In Vitro Activity of Cefiderocol on Multiresistant Bacterial Strains and Genomic Analysis of Two Cefiderocol Resistant Strains
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Activity of Cefiderocol against MDRGram-Negative Bacteria
2.2. Genomic Characteristics of Two Cefiderocol Resistant Klebsiella pneumoniae Isolates
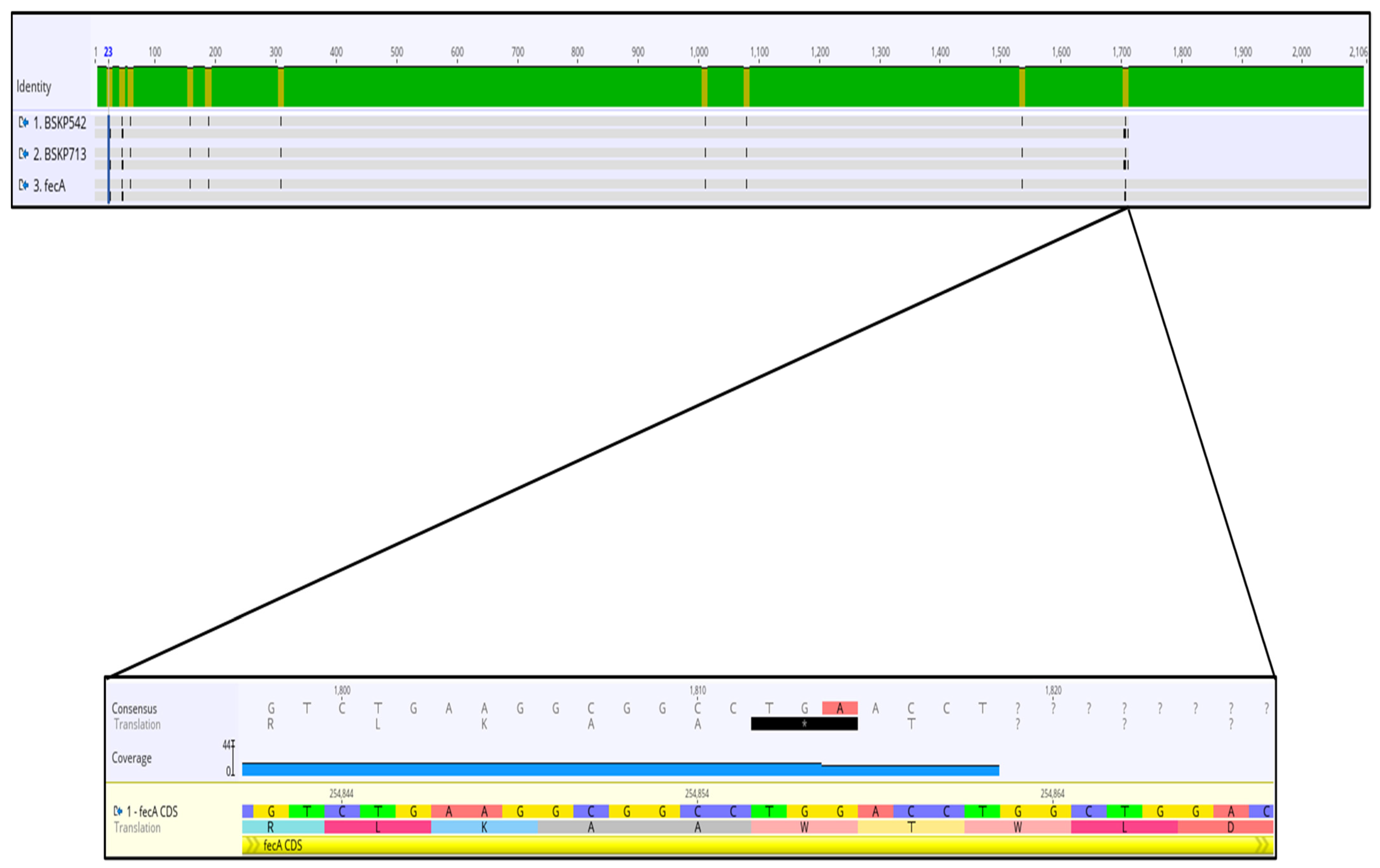
2.3. Analysis of Mutations and Alterations in Genes Involved in Iron Uptake and Transport Detected in Two Cefiderocol-Resistance Klebsiella pneumoniae Isolates
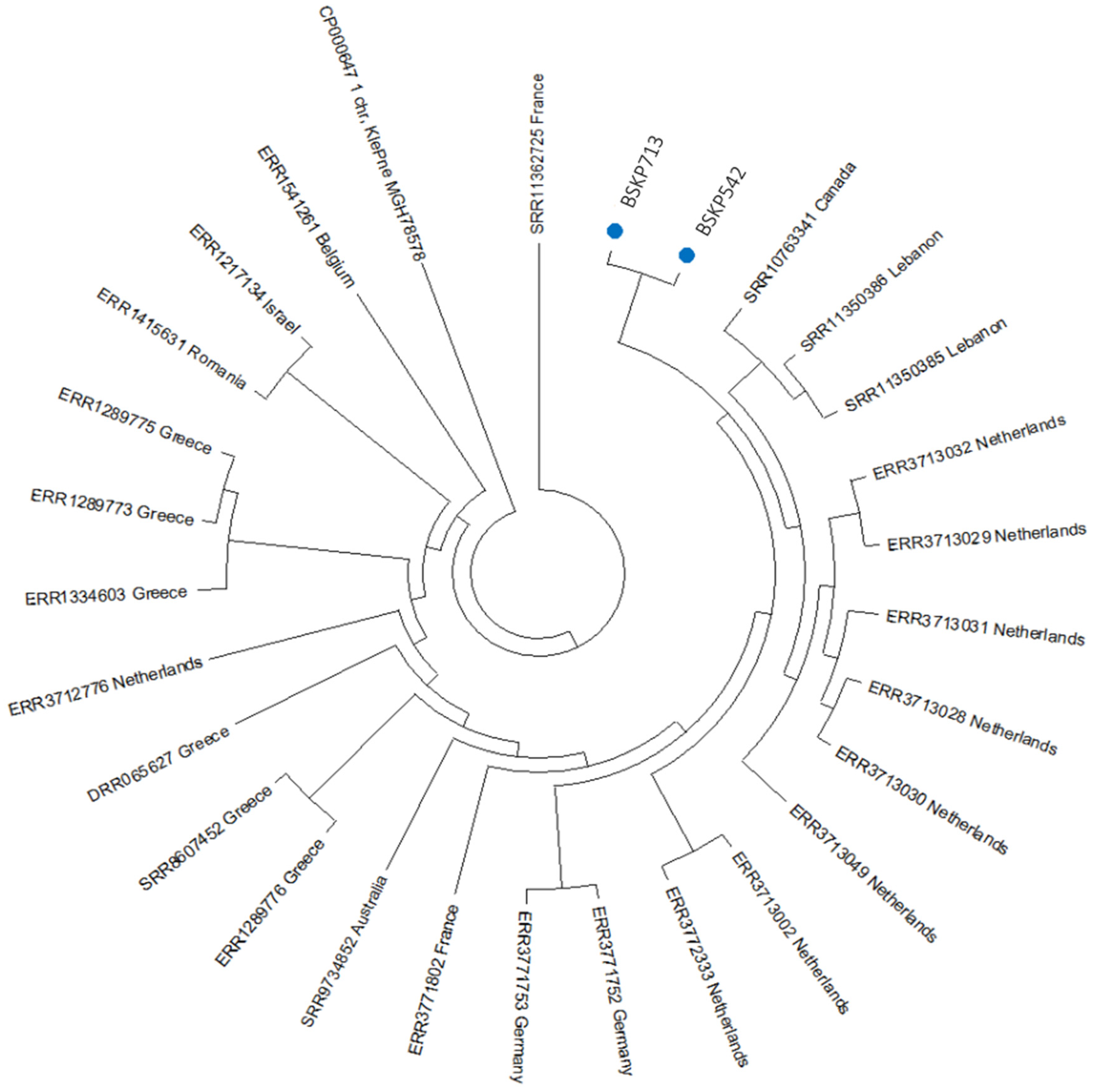
2.4. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Isolates
4.2. Antimicrobial Susceptibility Testing
4.3. Phenotypic Detection of Carbapenemases
4.4. Whole Genome Sequencing (WGS)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 17 November 2021).
- Jean, S.S.; Lee, W.S.; Lam, C.; Hsu, C.W.; Chen, R.J.; Hsueh, P.R. Carbapenemase-producing Gram-negative bacteria: Current epidemics, antimicrobial susceptibility and treatment options. Future Microbiol. 2015, 10, 407–425. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Hsueh, P.R.; SMART Asia-Pacific Group. Distribution of ESBLs, AmpC b-lactamses and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008-14; results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J. Antimicrob. Chemother. 2017, 72, 166–171. [Google Scholar] [PubMed]
- Lee, C.H.; Su, T.Y.; Ye, J.J.; Hsu, P.C.; Kuo, A.J.; Chia, J.H.; Lee, M.H. Risk factors and clinical significance of bacteremia caused by Pseudomonas aeruginosa resistant only to carbapenems. J. Microbiol. Immunol. Infect. 2017, 50, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.P.; Wang, S.F.; Ma, L.; Wang, T.Y.; Yang, T.Y.; Siu, L.K.; Chuang, Y.C.; Lee, P.S.; Wang, J.T.; Wu, T.L.; et al. The plasmid-mediated fosfomycin resistance determinants and synergy of fosfomycin and meropenem in carbapenem-resistant Klebsiella pneumoniae isolates in Taiwan. J. Microbiol. Immunol. Infect 2017, 50, 653–661. [Google Scholar] [CrossRef]
- Ku, Y.H.; Chen, C.C.; Lee, M.F.; Chuang, Y.C.; Tang, H.J.; Yu, W.L. Comparison of synergism between colistin, fosfomycin and tigecycline against extended-spectrum b-lactamase-producing Klebsiella pneumoniae isolates or with carbapenem resistance. J. Microbiol. Immunol. Infect. 2017, 50, 931–939. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef]
- De la Calle, C.; Rodríguez, O.; Morata, L.; Marco, F.; Cardozo, C.; García-Vidal, C.; Río, A.D.; Feher, C.; Pellicé, M.; Puerta-Alcade, P.; et al. Clinical characteristics and prognosis of infections caused by OXA-48 carbapenemase producing Enterobacteriaceae in patients treated with ceftazidime-avibactam. Int. J. Antimicrob. Agents 2019, 53, 520–524. [Google Scholar] [CrossRef]
- Syue, L.S.; Chen, Y.H.; Ko, W.C.; Hsueh, P.R. New drugs for the treatment of complicated intra-abdominal infections in the era of increasing antimicrobial resistance. Int. J. Antimicrob. Agents 2016, 47, 250–258. [Google Scholar] [CrossRef]
- Cassini, A.; Diaz Högberg, L.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Skov Simonsen, G.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleeschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- ISS 2021. Available online: https://www.epicentro.iss.it/antibiotico-resistenza/epidemiologia-italia (accessed on 17 November 2022).
- El Labadidi, R.M.; Rizk, J.G. Cefiderocol: A siderophore cephalosporin. Ann. Pharmacother. 2020, 54, 1215–1231. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Scweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A siderophore cephalosporin with activity against carbapenem resistant and multi-drug resistant Gram-negative bacilli. Drugs 2019, 79, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Toba, S.; Sato, T.; Ota, M.; Takemura, M.; Nishikawa, T.; Toba, S.; Kohira, N.; Miyagawa, S.; Ishibashi, N.; et al. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria. Antimicrob. Agents Chemother. 2018, 62, e01454-17. [Google Scholar] [CrossRef] [PubMed]
- McElheny, C.L.; Fowler, E.L.; Iovleva, A.; Shields, R.K.; Doy, Y. In vitro evolution of cefiderocol resistance in an NDM-producing Klebsiella pneumoniae due to functional loss of CirA. Microbiol. Spectr. 2021, 9, e0177921. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 132, 4188. [Google Scholar] [CrossRef]
- Tian, D.; Wang, B.; Zhang, H.; Pan, F.; Wang, C.; Shi, Y.; Sun, Y. Dissemination of the blaNDM-5 gene via IncX3-type plasmid among Enterobacteriaceae in children. mSphere 2020, 5, e00699-19. [Google Scholar] [CrossRef]
- Coppi, M.; Antonelli, A.; Niccolai, C.; Bartolini, A.; Bartolini, L.; Grazzini, M.; Mantengoli, E.; Farese, A.; Pieralli, F.; Mechi, M.T.; et al. Nosocomial outbreak by NDM-1-producing Klebsiella pneumoniae highly resistant to cefiderocol, Florence, Italy, August 2021 to June 2022. Euro. Surveill. 2022, 27, 2200795. [Google Scholar] [CrossRef]
- Simner, P.J.; Beisken, S.; Bergman, Y.; Ante, M.; Posch, A.E.; Tamma, P.D. Defining baseline mechanisms of cefiderocol resistance in the Enterobacterales. Microb. Drug Resist. 2022, 28, 161–170. [Google Scholar] [CrossRef]
- Lan, P.; Lu, Y.; Chen, Z.; Wu, X.; Hua, X.; Jiang, Y.; Zhou, J.; Yu, Y. Emergence of high-level cefiderocol resistance in carbapenem-resistant Klebsiella pneumonia from bloodstream infections in patients with hematologic malignancies in China. Microbiol. Spectr. 2022, 10, e0008422. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Rousaki, M.; Kritsotakis, E.I. Cefiderocol: Systematic review of mechanism of resistance, heteroresistance and in vivo emergence of resistance. Antibiotics 2022, 11, 723. [Google Scholar] [CrossRef]
- Bassetti, M.; Echols, R.; Matsunaga, Y.; Ariyasu, M.; Doi, Y.; Ferrer, R.; Lodise, T.P.; Naas, T.; Niki, Y.; Paterson, D.L.; et al. Efficacy and safety of cefiderocol or best available therapy for the treatment od serious infections caused by carbapenem-resistant Gram-negative bacteria (CREDIBLE-CR): A randomized, open-label, multicentre, pathogen focused, descriptive phase 3 trial. Lancet Infect. Dis. 2020, 21, 226–240. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Hackel, M.A.; Tsuji, M.; Yamano, Y.; Echols, R.; Sahm, D.F. In Vitro activity of cefiderocol, a siderophore cephalosporin, against Gram-negative bacilli isolated by clinical laboratories in North America and Europe in 2015–2016: SIDERO-WT-2015. Int. J. Antimicrob. Agents 2019, 53, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Yamano, Y. In vitro activity against a broad range of clinically important Gram-negative bacteria. Clin. Infect. Dis. 2019, 69, S544–S551. [Google Scholar] [CrossRef] [PubMed]
- Albano, M.; Karau, M.J.; Schuetz, A.N.; Patel, R. Comparison of agar dilution to broth microdilution for testing in vitro activity of cefiderocol against Gram-negative bacilli. J. Clin. Microbiol. 2020, 59, e00966-20. [Google Scholar] [CrossRef] [PubMed]
- Longshaw, C.; Manissero, D.; Tsuji, M.; Echols, R.; Yamano, Y. In Vitro activity of the siderophore cephalosporin, cefiderocol, against molecularly charcterized, carbapenem-non-susceptible Gram-negative bacteria from Europe. JAC Antimicrob. Resist. 2020, 2, dlaa060. [Google Scholar] [CrossRef]
- Mushtaq, S.; Sadouki, Z.; Vickers, A.; Livermore, D.M.; Woodford, N. In Vitro activity of cefiderocol, a siderophore cephalosporin, against multidrug-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 2020, 64, e01582-20. [Google Scholar] [CrossRef]
- Kohira, N.; West, J.; Ito-Horiyama, T.; Nakamura, R.; Sato, T.; Rittenhouse, S.; Tsuji, M.; Yamano, Y. In Vitro antimicrobial activity of a siderophore cephalosporin, s-649266, against enterobacteriaceae clinical isolates, including carbapenem-resistant strains. Antimicrob. Agents Chemother. 2016, 60, 729–734. [Google Scholar] [CrossRef]
- Kazmierczak, K.M.; Tsuji, M.; Wise, M.G.; Hackel, M.; Yamano, Y.; Echols, R.; Sahm, D.F. In Vitro activity of cefiderocol, a siderophore cephalosporin, against a recent collection of clinically relevant carbapenem-non-susceptibel Gram-negative bacilli, including serine carbapenemases-and metallo-beta-lactamase-producing isolates (Sidero-WT-2014 Study). Int. J. Antimicrob. Agents 2019, 53, 177–184. [Google Scholar]
- Iregui, A.; Khan, Z.; Landman, D.; Quale, J. Activity of cefiderocol against enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii endemic to medical centers in New York City. Microb. Drug Resist. 2020, 26, 722–726. [Google Scholar] [CrossRef]
- Golden, A.R.; Adam, H.J.; Baxter, M.; Walkty, A.; Lagacé-Wiens, P.; Karlowsky, J.A.; Zhanel, G.G. In Vitro activity of cefiderocol, a novel siderophore cephalosporin, against gram-negative bacilli isolated from patients in Canadian Intensive Care Units. Diagn. Microbiol. Infect Dis. 2020, 97, 115012. [Google Scholar] [CrossRef]
- Delgado-Valverde, M.; Conejo, M.D.C.; Serrano, L.; Fernández-Cuenca, F.; Pascual, Á. Activity of cefiderocol against high-risk clones of multidrug-resistant Enterobacterales, Acinetobacter baumannii, Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2020, 75, 1840–1849. [Google Scholar] [CrossRef]
- Hsueh, S.C.; Lee, Y.J.; Huang, Y.T.; Liao, C.H.; Tsuji, M.; Hsueh, P.R. In Vitro activities of cefiderocol, ceftolozane/tazobactam, ceftazidime/avibactam and other comparative drugs against imipenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii, and Stenotrophomonas maltophilia, all associated with bloodstream infections in Taiwan. J. Antimicrob. Chemother. 2019, 74, 380–386. [Google Scholar] [PubMed]
- Hackel, M.A.; Tsuji, M.; Yamano, Y.; Echols, R.; Karlowsky, J.A.; Sahm, D.F. In Vitro activity of the siderophore cephalosporin, cefiderocol, against carbapenem non susceptible and multidrug-resistant isolates of gram-negative bacilli collected worldwide in 2014 to 2016. Antimicrob. Agents Chemother. 2018, 62, 17. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.T.; Lin, C.Y.; Chen, Y.H.; Hsueh, P.R. Update on infections caused by Stenotrophomans maltophilia with particular attention to resistance mechanisms and therapeutic options. Front Microbiol. 2015, 6, 893. [Google Scholar] [CrossRef]
- Cho, S.Y.; Kang, C.I.; Ha, Y.E.; Chung, D.R.; Lee, N.Y.; Peck, K.R.; Song, J.H. Can levofloxacin be a useful alternatie to trimethoprim-sulfamethoxazole for treating Stenotrophomonas maltophilia bacteremia? Antimicrob. Agents Chemother. 2014, 58, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Kawai, A.; McElheny, C.L.; Iovleva, A.; Kline, E.G.; Slui-Cremer, N.; Shields, R.K.; Doi, Y. Structural basis of reduced susceptibility to ceftazidime-avibactam and cefiderocol in Enterobacter cloacae due to AmpC R2 loop deletion. Antimicrob. Agents Chemother. 2020, 64, e00198-20. [Google Scholar] [CrossRef]
- Shields, R.K.; Iovleva, A.; Kline, E.G.; Kawai, A.; McElheny, C.L.; Doi, Y. Clinical evolution of AmpC-mediated ceftazidime-avibactam and cefiderocol resistance in Enterobacter cloacae complex following exposure to cefepime. Clin. Infect Dis. 2020, 71, 2713–2716. [Google Scholar] [CrossRef]
- Streling, A.P.; Obaidi, M.M.; Lainhart, W.D.; Zangeneh, T.; Khan, A.; Dinh, A.Q.; Hanson, B.; Arias, C.A.; Millerm, W.R. Evolution of cefiderocol non-susceptibility in Pseudomonas aeruginosa in a patient without previous exposure to the antibiotic. Clin. Infect. Dis. 2021, 73, e4472–e4474. [Google Scholar] [CrossRef]
- Luscher, A.; Moynié, L.; Saint Auguste, P.; Bumann, D.; Mazza, L.; Pletzer, D.; Naismith, J.H.; Köhler, T. TonB-dependent receptor repertoire of Pseudomonas aeruginosa for uptake of siderophore-drug conjugate. Antimicrob. Agents Chemother. 2018, 62, e00097-18. [Google Scholar] [CrossRef]
- Kohira, N.; Hackel, M.A.; Ishioka, Y.; Kuroiwa, M.; Sahm, D.F.; Sato, T.; Maki, H.; Yamano, Y. Reduced susceptibility mechanism to cefiderocol, a siderophore cephalosporin, among clinical isolates from a global surveillance programme (SIDERO-WT-2014). J. Glob. Antimicrob. Resist. 2020, 22, 738–741. [Google Scholar] [CrossRef]
- Klein, S.; Boutin, S.; Kocer, K.; Fiedler, M.O.; Störzinger, D.; Weigand, M.A.; Tan, B.; Richter, D.; Rupp, C.; Mieth, M.; et al. Rapid development of cefiderocol resistance in carbapenem- resistant Enterobacter cloacae during therapy is associated with heterogeneous mutations in the catecholate siderophore receptor cirA. Clin. Infect. Dis. 2022, 74, 905–908. [Google Scholar] [CrossRef]
- Huang, W.C.; Wong, M.Y.; Wang, S.H.; Hashimoto, M.; Lin, M.H.; Lee, M.F.; Wu, J.J.; Wang, M.C.; Lin, W.H.; Jeng, S.L.; et al. The Ferric Citrate Uptake System Encoded in a Novel blaCTX–M–3- and blaTEM–1-Harboring Conjugative Plasmid Contributes to the Virulence of Escherichia coli. Front Microbiol. 2021, 12, 667782. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Recommendations for MIC Detemination of Colistin (Polymyxin E) as Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. EUCAST 2016; org 1-1. Available online: www.eucast.org (accessed on 22 March 2016).
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Seven Informational Supplement M100-S28; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoints for Cefiderocol from EUCAST. Addendum (May 20202) to EUCAST Breakpoint Tables V.10.0. Breakpoints to Be Included in EUCAST Breakpoint Tables v 11.0. January 2021. Available online: https://www.eucast.org/eucast_news/news_singleview?tx_ttnews%5Btt_news%5D=374&cHash=839808489fb6fb1033af6d8a1c5e7d07 (accessed on 26 September 2022).
- Brolund, A.; Lagerqvist, N.; Byfors, S.; Stuelens, M.J.; Monnet, D.L.; Albiger, B.; Kohlenberg, A. Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. EuroSurveill 2019, 9, 1900123. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Kang, D.; Kim, D. Performance evaluation of the newly developed in vitro rapid diagnostic test for detecting OXA-48-like, KPC-, NDM-, VIM- and IMP- type carbapenemases: The RESIST-5 O.K.N.V.I. multiplex lateral flow assay. Antibiotics 2021, 10, 460. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Gardner, S.N.; Slezak, T.; Hall, B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment of reference genome. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef]
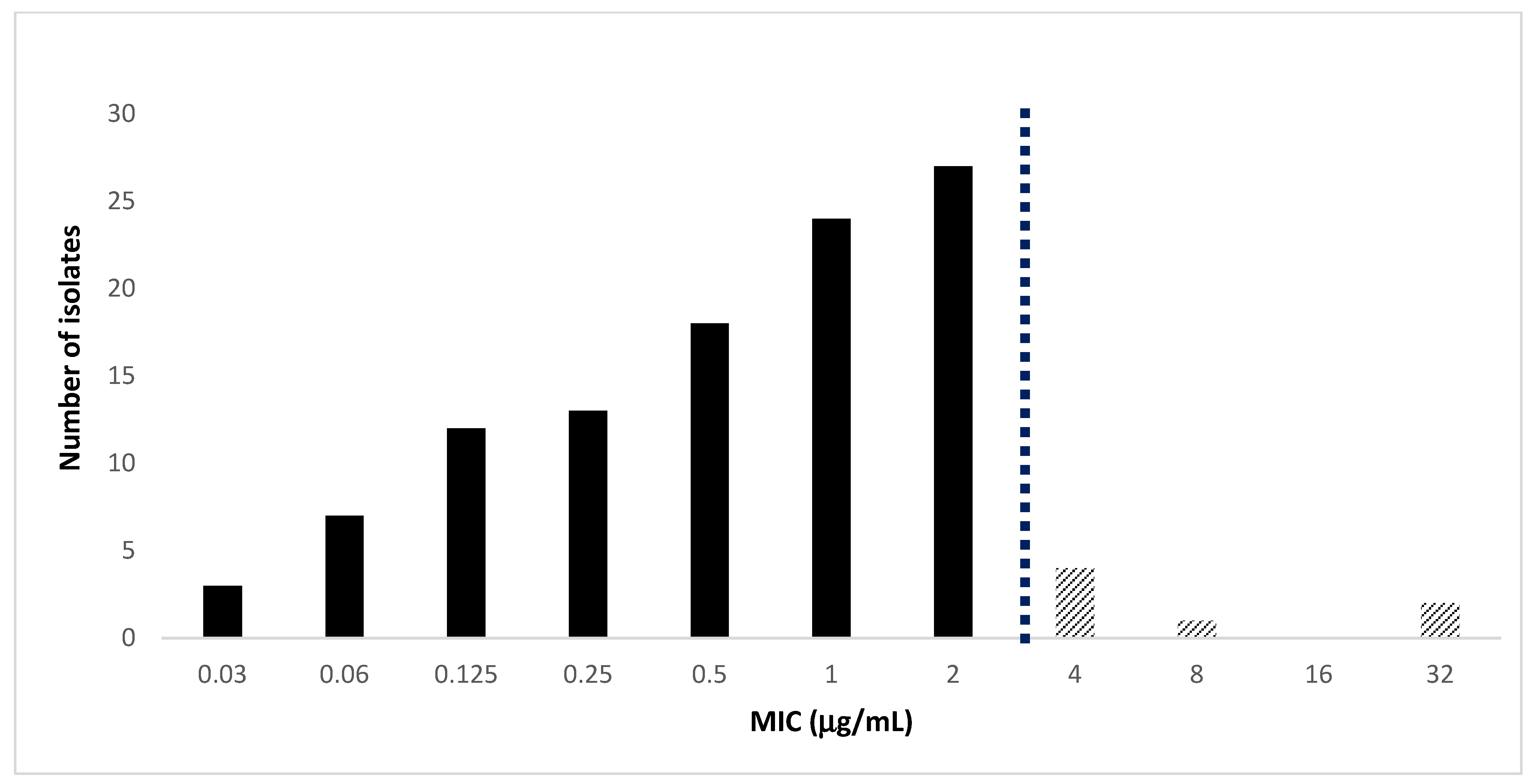
| Organisms (N° of Isolates) | Drug | MIC Range (μg/mL) | MIC50 | MIC90 | Susceptible a (%) | Resistant a (%) |
|---|---|---|---|---|---|---|
| K. pneumoniae (n = 56) | CFDC | 0.03–4 | 1 | 2 | 89 | 11 |
| AN | <1–32 | 32 | 32 | 27 | 73 | |
| AMC | >16 | >16 | >16 | 0 | 100 | |
| FEP | 0.12 to >16 | >16 | 32 | 0 | 100 | |
| CT | >8 to >16 | >16 | >16 | 0 | 100 | |
| CAZ | >32 | >32 | >32 | 0 | 100 | |
| CZA | 0.5 to >16 | >16 | >16 | 29 | 71 | |
| CIP | <0.06 to >2 | >2 | >2 | 4 | 96 | |
| GEM | <1 to >8 | 2 | >8 | 52 | 48 | |
| IPM | <0.25 to >8 | >8 | >8 | 2 | 98 | |
| MEM | 4 to >128 | >8 | >128 | 2 | 98 | |
| TZP | >64 | >64 | >64 | 0 | 100 | |
| CST | <0.5 to >8 | 0.5 | 1 | 98 | 2 | |
| SXT | <20 to >160 | >160 | >160 | 20 | 80 | |
| E. coli (n = 8) | CFDC | 0.03–32 | NC | NC | 88 | 12 |
| AN | 2–16 | NC | NC | 88 | 12 | |
| AMC | >16 | NC | NC | 0 | 100 | |
| FEP | 0.12 to >16 | NC | NC | 12 | 88 | |
| CT | <0.25 to >16 | NC | NC | 50 | 50 | |
| CAZ | 8 to >32 | NC | NC | 0 | 100 | |
| CZA | 0.12 to >8 | NC | NC | 50 | 50 | |
| CIP | >2 | NC | NC | 0 | 100 | |
| GEM | <1 to >8 | NC | NC | 63 | 37 | |
| IPM | <0.25 to >8 | NC | NC | 50 | 50 | |
| MEM | <0.25 to >8 | NC | NC | 50 | 50 | |
| TZP | <4 to >64 | NC | NC | 37 | 63 | |
| CST | 0.5 | NC | NC | 100 | 0 | |
| SXT | <20 to >160 | NC | NC | 12 | 88 | |
| Enterobacter spp. (n = 3) | CFDC | 0.12–2 | NC | NC | 100 | 0 |
| AN | <1 | NC | NC | 100 | 0 | |
| AMC | >16 | NC | NC | 0 | 100 | |
| FEP | >16 | NC | NC | 0 | 100 | |
| CT | >16 | NC | NC | 0 | 100 | |
| CAZ | >32 | NC | NC | 0 | 100 | |
| CZA | >8 | NC | NC | 0 | 100 | |
| CIP | 1 to >2 | NC | NC | 0 | 100 | |
| IPM | >8 | NC | NC | 0 | 100 | |
| MEM | >8 | NC | NC | 0 | 100 | |
| TZP | >64 | NC | NC | 0 | 100 | |
| CST | 0.5 | NC | NC | 100 | 0 | |
| SXT | >160 | NC | NC | 0 | 100 | |
| P. aeruginosa (n = 33) | CFDC | 0.03–2 | 1 | 2 | 100 | 0 |
| AN | <1 to >32 | 4 | >32 | 84 | 16 | |
| FEP | 4 to >16 | >16 | >16 | 0 | 100 | |
| CT | 0.5 to >64 | >8 | >16 | 40 | 60 | |
| CAZ | 2 to >32 | >32 | >32 | 0 | 100 | |
| CZA | 2 to >64 | >8 | >8 | 28 | 72 | |
| CIP | 0.5 to >2 | >2 | >2 | 0 | 100 | |
| IPM | 2 to >8 | >8 | >8 | 0 | 100 | |
| MEM | 1 to >8 | >8 | >8 | 0 | 100 | |
| TZP | 32 to >64 | >64 | >64 | 0 | 100 | |
| CST | <0.5 to >8 | 0.5 | 2 | 94 | 6 | |
| A. baumannii (n = 2) | CFDC | <=2 | NC | NC | 100 | 0 |
| AN | >32 | NC | NC | 0 | 100 | |
| FEP | >16 | NC | NC | 0 | 100 | |
| TIG | 1 | NC | NC | NA | NA | |
| CAZ | >32 | NC | NC | 0 | 100 | |
| ATM | >256 | NC | NC | NA | NA | |
| CIP | >2 | NC | NC | 0 | 100 | |
| IPM | >8 | NC | NC | 0 | 100 | |
| MEM | >8 to >256 | NC | NC | 0 | 100 | |
| TZP | >64 | NC | NC | 0 | 100 | |
| CST | 0.5 | NC | NC | 100 | 0 | |
| A. xylosoxidans (n = 1) | CFDC | 0.06 | NC | NC | 100 | 0 |
| AN | >16 | NC | NC | NA | NA | |
| AMC | >16 | NC | NC | NA | NA | |
| FEP | >16 | NC | NC | NA | NA | |
| CT | >64 | NC | NC | NA | NA | |
| CAZ | 16 | NC | NC | NA | NA | |
| CZA | 16 | NC | NC | NA | NA | |
| CIP | >1 | NC | NC | NA | NA | |
| GEM | >8 | NC | NC | NA | NA | |
| MEM | 8 | NC | NC | 0 | 100 | |
| TZP | >128 | NC | NC | 0 | 100 | |
| CST | 2 | NC | NC | NA | NA | |
| SXT | <1 | NC | NC | NA | NA | |
| S. maltophilia (n = 7) | CFDC | 0.5 to <2 | NC | NC | 100 | 0 |
| AN | >32 | NC | NC | NA | NA | |
| CT | >32 | NC | NC | NA | NA | |
| CAZ | >32 | NC | NC | 0 | 100 | |
| CZA | >16 | NC | NC | NA | NA | |
| MEM | >32 to >64 | NC | NC | NA | NA | |
| CST | >8 | NC | NC | NA | NA | |
| SXT | 0.01 to >4 | NC | NC | 71 | 29 |
| (a) | |||||
|---|---|---|---|---|---|
| Isolates | ST | Serotype | AMR Determinants | ||
| Gene Function | Detected Genes | Hypothetical Location | |||
| BSKP542 | 383 | HL30, O1V2 | aminoglycoside resistance | aac (6′)-1b’ | pNDM-MAR |
| aadA2 | pNDM-5-IT | ||||
| aph(3′)-VI | chromosome | ||||
| armA | chromosome | ||||
| beta-lactam resistance | blaNDM-1 | pNDM-MAR | |||
| blaCTX-M15 | pNDM-MAR | ||||
| blaTEM-1b | pKpQIL-IT | ||||
| blaTem-1c | pKpQIL-IT | ||||
| blaOXA-48 | pOXA-48 | ||||
| blaNDM-5 | pNDM-5-IT | ||||
| blaSHV-26 | chromosome | ||||
| blaSHV-78 | chromosome | ||||
| blaSHV-98 | chromosome | ||||
| blaSHV-179 | chromosome | ||||
| blaSHV-145 | chromosome | ||||
| blaSHV-194 | chromosome | ||||
| blaSHV-199 | chromosome | ||||
| blaOXA-9 | chromosome | ||||
| blaCTX-M14b | chromosome | ||||
| ompK36 | chromosome | ||||
| ompK37 | chromosome | ||||
| macrolide resistance | mph(A) | pNDM-5-IT | |||
| mph(E) | pNDM-5-IT | ||||
| msr(E) | chromosome | ||||
| quinolone resistance | qnrB19 | pHAD28 | |||
| qnrB1 | pNDM-MAR | ||||
| qnrS1 | chromosome | ||||
| parC | chromosome | ||||
| gyrA | chromosome | ||||
| tetracycline resistance | tet(A) | pNDM-5-IT | |||
| fosfomycin resistance | fosA | chromosome | |||
| phenicol resistance | catA1 | pNDM-MAR | |||
| trimethoprim resistance | dfrA5 | chromosome | |||
| sulphonamide resistance | sul1 | pNDM-5-IT | |||
| sul2 | chromosome | ||||
| BSKP713 | 6339 | HL30, O1V2 | aminoglycoside resistance | aac (6′)-1b’ | pNDM-MAR |
| aadA2 | pNDM-5-IT | ||||
| aph(3′)-VI | chromosome | ||||
| armA | chromosome | ||||
| beta-lactam resistance | blaNDM-1 | pNDM-MAR | |||
| blaCTX-M15 | pNDM-MAR | ||||
| blaTEM-1b | pKpQIL-IT | ||||
| blaTem-1c | pKpQIL-IT | ||||
| blaOXA-48 | pOXA-48 | ||||
| blaNDM-5 | pNDM-5-IT | ||||
| blaSHV-26 | chromosome | ||||
| blaSHV-78 | chromosome | ||||
| blaSHV-98 | chromosome | ||||
| blaSHV-179 | chromosome | ||||
| blaSHV-145 | chromosome | ||||
| blaSHV-194 | chromosome | ||||
| blaSHV-199 | chromosome | ||||
| blaOXA-9 | chromosome | ||||
| blaCTX-M14b | chromosome | ||||
| ompK36 | chromosome | ||||
| ompK37 | chromosome | ||||
| macrolide resistance | mph(A) | pNDM-5-IT | |||
| mph(E) | pNDM-5-IT | ||||
| msr(E) | chromosome | ||||
| quinolone resistance | qnrB19 | pHAD28 | |||
| qnrB1 | pNDM-MAR | ||||
| qnrS1 | chromosome | ||||
| tetracycline resistance | tet(A) | pNDM-5-IT | |||
| fosfomycin resistance | fosA | chromosome | |||
| phenicol resistance | catA1 | pNDM-MAR | |||
| trimethoprim resistance | dfrA5 | chromosome | |||
| sulphonamide resistance | sul1 | pNDM-5-IT | |||
| sul2 | chromosome | ||||
| (b) | |||||
| Isolates | Virulence Determinants | ||||
| Gene Function | Detected Genes | ||||
| BSKP542 and BSKP713 | Adhesion | ||||
| type 1 fimbriae | mrkABCDFHJ | ||||
| type 3 fimbriae | fimABCDEFGHIK | ||||
| type 4 pili | pilW | ||||
| fimbrial adherence determinants | stbABCDE | ||||
| Iron uptake | |||||
| aerobactin | iutA | ||||
| ent siderophore | entABCDEFS; fepABCDG; fes | ||||
| salmochelin | iroEN | ||||
| Regulatory system | rcsAB | ||||
| Secretion system | T6SS (I-III) | ||||
| Efflux pump genes | |||||
| RND efflux pump | arcAB | ||||
| Genes | Function | Mutations/Alterations | |
|---|---|---|---|
| BSKP542 | BSKP713 | ||
| fhuA | Iron uptake [18] | V176F, I178V, I212L, G269D, V609G | V176F, I178V, I212L, G269D, V609G |
| fepA | Iron uptake [18] | P531A | P531A |
| fbpA | Iron uptake [18] | WT | WT |
| efeO | Iron uptake [18] | WT | WT |
| exbB | Iron uptake [18] | WT | WT |
| exbD | TonB-dependent energy transduction system reported to affect the function of Iron transporters [18] | WT | WT |
| fiuA | Iron uptake [18] | Absent | Absent |
| fur | Iron uptake [18] | WT | WT |
| iutA | Iron uptake [18] | E160K, S285P | E160K, S285P |
| baeS | Encodes a sensor kinase protein of the two-component BaeSR signal transduction system [18,19] | L366H | L366H |
| envZ | Two-component transcriptional regulator reported to affect the expression of iron transporters [18,19] | R289C | R289C |
| cirA | Encodes receptor which preferentially transports catecholate siderophores [18,19] | A134V, N558D | A134V, N558D |
| feoA | Ferrous iron uptake [18] | WT | WT |
| sitC * | Iron/manganese ABC transporter permease subunit [18] | Y11H, R166C | Y11H, R166C |
| apbC | Iron-sulfur cluster carrier protein [18] | P334S, I339T | P334S, I339T |
| fepG | Iron-enterobactin ABC transporter permease [18] | V63M, G206S | V63M, G206S |
| fepC | Iron-enterobactin ABC transporter ATP-binding protein [18] | T9A, I182N | T9A, I182N |
| fetB | Iron export ABC transporter permease subunit FetB [18] | WT | WT |
| fetA | Iron ABC transporter ATP-binding protein FetA [18] | S27N | S27N |
| fecA | TonB energy transducing system-dependent ferric citrate uptake receptor [20] | A9V, V16L, premature STOP codon in position aa569 | A9V, V16L, premature STOP codon in position aa569 |
| tonB | Component of inner membrane protein complex providing energy to TonB dependent transporters [19] | G60A, A80V, insertion of 4 aa in position 103 (PKPK), P168A, E219Q | G60A, A80V, insertion of 4 aa in position 103 (PKPK), P168A, E219Q |
| fiu | Encodes receptor that preferentially transports catecholate siderophores [18,19] | D387N | D387N |
| ompR | Two-component transcriptional regulator reported to affect the expression of iron transporters [19] | A72V | A72V |
| yicI | Transporter family [18,19] | S30N, L323Q, K358N, G624C, H653R | S30N, L323Q, K358N, G624C, H653R |
| yicJ | Transporter family [18,19] | P102L, F181Y | P102L, F181Y |
| yicL | Transporter family [18,19] | WT | WT |
| chrA * | Heavy metals transporter [19,20] | A245V, G344A | A245V, G344A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padovani, M.; Bertelli, A.; Corbellini, S.; Piccinelli, G.; Gurrieri, F.; De Francesco, M.A. In Vitro Activity of Cefiderocol on Multiresistant Bacterial Strains and Genomic Analysis of Two Cefiderocol Resistant Strains. Antibiotics 2023, 12, 785. https://doi.org/10.3390/antibiotics12040785
Padovani M, Bertelli A, Corbellini S, Piccinelli G, Gurrieri F, De Francesco MA. In Vitro Activity of Cefiderocol on Multiresistant Bacterial Strains and Genomic Analysis of Two Cefiderocol Resistant Strains. Antibiotics. 2023; 12(4):785. https://doi.org/10.3390/antibiotics12040785
Chicago/Turabian StylePadovani, Michela, Anna Bertelli, Silvia Corbellini, Giorgio Piccinelli, Francesca Gurrieri, and Maria Antonia De Francesco. 2023. "In Vitro Activity of Cefiderocol on Multiresistant Bacterial Strains and Genomic Analysis of Two Cefiderocol Resistant Strains" Antibiotics 12, no. 4: 785. https://doi.org/10.3390/antibiotics12040785
APA StylePadovani, M., Bertelli, A., Corbellini, S., Piccinelli, G., Gurrieri, F., & De Francesco, M. A. (2023). In Vitro Activity of Cefiderocol on Multiresistant Bacterial Strains and Genomic Analysis of Two Cefiderocol Resistant Strains. Antibiotics, 12(4), 785. https://doi.org/10.3390/antibiotics12040785






