellow siderophore [81]. By facilitating iron acquisition, promoting bacterial growth, and stimulating the host immune response, it also plays an immense part in the pathogenesis of P. aeruginosa infections [82]. Furthermore, P. aeruginosa infections involve the production of a protease enzyme called Las A. Among the key factors in the pathogenesis of P. aeruginosa infection, it degrades host tissue proteins, interferes with host cell signaling, stimulates host immune responses, and promotes biofilm formation and detachment [83]. As for Las B, it is a zinc metalloprotease enzyme produced by P. aeruginosa that degrades host tissue proteins, including elastin, collagen, and fibronectin, during pathogenesis. This can result in tissue damage and destruction, which is then used by the bacterium to spread and colonize new sites within the host as a result of the damage [84]. Additionally, P. aeruginosa produces surface-active molecules known as rhamnolipids. Moreover, P. aeruginosa infections involve the activity of these molecules in a variety of ways. During infection by P. aeruginosa, it contributes to host cell lysis, promotes biofilm formation, stimulates the immune response, and aids bacterial adaptation to stress [85]. The QS is also responsible for the synthesis of prodigiosin, a bright red pigment produced by S. marcescens. The prodigiosin contributes to the pathogenesis of S. marcescens infections, as it is majorly responsible for the formation of biofilm, antimicrobial activity, immunomodulation, and cytotoxicity [86]. It is therefore possible to gain insights into the infections caused by pathogenic bacteria by regulating the activity of QS.
Furthermore, movement of bacteria by swimming is known as swimming motility that involves the rotation of flagella to propel the cell through liquid environments. Swimming motility plays a crucial role in the formation and spread of biofilms, as well as in the regulation of QS signaling pathways that are important for bacterial communication and virulence. Inhibition of swimming motility has therefore raised as a possible way for the development of antibiofilm and anti-QS agents. EPS inhibition is also an emerging strategy for development of antibiofilm and anti-QS agents [87]. The EPS matrix plays an important role for the development of biofilms, providing mechanical stability, protection against environmental stresses, and a barrier to antimicrobial agents. By targeting the EPS matrix or QS signaling pathways, it may be possible to prevent biofilm formation, make biofilms more susceptible to antimicrobial agents and disrupt a wide range of bacterial behaviors [88]. Accordingly, the results of this study clearly indicated that AgNPs-LR synthesized from L. rhamnosus played a broad-spectrum antibiofilm and anti-QS activity against Gram-negative pathogenic bacteria.
5. Conclusions
The biosynthesized AgNPs-LR demonstrated a remarkable reduction in multiple QS-regulated functions in Gram-negative bacteria, such as, C. violaceum, P. aeruginosa, and S. marcescens. In C. violaceum, there has been a significant decrease in the production of violacein pigments of more than 70%. Upon treatment with AgNPs-LR, S. marcescens virulent trait controlled by QS was also reduced by 70%. A dose-dependent inhibition of P. aeruginosa virulence factors was also observed with AgNPs-LR. All test bacteria were found to show a decrease in their ability to form biofilms at their respective sub-MICs by a dose-dependently manner. In addition, there was a notable reduction in the formation of biofilms on the surfaces of the coverslips, swimming motility as well as the production of EPS. As a result, it can be concluded that biosynthesized AgNPs-LR could be exploited for the treatment of skin infections resulting from topical application. In addition to this, medical implants/devices can also be coated with these materials in order to inhibit the bacterial adhesion and to prevent the formation of biofilms on the surfaces. However, there is a need to perform more in-vivo studies to determine the therapeutic efficacy of AgNPs-LR against infections caused by pathogens that are resistant to known antibiotics.
Author Contributions
Conceptualization, A.M.A., M.A. and M.P.; methodology, C.D., M.S., E.N., A.J.S., S.H. and S.A.A.; validation, C.D., M.S.A., M.S., F.B. and R.B.; formal analysis, C.D., M.P., M.A., M.S., F.B. and R.B.; investigation, C.D., S.A.A., A.J.S., M.S.A. and A.M.A.; data curation, M.P., F.B., A.M.A., S.A.A., S.H. and E.N.; writing—original draft preparation, A.M.A., M.P. and M.A.; writing—review and editing, F.B., M.S., M.S.A., S.H. and E.N.; software, M.A. and M.P.; visualization, M.P. and A.J.S.; supervision, M.A. and M.P.; project administration, M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been funded by Scientific Research Deanship at University of Ha’il-Saudi Arabia through project number RG-21093.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this article.
Acknowledgments
Authors are thankful to Scientific Research Deanship at University of Ha’il-Saudi Arabia for supporting this study through project number RG-21093.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Tanwar, J.; Das, S.; Fatima, Z.; Hameed, S. Multidrug Resistance: An Emerging Crisis. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 541340. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U. Antibiotic Resistance: A Rundown of a Global Crisis. Infect. Drug Resist. 2018, 11, 1645. [Google Scholar] [CrossRef] [PubMed]
- Sabtu, N.; Enoch, D.A.; Brown, N.M. Antibiotic Resistance: What, Why, Where, When and How? Br. Med. Bull. 2015, 116, 105–113. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef]
- Giurazza, R.; Mazza, M.C.; Andini, R.; Sansone, P.; Pace, M.C.; Durante-Mangoni, E. Emerging Treatment Options for Multi-Drug-Resistant Bacterial Infections. Life 2021, 11, 519. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E. The Global Threat of Antimicrobial Resistance: Science for Intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2021; World Health Organization: Geneva, Switzerland, 2021.
- Mshana, S.E.; Sindato, C.; Matee, M.I.; Mboera, L.E.G. Antimicrobial Use and Resistance in Agriculture and Food Production Systems in Africa: A Systematic Review. Antibiotics 2021, 10, 976. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.A.A.; Rahman, S.; Cohall, D.; Bharatha, A.; Singh, K.; Haque, M.; Gittens-St Hilaire, M. Antimicrobial Stewardship: Fighting Antimicrobial Resistance and Protecting Global Public Health. Infect. Drug Resist. 2020, 13, 4713–4738. [Google Scholar] [CrossRef]
- Ranjalkar, J.; Chandy, S.J. India’s National Action Plan for Antimicrobial Resistance–An Overview of the Context, Status, and Way Ahead. J. Fam. Med. Prim. Care 2019, 8, 1828. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Towse, A. New Drugs to Tackle Antimicrobial Resistance: Analysis of EU Policy Options. Available SSRN 2640028. 2010. Available online: https://www.ohe.org/publications/new-drugs-tackle-antimicrobial-resistance-analysis-eu-policy-options/ (accessed on 4 April 2023).
- Piewngam, P.; Chiou, J.; Chatterjee, P.; Otto, M. Alternative Approaches to Treat Bacterial Infections: Targeting Quorum-Sensing. Expert Rev. Anti. Infect. Ther. 2020, 18, 499–510. [Google Scholar] [CrossRef]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010, 2, a000398. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilm Formation: A Clinically Relevant Microbiological Process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef]
- Schachter, B. Slimy Business—The Biotechnology of Biofilms. Nat. Biotechnol. 2003, 21, 361–365. [Google Scholar] [CrossRef]
- Martins, N.; Rodrigues, C.F. Biomaterial-Related Infections. J. Clin. Med. 2020, 9, 722. [Google Scholar] [CrossRef] [PubMed]
- Lasa, I.; Del Pozo, J.L.; Penadés, J.R.; Leiva, J. Biofilms Bacterianos e Infección. In Anales del Sistema Sanitario de Navarra; Gobierno de Navarra. Departamento de Salud: Pamplona, Spain, 2005; Volume 28, pp. 163–175. [Google Scholar]
- Qais, F.A.; Khan, M.S.; Ahmad, I. Nanoparticles as Quorum Sensing Inhibitor: Prospects and Limitations. In Biotechnological Applications of Quorum Sensing Inhibitors; Springer: Berlin/Heidelberg, Germany, 2018; pp. 227–244. [Google Scholar]
- Nagajyothi, P.C.; Cha, S.J.; Yang, I.J.; Sreekanth, T.V.M.; Kim, K.J.; Shin, H.M. Antioxidant and Anti-Inflammatory Activities of Zinc Oxide Nanoparticles Synthesized Using Polygala Tenuifolia Root Extract. J. Photochem. Photobiol. B Biol. 2015, 146, 10–17. [Google Scholar] [CrossRef]
- Manikandan, R.; Manikandan, B.; Raman, T.; Arunagirinathan, K.; Prabhu, N.M.; Basu, M.J.; Perumal, M.; Palanisamy, S.; Munusamy, A. Biosynthesis of Silver Nanoparticles Using Ethanolic Petals Extract of Rosa Indica and Characterization of Its Antibacterial, Anticancer and Anti-Inflammatory Activities. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 138, 120–129. [Google Scholar] [CrossRef]
- Mohanpuria, P.; Rana, N.K.; Yadav, S.K. Biosynthesis of Nanoparticles: Technological Concepts and Future Applications. J. Nanoparticle Res. 2008, 10, 507–517. [Google Scholar] [CrossRef]
- Sre, P.R.R.; Reka, M.; Poovazhagi, R.; Kumar, M.A.; Murugesan, K. Antibacterial and Cytotoxic Effect of Biologically Synthesized Silver Nanoparticles Using Aqueous Root Extract of Erythrina Indica Lam. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 135, 1137–1144. [Google Scholar]
- Nishiyama, K.; Sugiyama, M.; Mukai, T. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms 2016, 4, 34. [Google Scholar] [CrossRef]
- Zhang, C.; Gui, Y.; Chen, X.; Chen, D.; Guan, C.; Yin, B.; Pan, Z.; Gu, R. Transcriptional homogenization of Lactobacillus rhamnosus hsryfm 1301 under heat stress and oxidative stress. Appl. Microbiol. Biotechnol. 2020, 104, 2611–2621. [Google Scholar] [CrossRef]
- Matsubara, V.H.; Wang, Y.; Bandara, H.M.H.N.; Mayer, M.P.A.; Samaranayake, L.P. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 2016, 100, 6415–6426. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Pang, B.; Li, J.; Liu, G.; Xu, X.; Shao, D.; Jiang, C.; Yang, B.; Shi, J. Mechanisms for Lactobacillus rhamnosus treatment of intestinal infection by drug-resistant Escherichia coli. Food Funct. 2020, 11, 4428–4445. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, Y.H.; Yang, G.Y.; Liu, X.; Xia, B.; Hu, X.; Su, J.H.; Wang, J.F. Lactobacillus rhamnosus GG affects microbiota and suppresses autophagy in the intestines of pigs challenged with Salmonella Infantis. Front. Microbiol. 2018, 8, 2705. [Google Scholar] [CrossRef]
- Assaf, J.C.; Khoury, A.E.; Chokr, A.; Louka, N.; Atoui, A. A novel method for elimination of aflatoxin M1 in milk using Lactobacillus rhamnosus GG biofilm. Int. J. Dairy Technol. 2019, 72, 248–256. [Google Scholar] [CrossRef]
- Syame, S.M.; Mansour, A.S.; Khalaf, D.D.; Ibrahim, E.S.; Gaber, E.S. Green Synthesis of Silver Nanoparticles Using Lactic Acid Bacteria: Assessment of Antimicrobial Activity. World’s Vet. J. 2020, 10, 625–633. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticles from UV−Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Adebayo-Tayo, B.C.; Popoola, A.O. Biogenic Synthesis and Antimicrobial Activity of Silver Nanoparticle Using Exopolysaccharides from Lactic Acid Bacteria. Int. J. Nano Dimens. 2017, 8, 61. [Google Scholar]
- Sintubin, L.; De Windt, W.; Dick, J.; Mast, J.; Van Der Ha, D.; Verstraete, W.; Boon, N. Lactic Acid Bacteria as Reducing and Capping Agent for the Fast and Efficient Production of Silver Nanoparticles. Appl. Microbiol. Biotechnol. 2009, 84, 741–749. [Google Scholar] [CrossRef]
- Shameli, K.; Ahmad, M.B.; Yunus, W.M.Z.W.; Ibrahim, N.A.; Rahman, R.A.; Jokar, M.; Darroudi, M. Silver/Poly (Lactic Acid) Nanocomposites: Preparation, Characterization, and Antibacterial Activity. Int. J. Nanomed. 2010, 7, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Biedenbach, D.; Lob, S.; Badal, R.; Sahm, D. Variability of Susceptibility and Multidrug Resistance among K. pneumoniae from IAI in Asia/Pacific Countries–SMART 2012–2013. Int. J. Antimicrob. Agents PO BOX 2015, 211, 1000. [Google Scholar]
- Matz, C.; Deines, P.; Boenigk, J.; Arndt, H.; Eberl, L.; Kjelleberg, S.; Jürgens, K. Impact of Violacein-Producing Bacteria on Survival and Feeding of Bacterivorous Nanoflagellates. Appl. Environ. Microbiol. 2004, 70, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Slater, H.; Crow, M.; Everson, L.; Salmond, G.P.C. Phosphate Availability Regulates Biosynthesis of Two Antibiotics, Prodigiosin and Carbapenem, in Serratia via Both Quorum-sensing-dependent And-independent Pathways. Mol. Microbiol. 2003, 47, 303–320. [Google Scholar] [CrossRef]
- Essar, D.W.; Eberly, L.E.E.; Hadero, A.; Crawford, I.P. Identification and Characterization of Genes for a Second Anthranilate Synthase in Pseudomonas aeruginosa: Interchangeability of the Two Anthranilate Synthases and Evolutionary Implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [PubMed]
- Ankenbauer, R.; Sriyosachati, S.; Cox, C.D. Effects of Siderophores on the Growth of Pseudomonas aeruginosa in Human Serum and Transferrin. Infect. Immun. 1985, 49, 132–140. [Google Scholar] [CrossRef]
- Kessler, E.; Safrin, M.; Olson, J.C.; Ohman, D.E. Secreted LasA of Pseudomonas aeruginosa is a Staphylolytic Protease. J. Biol. Chem. 1993, 268, 7503–7508. [Google Scholar] [CrossRef]
- Adonizio, A.; Kong, K.-F.; Mathee, K. Inhibition of Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa by South Florida Plant Extracts. Antimicrob. Agents Chemother. 2008, 52, 198–203. [Google Scholar] [CrossRef]
- Pinzon, N.M.; Ju, L.-K. Analysis of Rhamnolipid Biosurfactants by Methylene Blue Complexation. Appl. Microbiol. Biotechnol. 2009, 82, 975–981. [Google Scholar] [CrossRef]
- Kumar, L.; Chhibber, S.; Harjai, K. Zingerone Inhibit Biofilm Formation and Improve Antibiofilm Efficacy of Ciprofloxacin against Pseudomonas aeruginosa PAO1. Fitoterapia 2013, 90, 73–78. [Google Scholar] [CrossRef]
- Ghaima, K.K.; Rasheed, S.F.; Ahmed, E.F. Antibiofilm, Antibacterial and Antioxidant Activities of Water Extract of Calendula officinalis Flowers. Int. J. Biol. Pharm. Res. 2013, 4, 465–470. [Google Scholar]
- Musthafa, K.S.; Ravi, A.V.; Annapoorani, A.; Packiavathy, I.S.V.; Pandian, S.K. Evaluation of Anti-Quorum-Sensing Activity of Edible Plants and Fruits through Inhibition of the N-Acyl-Homoserine Lactone System in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy 2010, 56, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Borucki, M.K.; Krug, M.J.; Muraoka, W.T.; Call, D.R. Discrimination among Listeria monocytogenes Isolates Using a Mixed Genome DNA Microarray. Vet. Microbiol. 2003, 92, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Guandalini, S. Probiotics for Prevention and Treatment of Diarrhea. J. Clin. Gastroenterol. 2011, 45, S149–S153. [Google Scholar] [CrossRef]
- Ahmed, S.; Ikram, S. Silver Nanoparticles: One Pot Green Synthesis Using Terminalia arjuna Extract for Biological Application. J. Nanomed. Nanotechnol. 2015, 6, 309. [Google Scholar]
- Tsuji, T.; Iryo, K.; Watanabe, N.; Tsuji, M. Preparation of silver nanoparticles by laser ablation in solution: Influence of laser wavelength on particle size. Appl. Surface Sci. 2002, 202, 80–85. [Google Scholar] [CrossRef]
- Murphy, C.J.; Jana, N.R. Controlling the aspect ratio of inorganic nanorods and nanowires. Adv. Mat. 2002, 14, 80–82. [Google Scholar] [CrossRef]
- Sastry, M.; Mayya, K.S.; Bandyopadhyay, K. pH Dependent changes in the optical properties of carboxylic acid derivatized silver colloidal particles. Colloids Surf. A Physicochem. Eng. Asp. 1997, 127, 221–228. [Google Scholar] [CrossRef]
- Sondi, I.; Goia, D.V.; Matijević, E. Preparation of Highly Concentrated Stable Dispersions of Uniform Silver Nanoparticles. J. Colloid Interface Sci. 2003, 260, 75–81. [Google Scholar] [CrossRef]
- Torabiana, P.; Ghandeharia, F.; Fatemib, M. Asian Journal of Green Chemistry. Asian J. Green. Chem. 2017, 2, 89–97. [Google Scholar]
- Rajesh, S.; Dharanishanthi, V.; Kanna, A.V. Antibacterial Mechanism of Biogenic Silver Nanoparticles of Lactobacillus acidophilus. J. Exp. Nanosci. 2015, 10, 1143–1152. [Google Scholar] [CrossRef]
- Nithya, R.; Ragunathan, R. Synthesis of Silver Nanoparticles Using a Probiotic Microbe and Its Antibacterial Effect against Multidrug Resistant Bacteria. Afr. J. Biotechnol. 2012, 11, 11013–11021. [Google Scholar]
- Xu, C.; Guo, Y.; Qiao, L.; Ma, L.; Cheng, Y.; Roman, A. Biogenic Synthesis of Novel Functionalized Selenium Nanoparticles by Lactobacillus Casei ATCC 393 and Its Protective Effects on Intestinal Barrier Dysfunction Caused by Enterotoxigenic Escherichia coli K88. Front. Microbiol. 2018, 9, 1129. [Google Scholar] [CrossRef] [PubMed]
- Markus, J.; Mathiyalagan, R.; Kim, Y.-J.; Abbai, R.; Singh, P.; Ahn, S.; Perez, Z.E.J.; Hurh, J.; Yang, D.C. Intracellular Synthesis of Gold Nanoparticles with Antioxidant Activity by Probiotic Lactobacillus kimchicus DCY51T Isolated from Korean Kimchi. Enzym. Microb. Technol. 2016, 95, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.K.; Mahalakshmi, S.; Harikrishna, N.; Reddy, G. Production, characterization and antimicrobial activity of silver nanoparticles produced by Lactobacillus amylophilus GV6. European J. Pharm. Med. Res. 2016, 3, 236242. [Google Scholar]
- Garmasheva, I.; Kovalenko, N.; Voychuk, S.; Ostapchuk, A.; Livins’ka, O.; Oleschenko, L. Lactobacillus species mediated synthesis of silver nanoparticles and their antibacterial activity against opportunistic pathogens in vitro. BioImpacts BI 2016, 6, 219. [Google Scholar] [CrossRef]
- Naseer, Q.A.; Xue, X.; Wang, X.; Dang, S.; Din, S.U.; Jamil, J. Synthesis of silver nanoparticles using Lactobacillus bulgaricus and assessment of their antibacterial potential. Braz. J. Biol. 2021, 82, e232434. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, N.; Kaushal, N. Comparative Account of Biogenic Synthesis of Silver Nanoparticles Using Probiotics and Their Antimicrobial Activity Against Challenging Pathogens. BioNanoScience 2022, 12, 833–840. [Google Scholar] [CrossRef]
- Kalaiyarasan, T.; Bharti, V.K.; Chaurasia, O.P. Retracted Article: One Pot Green Preparation of Seabuckthorn Silver Nanoparticles (SBT@ AgNPs) Featuring High Stability and Longevity, Antibacterial, Antioxidant Potential: A Nano Disinfectant Future Perspective. RSC Adv. 2017, 7, 51130–51141. [Google Scholar] [CrossRef]
- Kora, A.J.; Arunachalam, J. Assessment of Antibacterial Activity of Silver Nanoparticles on Pseudomonas aeruginosa and Its Mechanism of Action. World J. Microbiol. Biotechnol. 2011, 27, 1209–1216. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. In Bacterial Biofilms; Springer: Berlin/Heidelberg, Germany, 2008; pp. 207–228. [Google Scholar]
- Gupta, P.; Chhibber, S.; Harjai, K. Subinhibitory Concentration of Ciprofloxacin Targets Quorum Sensing System of Pseudomonas aeruginosa Causing Inhibition of Biofilm Formation & Reduction of Virulence. Indian J. Med. Res. 2016, 143, 643. [Google Scholar]
- Pompilio, A.; Crocetta, V.; De Nicola, S.; Verginelli, F.; Fiscarelli, E.; Di Bonaventura, G. Cooperative Pathogenicity in Cystic Fibrosis: Stenotrophomonas Maltophilia Modulates Pseudomonas aeruginosa Virulence in Mixed Biofilm. Front. Microbiol. 2015, 6, 951. [Google Scholar] [CrossRef] [PubMed]
- Nickel, J.C.; Ruseska, I.; Wright, J.B.; Costerton, J.W. Tobramycin Resistance of Pseudomonas aeruginosa Cells Growing as a Biofilm on Urinary Catheter Material. Antimicrob. Agents Chemother. 1985, 27, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Ali, K.; Ahmed, B.; Dwivedi, S.; Saquib, Q.; Al-Khedhairy, A.A.; Musarrat, J. Microwave Accelerated Green Synthesis of Stable Silver Nanoparticles with Eucalyptus globulus Leaf Extract and Their Antibacterial and Antibiofilm Activity on Clinical Isolates. PLoS ONE 2015, 10, e0131178. [Google Scholar] [CrossRef]
- Ali, S.G.; Ansari, M.A.; Khan, H.M.; Jalal, M.; Mahdi, A.A.; Cameotra, S.S. Antibacterial and Antibiofilm Potential of Green Synthesized Silver Nanoparticles against Imipenem Resistant Clinical Isolates of P. aeruginosa. Bionanoscience 2018, 8, 544–553. [Google Scholar] [CrossRef]
- Chhibber, S.; Gondil, V.S.; Sharma, S.; Kumar, M.; Wangoo, N.; Sharma, R.K. A Novel Approach for Combating Klebsiella pneumoniae Biofilm Using Histidine Functionalized Silver Nanoparticles. Front. Microbiol. 2017, 8, 1104. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Jiang, Q.; Chen, J.; Yang, C.; Yin, Y.; Yao, K. Quorum Sensing: A Prospective Therapeutic Target for Bacterial Diseases. BioMed Res. Int. 2019, 2019, 2015978. [Google Scholar] [CrossRef]
- Gajdács, M.; Spengler, G. The Role of Drug Repurposing in the Development of Novel Antimicrobial Drugs: Non-Antibiotic Pharmacological Agents as Quorum Sensing-Inhibitors. Antibiotics 2019, 8, 270. [Google Scholar] [CrossRef]
- Abraham, W.-R. Controlling Pathogenic Gram-Negative Bacteria by Interfering with Their Biofilm Formation. Drug Des. Rev. 2005, 2, 13–33. [Google Scholar] [CrossRef]
- Vijay, K.; Sakshi, S.; Divya, P. Recent Research Advances on Chromobacterium violaceum. Asian Pac. J. Trop. Med. 2017, 10, 810–818. [Google Scholar]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The Role of Pyocyanin in Pseudomonas aeruginosa Infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef] [PubMed]
- De Vleesschauwer, D.; Cornelis, P.; Höfte, M. Redox-Active Pyocyanin Secreted by Pseudomonas aeruginosa 7NSK2 Triggers Systemic Resistance to Magnaporthe Grisea but Enhances Rhizoctonia solani Susceptibility in Rice. Mol. Plant-Microbe Interact. 2006, 19, 1406–1419. [Google Scholar] [CrossRef] [PubMed]
- Usher, L.R.; Lawson, R.A.; Geary, I.; Taylor, C.J.; Bingle, C.D.; Taylor, G.W.; Whyte, M.K.B. Induction of Neutrophil Apoptosis by the Pseudomonas aeruginosa Exotoxin Pyocyanin: A Potential Mechanism of Persistent Infection. J. Immunol. 2002, 168, 1861–1868. [Google Scholar] [CrossRef] [PubMed]
- Manago, A.; Becker, K.A.; Carpinteiro, A.; Wilker, B.; Soddemann, M.; Seitz, A.P.; Edwards, M.J.; Grassmé, H.; Szabo, I.; Gulbins, E. Pseudomonas aeruginosa Pyocyanin Induces Neutrophil Death via Mitochondrial Reactive Oxygen Species and Mitochondrial Acid Sphingomyelinase. Antioxid. Redox Signal. 2015, 22, 1097–1110. [Google Scholar] [CrossRef]
- Ringel, M.T.; Brüser, T. The Biosynthesis of Pyoverdines. Microb. Cell 2018, 5, 424. [Google Scholar] [CrossRef]
- Meyer, J.-M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, I.A. Pyoverdin Is Essential for Virulence of Pseudomonas aeruginosa. Infect. Immun. 1996, 64, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Coin, D.; Louis, D.; Bernillon, J.; Guinand, M.; Wallach, J. LasA, Alkaline Protease and Elastase in Clinical Strains of Pseudomonas Aeruginosa: Quantification by Immunochemical Methods. FEMS Immunol. Med. Microbiol. 1997, 18, 175–184. [Google Scholar] [CrossRef]
- Casilag, F.; Lorenz, A.; Krueger, J.; Klawonn, F.; Weiss, S.; Häussler, S. The LasB Elastase of Pseudomonas Aeruginosa Acts in Concert with Alkaline Protease AprA to Prevent Flagellin-Mediated Immune Recognition. Infect. Immun. 2016, 84, 162–171. [Google Scholar] [CrossRef]
- Caiazza, N.C.; Shanks, R.M.Q.; O’toole, G.A. Rhamnolipids Modulate Swarming Motility Patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 7351–7361. [Google Scholar] [CrossRef]
- Mun, I.R.A.; Hussin, M.S.; Kadum, M.M. Study of Prodigiosin and Virulence Factor Producing by Multi Drug Resistance Serratia marcescens Isolated from Some of Baghdad Hospitals Environment. Al-Mustansiriyah J. Sci. 2008, 19, 1–9. [Google Scholar]
- De La Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.M.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E.W. Inhibition of Bacterial Biofilm Formation and Swarming Motility by a Small Synthetic Cationic Peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, J.; Zhou, X.; Li, Y. Inhibition of Streptococcus mutans Biofilm Formation by Strategies Targeting the Metabolism of Exopolysaccharides. Crit. Rev. Microbiol. 2021, 47, 667–677. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).