Screening of Antibiotic and Virulence Genes from Whole Genome Sequenced Cronobacter sakazakii Isolated from Food and Milk-Producing Environments
Abstract
1. Introduction
2. Results
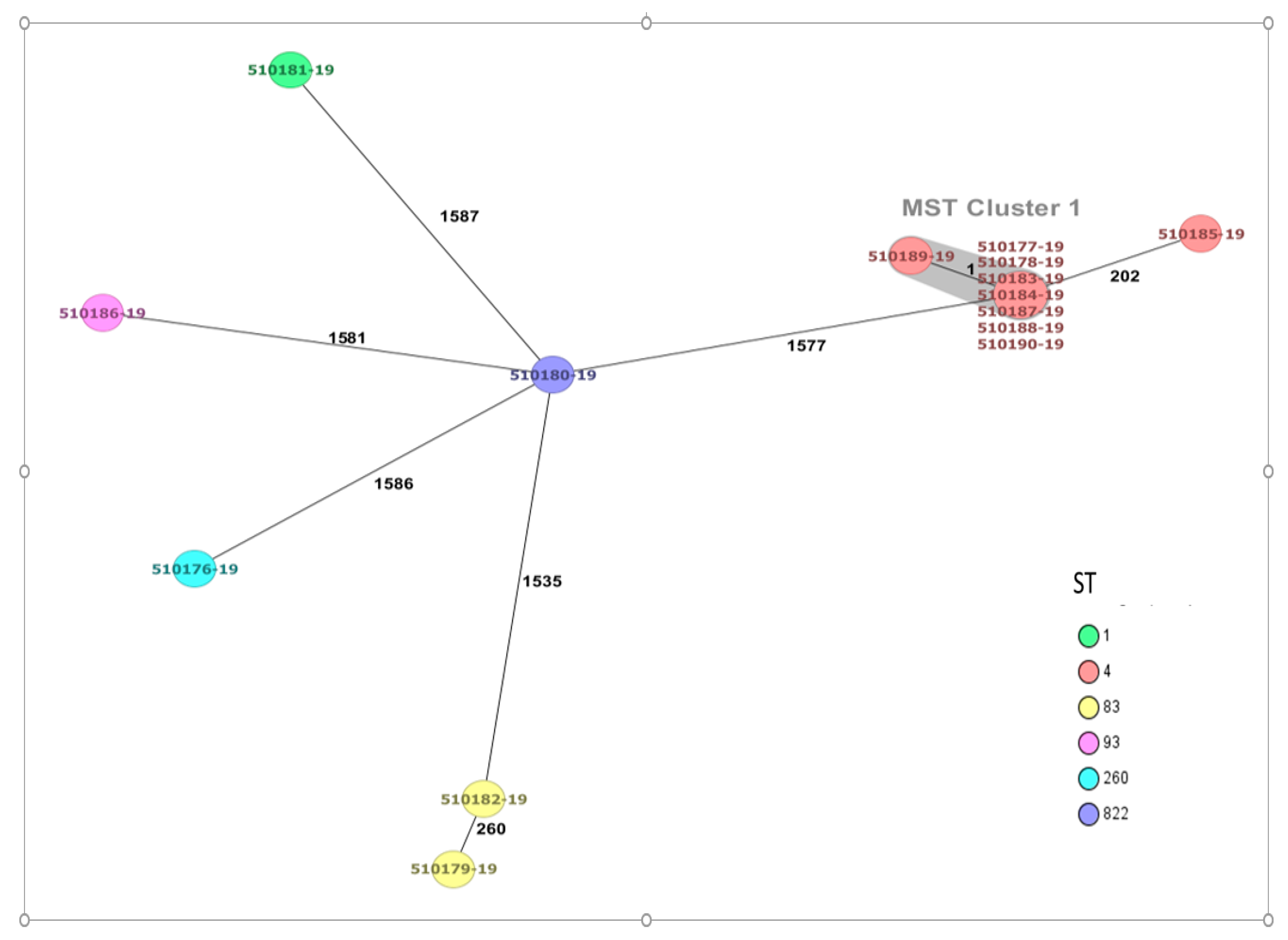
2.1. Sampling and Identification of Isolates
2.2. Antibiotic Resistance Profile
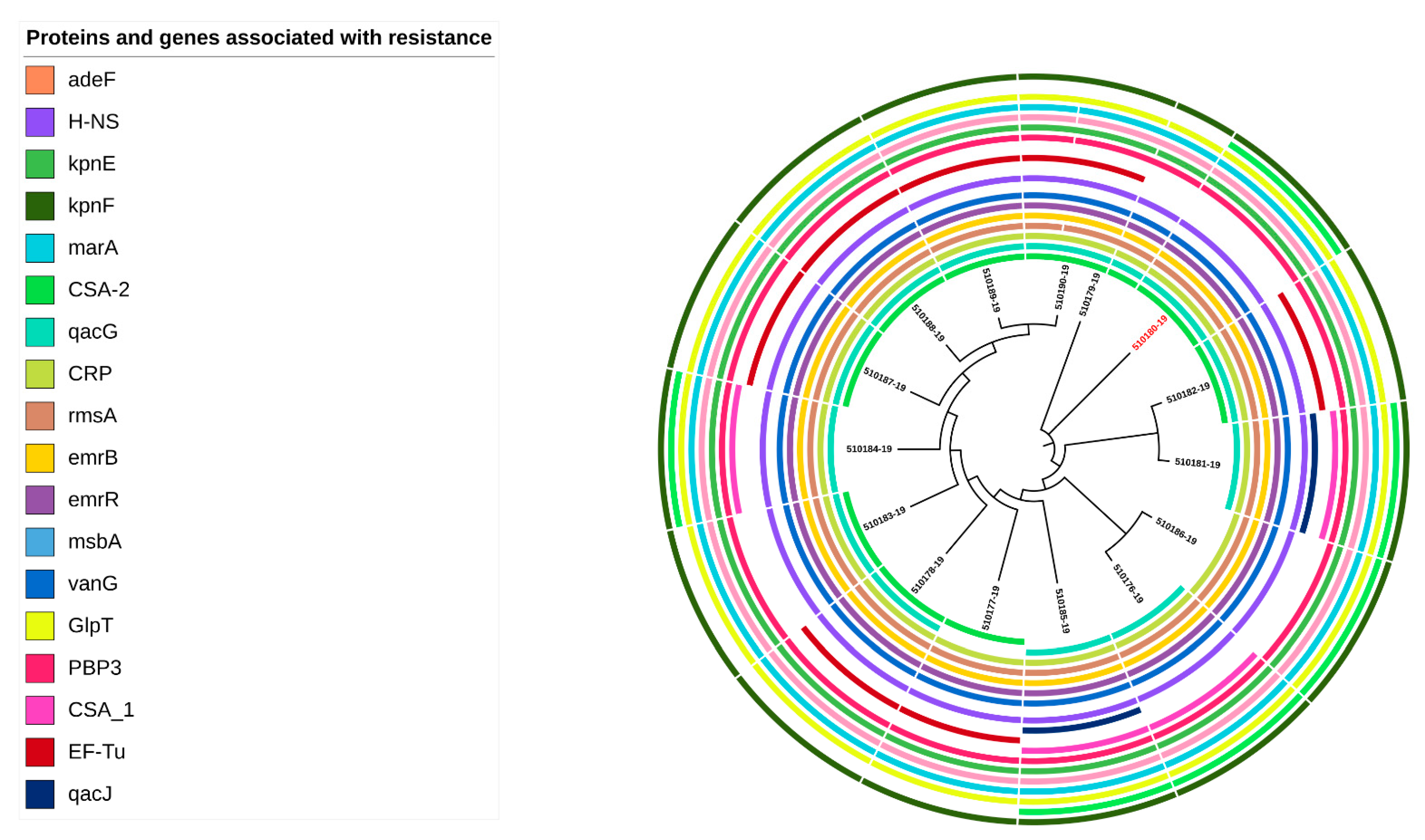
2.3. Antibiotic Genes Detected in C. sakazakii Genomes
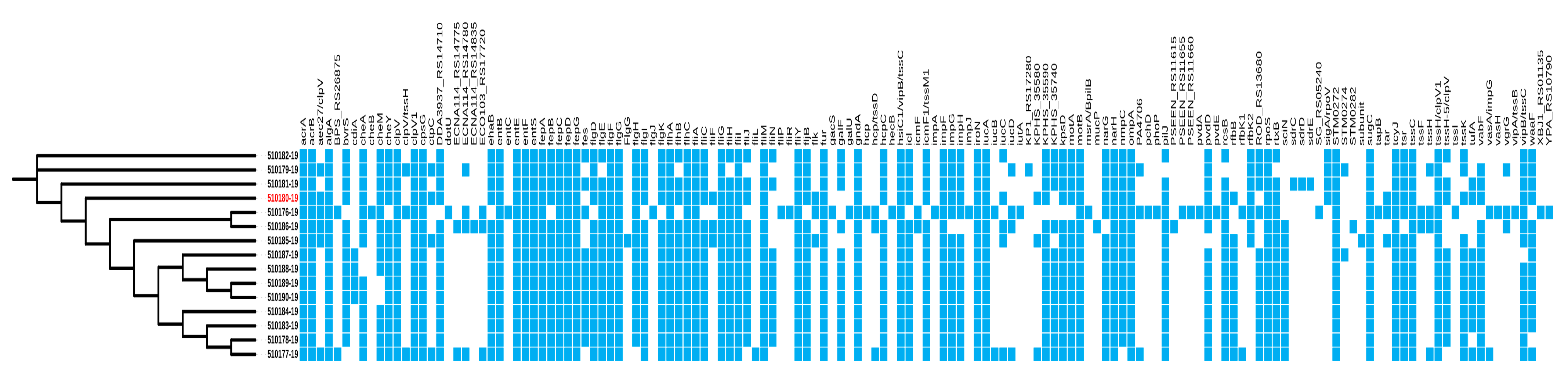
2.4. Virulence Genes in C. sakazakii Genomes
2.5. Plasmids and Mobile Genetic Elements in C. sakazakii Genomes
3. Discussion
4. Materials and Methods
4.1. Sampling
4.2. Isolation and Identification Methods of Cronobacter spp.
4.3. Whole-Genome Sequencing (WGS)
4.4. Sequence Type (ST) and Core Genome Multilocus Sequence Typing (cgMLST) of Cronobacter sakazakii
4.5. Determination of Serotypes
4.6. Antibiotic Susceptibility
4.7. Detection of Antibiotic Resistance and Virulence Genes
4.8. Detection of Plasmids and Mobile Genetic Elements (MGEs)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iversen, C.; Mullane, N.; Mc Cardell, B.; Tall, B.; Lehner, A.; Fanning, S.; Stephan, R.; Joosten, H. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov.comb. nov., C. malonaticus sp. nov., C. turicensis sp. nov., C. muytjensii sp. nov., C. dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, C. dublinensis sp. nov.subsp. dublinensis subsp. nov., C. dublinensis sp. nov.subsp. lausannensis subsp. nov., and C. dublinensis sp. nov.subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 1442–1447. [Google Scholar]
- Joseph, S.; Cetinkaya, E.; Drahovska, H.; Levican, A.; Figueras, M.; Forsythe, S. Cronobacter condimenti sp. Nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomoespecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 2012, 62, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Grim, C.; Gopinath, G.; Mammel, M.; Sathyamoorthy, V.; Trach, L.; Chase, H.; Fanning, S.; Tall, B. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2014, 64, 3402–3410. [Google Scholar] [PubMed]
- Forsythe, S.J. Updates on the Cronobacter Genus. Annu. Rev. Food Sci. Technol. 2018, 25, 23–44. [Google Scholar] [CrossRef] [PubMed]
- Hariri, S.; Joseph, S.; Forsythe, S.J. Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg. Infect. Dis. 2013, 19, 175–177. [Google Scholar] [CrossRef]
- Patrick, M.; Mahon, B.; Greene, S.; Rounds, J.; Conquist, A.; Wymore, K.; Boothe, E.; Lathrop, S.; Palmer, A.; Bowen, A. Incidence of Cronobacter spp. infections, United States, 2003–2009. Emerg. Inf. Dis. 2014, 20, 1520–1523. [Google Scholar] [CrossRef]
- Holý, O.; Cruz-Cordova, A.; Xicohtencatl-Cortés, J.; Hochel, I.; Parra-Flores, J.; Petrzelova, J.; Fačevicová, K.; Forsythe, S.; Alsonosi, A. Occurrence of virulence factors in Cronobacter sakazakii and Cronobacter malonaticus originated from clinical samples. Microb. Pathog. 2019, 127, 250–256. [Google Scholar] [CrossRef]
- Holý, O.; Forsythe, S. Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 2014, 86, 169–177. [Google Scholar] [CrossRef]
- Baumgartner, A.; Grand, M.; Liniger, M.; Iversen, C. Detection and frequency of Cronobacter spp. (Enterobacter sakazakii) in different categories of ready-to-eat foods other than infant formula. Int. J. Food Microbiol. 2009, 136, 189–192. [Google Scholar] [CrossRef]
- Vojkovska, H.; Karpiskova, R.; Orieskova, M.; Drahovska, H. Characterization of Cronobacter spp. isolated from food of plant origin and environmental samples collected from farms and from supermarkets in the Czech Republic. Int. J. Food Microbiol. 2016, 217, 130–136. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Maury-Sintjago, E.; Rodriguez-Fernández, A.; Acuña, S.; Cerda, F.; Aguirre, J.; Holý, O. Microbiological quality of powdered infant formula in Latin America. J. Food. Prot. 2020, 83, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, D.; Laing, S.; Fouhy, K.I.; Souhoka, L.; Beaven, A.; Soboleva, T.; Malakar, P. Quantifying the uncertainty of transfer of Cronobacter spp. between fomites and floors and touch points in dairy processing plants. Food Microbiol. 2019, 84, 103256. [Google Scholar] [CrossRef] [PubMed]
- El-Sharoud, W.; O’Brien, S.; Negredo, C.; Iversen, C.; Fanning, S.; Healy, B. Characterization of Cronobacter recovered from dried milk and related products. BMC Microbiol. 2009, 9, 24. [Google Scholar] [CrossRef]
- Craven, H.H.; McAuley, C.M.; Duffy, L.L.; Fegan, N. Distribution, prevalence and persistence of Cronobacter (Enterobacter sakazakii) in the non-processing and processing environments of five milk powder factories. J. Appl. Microbiol. 2010, 109, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.; Kothary, M.; Gopinath, G.; Jarvis, K.; Grim, C.; Hu, L.; Datta, A.; McCardell, B.A.; Tall, B.D. Cpa, the outer membrane protease of Cronobacter sakazakii, activates plasminogen and mediates resistance to serum bactericidal activity. Infect. Immun. 2011, 79, 1578–1587. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Córdova, A.; Rocha-Ramírez, L.; Ochoa, S.; Gónzalez-Pedrajo, B.; Espinosa, N.; Eslava, C.; Hernández-Chiñas, U.; Mendoza-Hernández, G.; Rodríguez-Leviz, A.; Valencia-Mayoral, P.; et al. Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PLoS ONE 2012, 7, e52091. [Google Scholar] [CrossRef]
- Aldubyan, M.; Almami, I.; Benslimane, F.; Alsonosi, A.; Forsythe, S. Comparative outer membrane protein analysis of high and low-invasive strains of Cronobacter malonaticus. Front. Microbiol. 2017, 8, 2268. [Google Scholar] [CrossRef]
- Ogrodzki, P.; Forsythe, S. Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genom. 2015, 16, 758. [Google Scholar] [CrossRef]
- Lee, Y.-D.; Park, J.; Chang, H. Detection, antibiotic susceptibility and biofilm formation of Cronobacter spp. from various foods in Korea. Food Control 2012, 24, 225–230. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Holý, O.; Riffo, F.; Lepuschitz, S.; Maury-Sintjago, E.; Rodríguez-Fernández, A.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J.; Mancilla-Rojano, J.; Troncoso, M.; et al. Profiling the virulence and antibiotic resistance genes of Cronobacter sakazakii strains isolated from powdered and dairy formulas by whole-genome sequencing. Front. Microbiol. 2021, 12, 694922. [Google Scholar] [CrossRef]
- Shi, L.; Liang, Q.; Zhan, Z.; Feng, J.; Zhao, Y.; Chen, Y.; Huang, M.; Tong, Y.; Wu, W.; Chen, W.; et al. Co-occurrence of 3 different resistance plasmids in a multi-drug resistant Cronobacter sakazakii isolate causing neonatal infections. Virulence 2018, 9, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Aly, M.A.; Domig, K.J.; Kneifel, W.; Reimhult, E. Whole genome sequencing-based comparison of food isolates of Cronobacter sakazakii. Front. Microbiol. 2019, 10, 1464. [Google Scholar] [CrossRef]
- Leopold, S.; Goering, R.; Witten, A.; Harmsen, D.; Mellmann, A. Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef] [PubMed]
- Lehner, A.; Tall, B.D.; Fanning, S.; Srikumar, S. Cronobacter spp.—Opportunistic foodborne pathogens: An update on evolution, osmotic adaptation and pathogenesis. Curr. Clin. Micro. Rpt. 2018, 5, 97–105. [Google Scholar] [CrossRef]
- Joseph, S.; Forsythe, S. Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front. Food Microbiol. 2012, 3, 397. [Google Scholar] [CrossRef] [PubMed]
- Fei, P.; Jiang, Y.; Jiang, Y.; Yuan, X.; Yang, T.; Chen, J.; Wang, Z.; Kang, H.; Forsythe, S.J. Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter sakazakii isolates from powdered infant formula collected from Chinese retail markets. Front. Microbiol. 2017, 8, 2026. [Google Scholar] [CrossRef]
- Sonbol, H.; Joseph, S.; McAuley, C.; Craven, H.; Forsythe, S. Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk powder production factories. Int. Dairy J. 2013, 30, 1–7. [Google Scholar] [CrossRef]
- Csorba, C.; Pajić, M.; Blagojević, B.; Forsythe, S.; Radinović, M.; Velebit, B. Prevalence, characterization, and antibiotic susceptibility of Cronobacter spp. in a milk powder processing environment: The first reported case in Serbia. Food Sc. Nutr. 2022, 10, 554–563. [Google Scholar] [CrossRef]
- Grim, C.J.; Gopinath, G.R.; Jarvis, K.G.; Sathyamoorthy, V.; Trach, L.H.; Chase, H.R.; Tall, B.D. Genome sequence of Cronobacter sakazakii serogroup O:4, sequence type 4 strain CDC 2009-03746, isolated from a fatal case of infantile meningitis. Genome Announc. 2015, 3, e00492-15. [Google Scholar] [CrossRef]
- Costa, P.V.; Vasconcellos, L.; Forsythe, S.J.; Brandão, M.L. Diversity of Cronobacter genus isolated between 1970 and 2019 on the American continent and genotyped using multi-locus sequence typing. FEMS Microbiol. Lett. 2021, 368, 1–9. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Ruppitsch, W.; Pekard-Amenitsch, S.; Forsythe, S.J.; Cormican, M.; Mach, R.L.; Piérard, D.; Allerberger, F.; The EUCRONI Study Group. Multicenter study of Cronobacter sakazakii infections in humans, Europe, 2017. Emerg. Infect. Dis. 2019, 25, 515–522. [Google Scholar] [CrossRef]
- Hu, J.; Li, X.; Du, X.; Cui, Z.; Cui, J. Identification and characterization of Cronobacter strains isolated from environmental samples. Curr. Microbiol. 2019, 76, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Pakbin, B.; Brück, W.M.; Allahyari, S.; Rossen, J.W.A.; Mahmoudi, R. Antibiotic resistance and molecular characterization of Cronobacter sakazakii strains isolated from powdered infant formula milk. Foods 2022, 11, 1093. [Google Scholar] [CrossRef]
- Fei, P.; Jiang, Y.; Feng, J.; Forsythe, S.J.; Li, R.; Zhou, Y.; Man, C. Antibiotic and desiccation resistance of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and processing environments. Front. Microbiol. 2017, 8, 316. [Google Scholar] [CrossRef]
- Kilonzo-Nthenge, A.; Rotich, E.; Godwin, S.; Nahashon, S.; Chen, F. Prevalence and antimicrobial resistance of Cronobacter sakazakii isolated from domestic kitchens in middle Tennessee, United States. J. Food Prot. 2012, 75, 1512–1517. [Google Scholar] [CrossRef]
- Zurfluh, K.; Nuësch-Inderbinen, M.; Morach, M.; Zihler Berner, A.; Hächler, H.; Stephan, R. Extended-spectrum-β-lactamaseproducing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl. Environ. Microbiol. 2015, 81, 3115–3120. [Google Scholar] [CrossRef] [PubMed]
- Holý, O.; Alsonosi, A.; Hochel, I.; Röderová, M.; Zatloukalova, S.; Mlynárčik, P.; Kolář, M.; Petrželová, J.; Alazraq, A.; Chmelař, D. Antibiotic susceptibility of Cronobacter spp. isolated from clinical samples. Pol. J. Microbiol. 2019, 68, 5–14. [Google Scholar] [CrossRef]
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The diverse functional roles of elongation Factor Tu (EF-Tu) in microbial pathogenesis. Front Microbiol. 2019, 10, 2351. [Google Scholar] [CrossRef]
- Müller, A.; Hächler, H.; Stephan, R.; Lehner, A. Presence of AmpC beta-lactamases, CSA-1, CSA-2, CMA-1, and CMA-2 conferring an unusual resistance phenotype in Cronobacter sakazakii and Cronobacter malonaticus. Microb. Drug Resist. 2014, 20, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.; Cambillau, C.; Cascales, E.; Journet, L. VgrG, Tae, Tle, and beyond: The versatile arsenal of type VI secretion effectors. Trends Microbiol. 2014, 22, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Gopinath, G.R.; Eshwar, A.; Srikumar, S.; Nguyen, S.; Gangiredla, J.; Patel, I.R.; Finkelstein, S.B.; Negrete, F.; Woo, J.; et al. The Secretion of toxins and other exoproteins of Cronobacter: Role in virulence, adaption, and persistence. Microorganisms 2020, 8, 229. [Google Scholar] [CrossRef]
- Coulthurst, S. The Type VI secretion system: A versatile bacterial weapon. Microbiology (Reading) 2019, 165, 503–515. [Google Scholar] [CrossRef]
- Singh, N.; Goel, G.; Raghav, M. Insights into virulence factors determining the pathogenicity of Cronobacter sakazakii. Virulence 2015, 6, 433–440. [Google Scholar] [CrossRef]
- Mohan Nair, M.K.; Venkitanarayanan, K. Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr. Res. 2007, 62, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Hariri, S.; Masood, N.; Forsythe, S. Sialic acid utilization by Cronobacter sakazakii. Microb. Inform. Exp. 2013, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; McVeagh, P.; Petocz, P.; Brand-Miller, J. Brain ganglioside and glycoprotein sialic acid in breastfed compared with formula-fed infants. Am. J. Clin. Nutr. 2003, 78, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Severi, E.; Hood, D.W.; Thomas, G.H. Sialic acid utilization by bacterial pathogens. Microbiology (Reading) 2007, 153, 2817–2822. [Google Scholar] [CrossRef]
- Lee, C.; Wigren, E.; Lünsdorf, H.; Römling, U. Protein homeostasis-more than resisting a hot bath. Curr. Opin. Microbiol. 2016, 30, 147–154. [Google Scholar] [CrossRef]
- Schellhorn, H.E. Function, evolution, and composition of the RpoS regulon in Escherichia coli. Front. Microbiol. 2020, 11, 560099. [Google Scholar] [CrossRef]
- Bojer, M.S.; Struve, C.; Ingmer, H.; Hansen, D.S.; Krogfelt, K.A. Heat resistance mediated by a new plasmid encoded Clp ATPase, ClpK, as a possible novel mechanism for nosocomial persistence of Klebsiella pneumoniae. PLoS ONE 2010, 5, e15467. [Google Scholar] [CrossRef]
- Boll, E.J.; Frimodt-Møller, J.; Olesen, B.; Krogfelt, K.A.; Struve, C. Heat resistance in extended-spectrum beta-lactamase-producing Escherichia coli may favor environmental survival in a hospital setting. Res. Microbiol. 2016, 167, 345–349. [Google Scholar] [CrossRef]
- Niu, H.; Mingzhe, Y.; Qi, Y.; Liu, Y.; Wang, X.; Dong, Q. Heat shock in Cronobacter sakazakii induces direct protection and cross-protection against simulated gastric fluid stress. Food Microbiol. 2022, 103, 10394. [Google Scholar] [CrossRef]
- Brooks, K.; Eze, J.; Onalenna, O.; Rahube, T.O. Analysis of antibiotic resistance from a rural community and wastewater contaminated environment linked to human and animal activities. J. Hazard Mater. Adv. 2023, 9, 100232. [Google Scholar] [CrossRef]
- Jang, H.; Eshwar, A.; Lehner, A.; Gangiredla, J.; Patel, I.R.; Beaubrun, J.J.-G.; Chase, H.R.; Negrete, F.; Finkelstein, S.; Weinstein, L.M.; et al. Characterization of Cronobacter sakazakii strains originating from plant-origin foods using comparative genomic analyses and zebrafish infectivity studies. Microorganisms 2022, 10, 1396. [Google Scholar] [CrossRef]
- Iversen, C.; Forsythe, S.J. Isolation of Enterobacter sakazakii and other Enterobacteriaceae from powdered infant formula milk and related products. Food Microbiol. 2004, 21, 771–776. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Sorschag, S.; Springer, B.; Allerberger, F.; Ruppitsch, W. Draft genome sequence of carbapenemase-producing Serratia marcescens isolated from a patient with chronic obstructive pulmonary disease. Genome Announc. 2017, 5, e01288-17. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology (Reading) 2012, 158, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef]
- Baldwin, A.; Loughlin, M.; Caubilla-Barron, J.; Kucerova, E.; Manning, G.; Dowson, C.; Forsythe, S. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance, which do not correlate with biotypes. BMC Microbiol. 2009, 9, 223. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standars Institute Publisher Supplement M100: Wayne, PA, USA, 2020; pp. 1–263. [Google Scholar]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.; Lago, B.; Dave, B.; Pereira, S.; Sharma, A.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby-Larsen, M.; Lund, O.; Villa, L.; Møller-Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Johansson, M.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Cechin, C.D.F.; Carvalho, G.G.; Bastos, C.P.; Kabuki, D.Y. Cronobacter spp. in foods of plant origin: Occurrence, contamination routes, and pathogenic potential. Crit. Rev. Food Sci. Nutr. 2022, 22, 1–15. [Google Scholar] [CrossRef]
- Gan, X.; Li, M.; Xu, J.; Yan, S.; Wang, W.; Li, F. Emerging of multidrug-resistant Cronobacter sakazakii isolated from infant supplementary food in China. Microbiol. Spectr. 2022, 10, 0119722. [Google Scholar] [CrossRef]
| Group of Samples | Specific Commodity | Number of Examined Samples |
|---|---|---|
| Swabs from the food production environment | Swabs | 855 |
| Poultry | Feaces | 148 |
| Food | Chockolate | 72 |
| Caramel | 55 | |
| Spice (dill, pepper) | 260 | |
| Poultry meat | 77 | |
| Dried cow milk | 244 | |
| Wheat flour | 91 | |
| Seeds (soy, pistachio, barley, mustard) | 198 | |
| Total | 2000 |
| Sample ID | * PubMLST ID | Source | WGS rMLST Result | ST | CC | Serotype (gnd-galF Alleles) | Collection Date |
|---|---|---|---|---|---|---|---|
| 510177-19 | 3823 | Food | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510178-19 | 3824 | Food | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510183-19 | 3828 | Production environment | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510184-19 | 3829 | Production environment | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510185-19 | 3830 | Food | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510187-19 | 3831 | Food | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510188-19 | 3832 | Production environment | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510189-19 | 3833 | Production environment | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510190-19 | 3834 | Production environment | Cronobacter sakazakii | 4 | 4 | Csak O:2 | 2015 |
| 510181-19 | 3826 | Food | Cronobacter sakazakii | 1 | 1 | Csak O:1 | 2015 |
| 510179-19 | 3825 | Production environment | Cronobacter sakazakii | 83 | 83 | Csak O:7 | 2015 |
| 510182-19 | 3827 | Food | Cronobacter sakazakii | 83 | 83 | Csak O:7 | 2015 |
| 510176-19 | 3695 | Food | Cronobacter sakazakii | 260 | - | Csak O:1 | 2015 |
| 510186-19 | 3697 | Hen | Cronobacter sakazakii | 93 | - | Csak O:7 | 2015 |
| 510180-19 | 3696 | Food | Cronobacter dublinensis | 822 | - | ND | 2015 |
| ST | Strains a | AM (10 µg) | AMC (20/10 µg) | CAZ (30 µg) | CIP (5 µg) | CL (30 µg) | CTX (30 µg) | GE (10 µg) | KF (30 µg) | TE (30 µg) | W (30 µg) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 510181-19 | S | S | S | S | S | S | S | R | S | S |
| 4 | 510177-19 | R | S | S | S | S | S | S | R | R | S |
| 4 | 510178-19 | S | S | S | S | S | S | S | R | S | S |
| 4 | 510183-19 | S | R | S | S | S | S | S | R | S | S |
| 4 | 510184-19 | S | S | S | S | S | S | S | R | R | S |
| 4 | 510185-19 | S | S | S | S | S | S | S | R | S | S |
| 4 | 510187-19 | S | S | S | S | S | S | S | R | S | S |
| 4 | 510188-19 | R | S | S | S | S | S | S | R | S | S |
| 4 | 510189-19 | S | S | S | S | S | S | S | R | S | S |
| 4 | 510190-19 | S | S | S | S | S | S | S | R | S | S |
| 83 | 510179-19 | R | S | S | S | S | S | S | R | S | S |
| 83 | 510182-19 | R | S | R | S | S | S | S | R | S | S |
| 260 | 510176-19 | R | S | S | S | S | S | S | R | R | S |
| 822 | 510180-19 * | S | S | S | S | S | S | S | S | S | S |
| ID Strain a | ST | Plasmid | Plasmid Accession Number | Mobile Genetic Elements |
|---|---|---|---|---|
| 510177-19 | 4 | IncFIB(pCTU3) | FN543096 | ISEhe3, ISEsa1, ISEc52 |
| 510178-19 | 4 | IncFIB(pCTU3) | FN543096 | ISEhe3, ISEsa1, ISEc52 |
| 510183-19 | 4 | IncFIB(pCTU3) | FN543096 | ISEc52 |
| 510184-19 | 4 | IncFIB(pCTU3) | FN543096 | ISEhe3, ISEsa1 |
| 510185-19 | 4 | ----- | ----- | ISEhe3 |
| 510187-19 | 4 | IncFIB(pCTU3) | FN543096 | ISEhe3, ISEsa1, IS26, ISEc52 |
| 510188-19 | 4 | IncFIB(pCTU3) | FN543096 | ISEsa1,IS26, ISEc52 |
| 510189-19 | 4 | IncFIB(pCTU3) | FN543096 | ISEhe3, ISEsa1 |
| 510190-19 | 4 | IncFIB(pCTU3) | FN543096 | ISEhe3, ISEsa1, ISEc52 |
| 510179-19 | 83 | rep7a | SAU83488 | ----- |
| 510180-19 | 822 | IncFII(pCTU2) pESA2 | FN543095 CP000784 | ISEch12 |
| 510186-19 | 93 | Col440l | Cp023920.1 | ISEsa1, ISKpn34 |
| 510176-19 | 260 | IncFIB(pCTU3) | FN543096 | ISEsa1, IS26, cn_6897_IS26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holý, O.; Parra-Flores, J.; Bzdil, J.; Cabal-Rosel, A.; Daza-Prieto, B.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J.; Rodríguez-Martínez, R.; Acuña, S.; Forsythe, S.; et al. Screening of Antibiotic and Virulence Genes from Whole Genome Sequenced Cronobacter sakazakii Isolated from Food and Milk-Producing Environments. Antibiotics 2023, 12, 851. https://doi.org/10.3390/antibiotics12050851
Holý O, Parra-Flores J, Bzdil J, Cabal-Rosel A, Daza-Prieto B, Cruz-Córdova A, Xicohtencatl-Cortes J, Rodríguez-Martínez R, Acuña S, Forsythe S, et al. Screening of Antibiotic and Virulence Genes from Whole Genome Sequenced Cronobacter sakazakii Isolated from Food and Milk-Producing Environments. Antibiotics. 2023; 12(5):851. https://doi.org/10.3390/antibiotics12050851
Chicago/Turabian StyleHolý, Ondrej, Julio Parra-Flores, Jaroslav Bzdil, Adriana Cabal-Rosel, Beatriz Daza-Prieto, Ariadnna Cruz-Córdova, Juan Xicohtencatl-Cortes, Ricardo Rodríguez-Martínez, Sergio Acuña, Stephen Forsythe, and et al. 2023. "Screening of Antibiotic and Virulence Genes from Whole Genome Sequenced Cronobacter sakazakii Isolated from Food and Milk-Producing Environments" Antibiotics 12, no. 5: 851. https://doi.org/10.3390/antibiotics12050851
APA StyleHolý, O., Parra-Flores, J., Bzdil, J., Cabal-Rosel, A., Daza-Prieto, B., Cruz-Córdova, A., Xicohtencatl-Cortes, J., Rodríguez-Martínez, R., Acuña, S., Forsythe, S., & Ruppitsch, W. (2023). Screening of Antibiotic and Virulence Genes from Whole Genome Sequenced Cronobacter sakazakii Isolated from Food and Milk-Producing Environments. Antibiotics, 12(5), 851. https://doi.org/10.3390/antibiotics12050851










