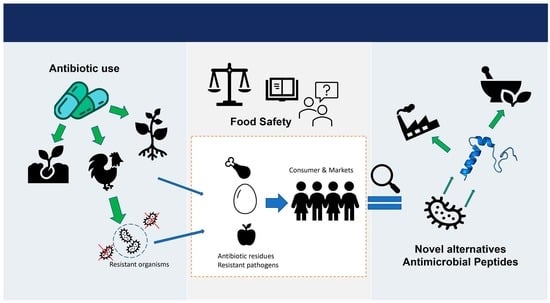
Antibiotic Resistance and Food Safety: Perspectives on New Technologies and Molecules for Microbial Control in the Food Industry
Abstract
1. Introduction
2. Methodology
3. Antibiotic Resistance in the Food Chain
3.1. Acquisition of Antibiotic Resistance and Mechanisms of Transfer
3.2. Mechanisms of Antibiotic Resistance
3.2.1. Drug Uptake Limitation
3.2.2. Drug Target Modification
3.2.3. Drug Inactivation
3.2.4. Drug Efflux
ABC Transporter Family
MATE Transporter Family
SMR Transporter Family
MFS Transporter Family
RND Transport Family
4. Potential Routes of Transmission and Prevalence of ABR in the Food Chain
5. Antibiotic Resistance and Food Safety: Implications for Public Health
6. New Alternatives to Antibiotics: Bacteriocins and Their Physicochemical Properties
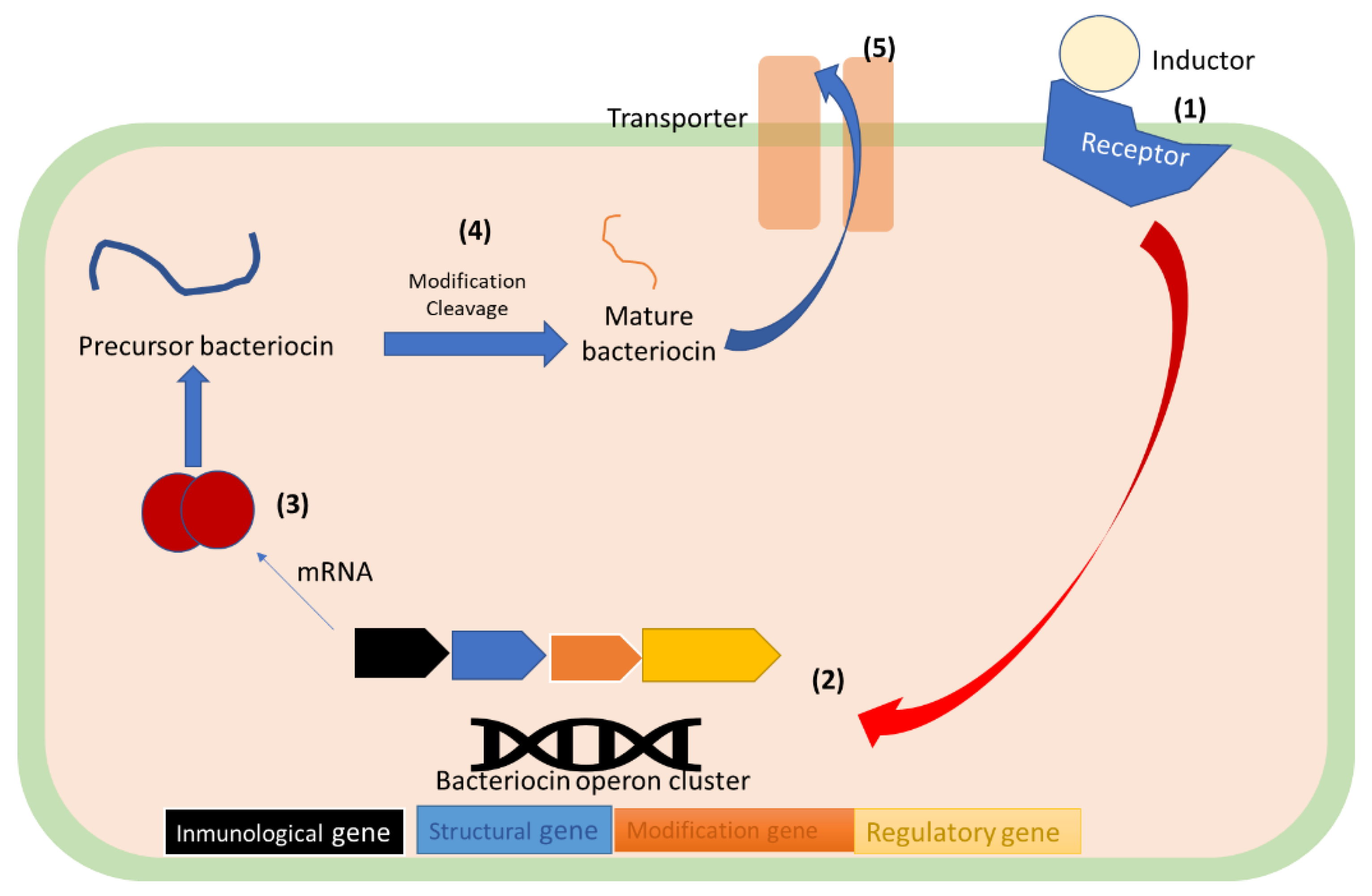
6.1. Bacteriocins
6.2. Bacteriocin Classification
6.2.1. Classification of Gram-Negative Bacteriocins
6.2.2. Classification of Gram-Positive Bacteriocins
6.2.3. Classification of Archaea Bacteriocins
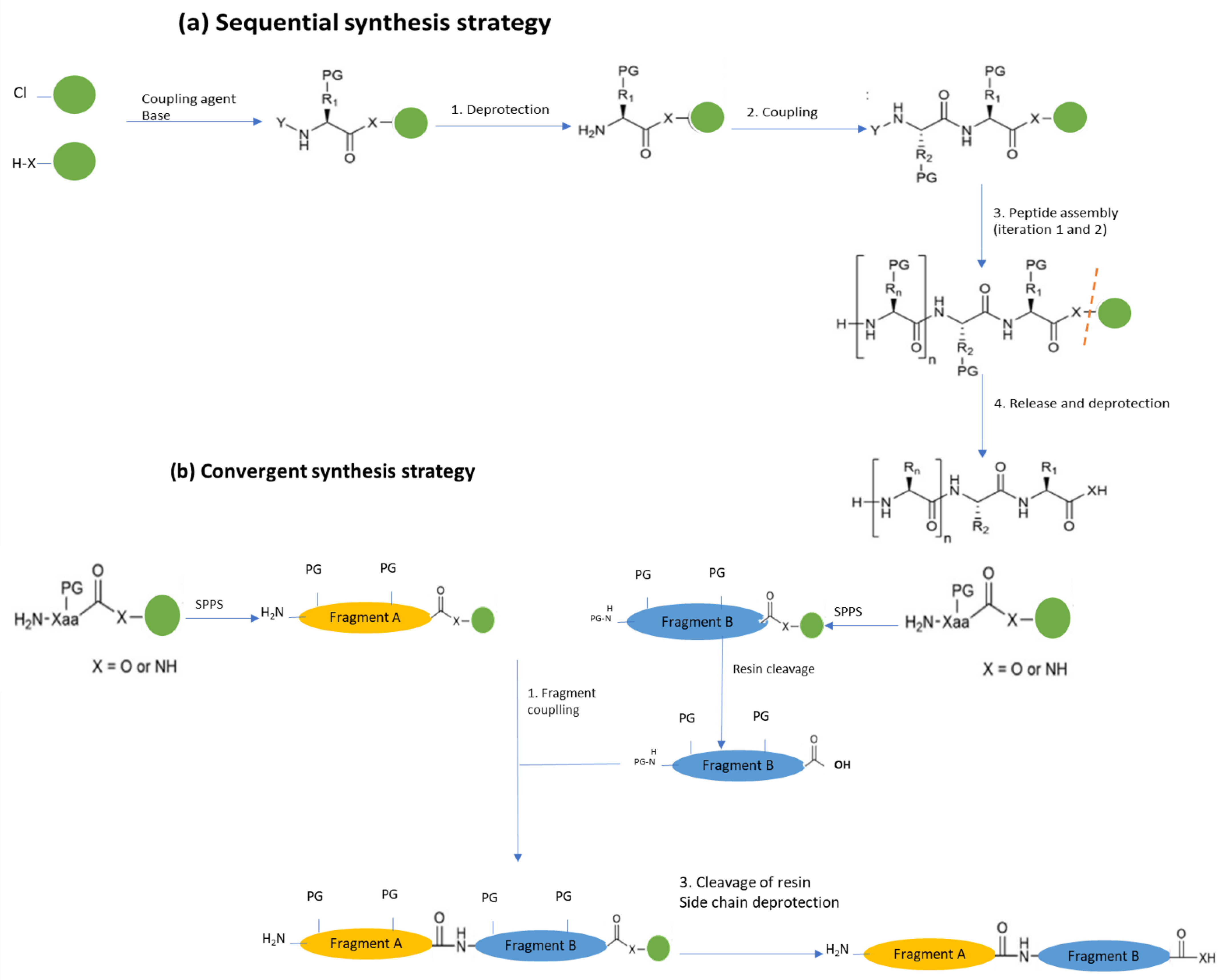
6.3. Bacteriocin Synthesis
Solid-Phase Chemical Synthesis (SPPS)
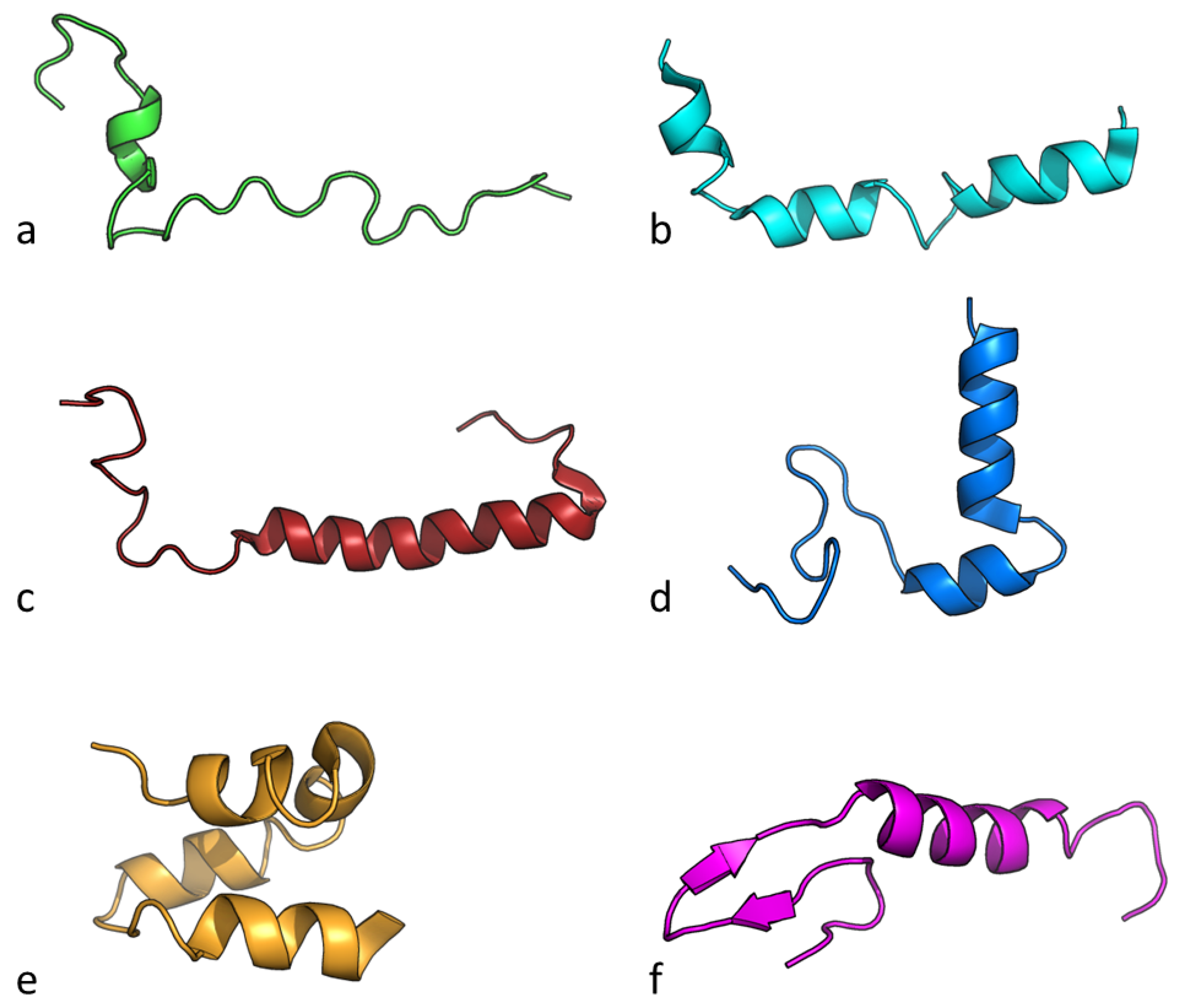
6.4. Bacteriocin Chemical Structure and Physicochemical Properties
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016, 7, 1881. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.; Roe, E.; Hocknell, S. Food Supply Chains and the Antimicrobial Resistance Challenge: On the Framing, Accomplishments and Limitations of Corporate Responsibility. Environ. Plan. A Econ. Space 2021, 53, 1373–1390. [Google Scholar] [CrossRef]
- Nelson, D.W.; Moore, J.E.; Rao, J.R. Antimicrobial Resistance (AMR): Significance to Food Quality and Safety. Food Qual. Saf. 2019, 3, 15–22. [Google Scholar] [CrossRef]
- Nji, E.; Kazibwe, J.; Hambridge, T.; Joko, C.A.; Larbi, A.A.; Damptey, L.A.O.; Nkansa-Gyamfi, N.A.; Lundborg, C.S.; Lien, L.T.Q. High Prevalence of Antibiotic Resistance in Commensal Escherichia coli from Healthy Human Sources in Community Settings. Sci. Rep. 2021, 11, 3372. [Google Scholar] [CrossRef]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The Threat of Antimicrobial Resistance in Developing Countries: Causes and Control Strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef]
- Njoga, E.O.; Onunkwo, J.I.; Okoli, C.E.; Ugwuoke, W.I.; Nwanta, J.A.; Chah, K.F. Assessment of Antimicrobial Drug Administration and Antimicrobial Residues in Food Animals in Enugu State, Nigeria. Trop. Anim. Health Prod. 2018, 50, 897–902. [Google Scholar] [CrossRef]
- Ahmed, T.A.E.; Hammami, R. Recent Insights into Structure–Function Relationships of Antimicrobial Peptides. J. Food Biochem. 2019, 43, e12546. [Google Scholar] [CrossRef]
- Rima, M.; Rima, M.; Fajloun, Z.; Sabatier, J.-M.; Bechinger, B.; Naas, T. Antimicrobial Peptides: A Potent Alternative to Antibiotics. Antibiotics 2021, 10, 1095. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial Peptides: Mechanism of Action, Activity and Clinical Potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Tackling Antibiotic from a Food Safety Perspective in Europe; World Health Organization: Geneva, Switzerland, 2011; pp. 8–12.
- World Health Organization. WHO Library Cataloguing-in-Publication Data Global Action Plan on Antimicrobial Resistance; WHO Document Production Services: Geneva, Switzerland, 2015; ISBN 9789241509763.
- Cahill, S.M.; Desmarchelier, P.; Fattori, V.; Bruno, A.; Cannavan, A. Techniques in Food and Agriculture Global Perspectives on Antimicrobial Resistance in the Food Chain. Food Prot. Trends 2017, 37, 353–360. [Google Scholar]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Harbarth, S.; Participants, F.T.W.H.-A.I.R.F.; Goossens, H.; Jarlier, V.; Kluytmans, J.; Laxminarayan, R.; Saam, M.; van Belkum, A.; Pittet, D. Antimicrobial Resistance: One World, One Fight! Antimicrob. Resist. Infect. Control 2015, 4, 49. [Google Scholar] [CrossRef]
- Lorenzo-Díaz, F.; Fernández-López, C.; Lurz, R.; Bravo, A.; Espinosa, M. Crosstalk between Vertical and Horizontal Gene Transfer: Plasmid Replication Control by a Conjugative Relaxase. Nucleic Acids Res. 2017, 45, 7774–7785. [Google Scholar] [CrossRef]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral Gene Transfer and the Nature of Bacterial Innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef]
- Croucher, N.J.; Mostowy, R.; Wymant, C.; Turner, P.; Bentley, S.D.; Fraser, C. Horizontal DNA Transfer Mechanisms of Bacteria as Weapons of Intragenomic Conflict. PLoS Biol. 2016, 14, e1002394. [Google Scholar] [CrossRef]
- Chiang, Y.N.; Penadés, J.R.; Chen, J. Genetic Transduction by Phages and Chromosomal Islands: The New and Noncanonical. PLoS Pathog. 2019, 15, e1007878. [Google Scholar] [CrossRef]
- Gillings, M.R. Lateral Gene Transfer, Bacterial Genome Evolution, and the Anthropocene. Ann. N. Y. Acad. Sci. 2017, 1389, 20–36. [Google Scholar] [CrossRef]
- Haudiquet, M.; de Sousa, J.M.; Touchon, M.; Rocha, E.P.C. Selfish, Promiscuous and Sometimes Useful: How Mobile Genetic Elements Drive Horizontal Gene Transfer in Microbial Populations. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20210234. [Google Scholar] [CrossRef]
- Raleigh, E.A.; Low, K.B. Conjugation. In Brenner’s Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 144–151. [Google Scholar]
- Hall, R.M. Mobile Gene Cassettes and Integrons: Moving Antibiotic Resistance Genes in Gram-Negative Bacteria. In Ciba Foundation Symposium 207-Antibiotic Resistance: Origins, Evolution, Selection and Spread: Antibiotic Resistance: Origins, Evolution, Selection and Spread: Ciba Foundation Symposium 207; John Wiley & Sons, Ltd.: Chichester, UK, 2007; pp. 192–205. [Google Scholar]
- Blot, M. Transposable Elements and Adaptation of Host Bacteria. Genetica 1994, 93, 5–12. [Google Scholar] [CrossRef]
- Cambray, G.; Guerout, A.-M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef]
- Sabbagh, P.; Rajabnia, M.; Maali, A.; Ferdosi-Shahandashti, E. Spectroscopic Investigation on the Interaction of DNA with Superparamagnetic Iron Oxide Nanoparticles Doped with Chromene via Dopamine as Cross Linker. Iran. J. Basic Med. Sci. 2021, 24, 136–142. [Google Scholar] [CrossRef]
- Wistrand-Yuen, E.; Knopp, M.; Hjort, K.; Koskiniemi, S.; Berg, O.G.; Andersson, D.I. Evolution of High-Level Resistance during Low-Level Antibiotic Exposure. Nat. Commun. 2018, 9, 1599. [Google Scholar] [CrossRef]
- Van Bambeke, F.; Balzi, E.; Tulkens, P.M. Antibiotic Efflux Pumps. Biochem. Pharmacol. 2000, 60, 457–470. [Google Scholar] [CrossRef]
- Blair, J.M.; E Richmond, G.E.; Piddock, L.J. Multidrug Efflux Pumps in Gram-Negative Bacteria and Their Role in Antibiotic Resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Reygaert, W.C. An Overview of the Antimicrobial Resistance Mechanisms of Bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Mahon, C.; Lehman, D.; Manuselis, G. Antimicrobial Agent Mechanism of Action and Resistance. In Textbook of Diagnostic Microbiology; Saunders: St. Louis, MO, USA, 2014; pp. 254–273. [Google Scholar]
- Schaenzer, A.J.; Wright, G.D. Antibiotic Resistance by Enzymatic Modification of Antibiotic Targets. Trends Mol. Med. 2020, 26, 768–782. [Google Scholar] [CrossRef]
- Andreoletti, O.; Budka, H.; Buncic, S.; Colin, P.; Collins, J.D.; de Koeijer, A.; Griffin, J.; Havelaar, A.; Hope, J.; Klein, G.; et al. Foodborne Antimicrobial Resistance as a Biological Hazard—Scientific Opinion of the Panel on Biological Hazards. EFSA J. 2008, 6, 765. [Google Scholar] [CrossRef]
- McDermott, P.F.; Zhao, S.; Wagner, D.D.; Simjee, S.; Walker, R.D.; White, D.G. The Food Safety Perspective of Antibiotic Resistance. Anim. Biotechnol. 2002, 13, 71–84. [Google Scholar] [CrossRef]
- You, Y.; Silbergeld, E.K. Learning from Agriculture: Understanding Low-Dose Antimicrobials as Drivers of Resistome Expansion. Front. Microbiol. 2014, 5, 284. [Google Scholar] [CrossRef]
- FAO-WHO. Foodborne Antimicrobial Resistance; FAO; WHO: Rome, Italy, 2022; ISBN 978-92-5-135734-7.
- Kumar, K.C.; Gupta, S.; Chander, Y.; Singh, A.K. Antibiotic Use in Agriculture and Its Impact on the Terrestrial Environment. Adv. Agron. 2005, 87, 1–54. [Google Scholar]
- Raison-Peyron, N.; Messaad, D.; Bousquet, J.; Demoly, P. Anaphylaxis to Beef in Penicillin-Allergic Patient. Allergy 2001, 56, 796–797. [Google Scholar] [CrossRef] [PubMed]
- Baynes, R.E.; Dedonder, K.; Kissell, L.; Mzyk, D.; Marmulak, T.; Smith, G.; Tell, L.; Gehring, R.; Davis, J.; Riviere, J.E. Health Concerns and Management of Select Veterinary Drug Residues. Food Chem. Toxicol. 2016, 88, 112–122. [Google Scholar] [CrossRef]
- Treiber, F.M.; Beranek-Knauer, H. Antimicrobial Residues in Food from Animal Origin—A Review of the Literature Focusing on Products Collected in Stores and Markets Worldwide. Antibiotics 2021, 10, 534. [Google Scholar] [CrossRef] [PubMed]
- Andrei, S.; Droc, G.; Stefan, G. FDA Approved Antibacterial Drugs: 2018—2019. Discoveries 2019, 7, e102. [Google Scholar] [CrossRef] [PubMed]
- Kyuchukova, R. Antibiotic Residues and Human Health Hazard—Review. Bulg. J. Agric. Sci. 2020, 26, 664–668. [Google Scholar]
- Bacanlı, M.; Başaran, N. Importance of Antibiotic Residues in Animal Food. Food Chem. Toxicol. 2019, 125, 462–466. [Google Scholar] [CrossRef]
- Kimera, Z.I.; Mdegela, R.H.; Mhaiki, C.J.N.; Karimuribo, E.D.; Mabiki, F.; Nonga, H.E.; Mwesongo, J. Determination of Oxytetracycline Residues in Cattle Meat Marketed in the Kilosa District, Tanzania. Onderstepoort J. Vet. Res. 2015, 82. [Google Scholar] [CrossRef]
- Olatoye, I.O.; Ehinmowo, A.A. Oxytetracycline Residues in Edible Tissues of Cattle Slaughtered in Akure, Nigeria. Niger. Vet. J. 2011, 31. [Google Scholar] [CrossRef]
- Tavakoli, H.R.; Safaeefirouzabadi, M.S.; Afsharfarnia, S.; Joneidijafari, N.; Saadat, S. Detecting Antibiotic Residues by HPLC Method in Chicken and Calves Meat in Diet of a Military Center in Tehran. Acta Med. Mediterr. 2015, 31, 1427–1433. [Google Scholar]
- Er, B.; Onurdağ, F.K.; Demirhan, B.; Ozgacar, S.Ö.; Öktem, A.B.; Abbasoğlu, U. Screening of Quinolone Antibiotic Residues in Chicken Meat and Beef Sold in the Markets of Ankara, Turkey. Poult. Sci. 2013, 92, 2212–2215. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Hassan, M.M.; Alam, M.; Sattar, S.; Bari, S.; Saifuddin, A.K.M.; Hoque, A. Antibiotic Residues in Milk and Eggs of Commercial and Local Farms at Chittagong, Bangladesh. Vet. World 2015, 8, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Wang, J.; Han, R.; Xu, X.; Zhen, Y.; Qu, X.; Sun, P.; Li, S.; Yu, Z. Occurrence of Several Main Antibiotic Residues in Raw Milk in 10 Provinces of China. Food Addit. Contam. Part B 2013, 6, 84–89. [Google Scholar] [CrossRef]
- Ghimpețeanu, O.M.; Pogurschi, E.N.; Popa, D.C.; Dragomir, N.; Drăgotoiu, T.; Mihai, O.D.; Petcu, C.D. Antibiotic Use in Livestock and Residues in Food—A Public Health Threat: A Review. Foods 2022, 11, 1430. [Google Scholar] [CrossRef]
- Tao, Q.; Wu, Q.; Zhang, Z.; Liu, J.; Tian, C.; Huang, Z.; Malakar, P.K.; Pan, Y.; Zhao, Y. Meta-Analysis for the Global Prevalence of Foodborne Pathogens Exhibiting Antibiotic Resistance and Biofilm Formation. Front. Microbiol. 2022, 13, 906490. [Google Scholar] [CrossRef] [PubMed]
- Carlie, S.M.; Boucher, C.E.; Bragg, R.R. Molecular Basis of Bacterial Disinfectant Resistance. Drug Resist. Update 2020, 48, 100672. [Google Scholar] [CrossRef]
- Hassani, S.; Moosavy, M.-H.; Gharajalar, S.N.; Khatibi, S.A.; Hajibemani, A.; Barabadi, Z. High Prevalence of Antibiotic Resistance in Pathogenic Foodborne Bacteria Isolated from Bovine Milk. Sci. Rep. 2022, 12, 3878. [Google Scholar] [CrossRef] [PubMed]
- Boonyasiri, A.; Tangkoskul, T.; Seenama, C.; Saiyarin, J.; Tiengrim, S.; Thamlikitkul, V. Prevalence of Antibiotic Resistant Bacteria in Healthy Adults, Foods, Food Animals, and the Environment in Selected Areas in Thailand. Pathog. Glob. Health 2014, 108, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Frana, T.S.; Beahm, A.R.; Hanson, B.M.; Kinyon, J.M.; Layman, L.L.; Karriker, L.A.; Ramirez, A.; Smith, T.C. Isolation and Characterization of Methicillin-Resistant Staphylococcus Aureus from Pork Farms and Visiting Veterinary Students. PLoS ONE 2013, 8, e53738. [Google Scholar] [CrossRef]
- Hiroi, M.; Kawamori, F.; Harada, T.; Sano, Y.; Miwa, N.; Sugiyama, K.; Hara-Kudo, Y.; Masuda, T. Antibiotic Resistance in Bacterial Pathogens from Retail Raw Meats and Food-Producing Animals in Japan. J. Food Prot. 2012, 75, 1774–1782. [Google Scholar] [CrossRef]
- Food and Drug Administration (FDA) HACCP Principles & Application Guidelines. Available online: https://www.fda.gov/food/hazard-analysis-critical-control-point-haccp/haccp-principles-application-guidelines (accessed on 28 December 2022).
- Brenes, A. Gestión Del Riesgo; Costa Rica. 2013; Available online: https://hdl.handle.net/20.500.12337/425 (accessed on 27 December 2022).
- Fernández, A.; Sibera, M.; Vidal Ortega, C.; Premiumlab, S. Guía Para La Prevención Del Fraude En La Indústria Agroalimentaria. Premiumlab 2017, 7–38. [Google Scholar]
- Strayer, S.E.; Everstine, K.; Kennedy, S. Economically Motivated Adulteration of Honey: Quality Control Vulnerabilities in the International Honey Market. Food Prot. Trends 2014, 34, 8–14. [Google Scholar]
- Arsène, M.M.J.; Davares, A.K.L.; Viktorovna, P.I.; Andreevna, S.L.; Sarra, S.; Khelifi, I.; Sergueïevna, D.M. The Public Health Issue of Antibiotic Residues in Food and Feed: Causes, Consequences, and Potential Solutions. Vet. World 2022, 15, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Okocha, R.C.; Olatoye, I.O.; Adedeji, O.B. Food Safety Impacts of Antimicrobial Use and Their Residues in Aquaculture. Public Health Rev. 2018, 39, 21. [Google Scholar] [CrossRef] [PubMed]
- Silveira, R.F.; Roque-Borda, C.A.; Vicente, E.F. Antimicrobial Peptides as a Feed Additive Alternative to Animal Production, Food Safety and Public Health Implications: An Overview. Anim. Nutr. 2021, 7, 896–904. [Google Scholar] [CrossRef]
- Zwietering, M.H.; Jacxsens, L.; Membré, J.-M.; Nauta, M.; Peterz, M. Relevance of Microbial Finished Product Testing in Food Safety Management. Food Control 2016, 60, 31–43. [Google Scholar] [CrossRef]
- Chen, J.; Ying, G.-G.; Deng, W.-J. Antibiotic Residues in Food: Extraction, Analysis, and Human Health Concerns. J. Agric. Food Chem. 2019, 67, 7569–7586. [Google Scholar] [CrossRef] [PubMed]
- Ben, Y.; Hu, M.; Zhong, F.; Du, E.; Li, Y.; Zhang, H.; Andrews, C.B.; Zheng, C. Human Daily Dietary Intakes of Antibiotic Residues: Dominant Sources and Health Risks. Environ. Res. 2022, 212, 113387. [Google Scholar] [CrossRef] [PubMed]
- Beyene, T. Veterinary Drug Residues in Food-Animal Products: Its Risk Factors and Potential Effects on Public Health. J. Vet. Sci. Technol. 2015, 7, 285. [Google Scholar] [CrossRef]
- Donkor, E.S.; Newman, M.J.; Tay, S.C.K.; Dayie, N.T.K.D.; Bannerman, E.; Olu-Taiwo, M. Investigation into the Risk of Exposure to Antibiotic Residues Contaminating Meat and Egg in Ghana. Food Control 2011, 22, 869–873. [Google Scholar] [CrossRef]
- Thiim, M.; Friedman, L.S. Hepatotoxicity of Antibiotics and Antifungals. Clin. Liver Dis. 2003, 7, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cheng, L.; Wang, J.; Hao, M.; Che, H. Antibiotic-Induced Gut Microbiota Dysbiosis Damages the Intestinal Barrier, Increasing Food Allergy in Adult Mice. Nutrients 2021, 13, 3315. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The Antimicrobial Peptides and Their Potential Clinical Applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Rai, M.; Pandit, R.; Gaikwad, S.; Kövics, G. Antimicrobial Peptides as Natural Bio-Preservative to Enhance the Shelf-Life of Food. J. Food Sci. Technol. 2016, 53, 3381–3394. [Google Scholar] [CrossRef] [PubMed]
- Borah, A.; Deb, B.; Chakraborty, S. A Crosstalk on Antimicrobial Peptides. Int. J. Pept. Res. Ther. 2021, 27, 229–244. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell. Infect. Microbiol. 2021, 11, 453. [Google Scholar] [CrossRef]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a New Generation of Antimicrobials: Toxicity Aspects and Regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, Synthesis, Mechanism of Action and Resistance Development in Food Spoilage Causing Bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef]
- Benítez-Chao, D.F.; León-Buitimea, A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. Bacteriocins: An Overview of Antimicrobial, Toxicity, and Biosafety Assessment by in Vivo Models. Front. Microbiol. 2021, 12, 630695. [Google Scholar] [CrossRef] [PubMed]
- Dijksteel, G.S.; Ulrich, M.M.W.; Middelkoop, E.; Boekema, B.K.H.L. Review: Lessons Learned From Clinical Trials Using Antimicrobial Peptides (AMPs). Front. Microbiol. 2021, 12, 616979. [Google Scholar] [CrossRef]
- Cao, L.T.; Wu, J.Q.; Xie, F.; Hu, S.H.; Mo, Y. Efficacy of Nisin in Treatment of Clinical Mastitis in Lactating Dairy Cows. J. Dairy Sci. 2007, 90, 3980–3985. [Google Scholar] [CrossRef] [PubMed]
- De Kwaadsteniet, M.; Doeschate, K.T.; Dicks, L.M.T. Nisin F in the Treatment of Respiratory Tract Infections Caused by Staphylococcus aureus. Lett. Appl. Microbiol. 2009, 48, 65–70. [Google Scholar] [CrossRef]
- Heunis, T.D.J.; Smith, C.; Dicks, L.M.T. Evaluation of a Nisin-Eluting Nanofiber Scaffold To Treat Staphylococcus Aureus-Induced Skin Infections in Mice. Antimicrob. Agents Chemother. 2013, 57, 3928–3935. [Google Scholar] [CrossRef]
- Sharma, A. Importance of Probiotics in Cancer Prevention and Treatment. In Recent Developments in Applied Microbiology and Biochemistry; Elsevier: Amsterdam, The Netherlands, 2019; pp. 33–45. [Google Scholar]
- Leisner, J.J.; Laursen, B.G.; Prévost, H.; Drider, D.; Dalgaard, P. Carnobacterium: Positive and Negative Effects in the Environment and in Foods. FEMS Microbiol. Rev. 2007, 31, 592–613. [Google Scholar] [CrossRef] [PubMed]
- O’Bryan, C.A.; Koo, O.K.; Sostrin, M.L.; Ricke, S.C.; Crandall, P.G.; Johnson, M.G. Characteristics of Bacteriocins and Use as Food Antimicrobials in the United States. In Food and Feed Safety Systems and Analysis; Elsevier: Amsterdam, The Netherlands, 2018; pp. 273–286. [Google Scholar]
- Kasimin, M.E.; Shamsuddin, S.; Molujin, A.M.; Sabullah, M.K.; Gansau, J.A.; Jawan, R. Enterocin: Promising Biopreservative Produced by Enterococcus sp. Microorganisms 2022, 10, 684. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Microbial Production of Bacteriocins: Latest Research Development and Applications. Biotechnol. Adv. 2018, 36, 2187–2200. [Google Scholar] [CrossRef]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial Activities of Bacteriocins: Application in Foods and Pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [CrossRef]
- Zouhir, A.; Hammami, R.; Fliss, I.; Ben Hamida, J. A New Structure-Based Classification of Gram-Positive Bacteriocins. Protein J. 2010, 29, 432–439. [Google Scholar] [CrossRef]
- Charlesworth, J.; Burns, B.P. Untapped Resources: Biotechnological Potential of Peptides and Secondary Metabolites in Archaea. Archaea 2015, 2015, 282035. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, P.; Bhat, S.G.; Chandrasekaran, M. Halocin SH10 Production by an Extreme Haloarchaeon Natrinema Sp. BTSH10 Isolated from Salt Pans of South India. Saudi J. Biol. Sci. 2013, 20, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Uzelac, G.; Miljkovic, M.; Lozo, J.; Radulovic, Z.; Tosic, N.; Kojic, M. Expression of Bacteriocin LsbB Is Dependent on a Transcription Terminator. Microbiol. Res. 2015, 179, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Risøen, P.A.; Brurberg, M.B.; Eijsink, V.G.H.; Nes, I.F. Functional Analysis of Promoters Involved in Quorum Sensing-Based Regulation of Bacteriocin Production in Lactobacillus. Mol. Microbiol. 2002, 37, 619–628. [Google Scholar] [CrossRef]
- Morton, J.T.; Freed, S.D.; Lee, S.W.; Friedberg, I. A Large Scale Prediction of Bacteriocin Gene Blocks Suggests a Wide Functional Spectrum for Bacteriocins. BMC Bioinform. 2015, 16, 381. [Google Scholar] [CrossRef]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Novel Bacteriocins from Lactic Acid Bacteria (LAB): Various Structures and Applications. Microb. Cell Fact. 2014, 13 (Suppl. S1), S3. [Google Scholar] [CrossRef]
- Bédard, F.; Biron, E. Recent Progress in the Chemical Synthesis of Class II and S-Glycosylated Bacteriocins. Front. Microbiol. 2018, 9, 1048. [Google Scholar] [CrossRef]
- Collins, J.M.; Porter, K.A.; Singh, S.K.; Vanier, G.S. High-Efficiency Solid Phase Peptide Synthesis (HE-SPPS). Org. Lett. 2014, 16, 940–943. [Google Scholar] [CrossRef]
- Mueller, L.K.; Baumruck, A.C.; Zhdanova, H.; Tietze, A.A. Challenges and Perspectives in Chemical Synthesis of Highly Hydrophobic Peptides. Front. Bioeng. Biotechnol. 2020, 8, 162. [Google Scholar] [CrossRef]


| Antimicrobial Group | Resistance Mechanism |
|---|---|
| Aminoglycosides Gentamicin Streptomycin Kanamycin | Enzyme modification Decreased permeability Target resistance (ribosome) Efflux pumps |
| β-Lactams Cephalothin Cefoxitin Ceftiofur Cefquinome | Reduced permeability Altered penicillin-binding proteins (PBPs) β-Lactamases, cephalosporinases Efflux pumps |
| Folate pathway inhibitors Sulfonamides | Decreased permeability Production of drug-insensitive enzymes |
| Macrolide-lincosamide-streptgramin B Erythromycin Lincomycin Virginiamycin | Enzyme modification Decreased permeability Decreased ribosomal binding |
| Phenicols Chloramphenicol Florfenicol | Enzyme modification Decreased permeability Decreased ribosomal binding Efflux pumps |
| Quinolones and fluoroquinolones Nalidixic acid Ciprofloxacin Enrofloxacin | Target resistance (DNA gyrase, topoisomerase IV) Efflux pumps Decreased permeability |
| Tetracyclines Chlortetracycline Tetracycline Doxycycline | Target resistance (ribosome) Drug detoxification Efflux pumps |
| Antibiotic Residue | Concentration | Food Product | Associated Health Risks | Source |
|---|---|---|---|---|
| Oxytetracycline | 2604.1 ± 703.7 μg/kg | Chicken muscle | Allergic hypersensitivity reactions or toxic effects (phototoxic skin reactions, chondrotoxic) | [44] |
| 3434.4 ± 604.4 μg/kg | Chicken liver | Carcinogenicity, cytotoxicity | ||
| 51.8 ± 90.53 μg/kg | Beef | Carcinogenicity, cytotoxicity | [45] | |
| Enrofloxacin | 0.73–2.57 μg/kg | Chicken meat | Allergic hypersensitivity reactions or toxic effects, phototoxic skin reactions, chondrotoxic. | [47] |
| Chloramphenicol | 1.34–13.9 μg/kg | Chicken | Bone marrow toxicity, optic neuropathy, brain abscess | |
| Penicillin | 0.87–1.3 μg/kg | Veal | Allergy, affects starter cultures for fermented milk products | |
| Oxytetracycline | 3.5–4.61 μg/kg | Chicken meat | Carcinogenicity, cytotoxicity in the bones of broiler chickens | |
| Quinolones | 30.81 ± 0.45 μg/kg | Chicken meat | Allergic hypersensitivity reactions or toxic effects (phototoxic skin reactions, chondrotoxic) | [47] |
| 6.64 ± 1.11 μg/kg | Beef | |||
| Amoxicillin | 9.8–56.16 μg/mL | Milk | Carcinogenic, teratogenic, and mutagenic effects | [48] |
| 10.46–48.8 μg/g | Eggs | |||
| Sulfonamides | 16.28 μg/g | Raw milk | Carcinogenicity, allergic reaction | [49] |
| Quinolones | 23.25 μg/g | Allergic hypersensitivity reactions or toxic effects (phototoxic skin reactions, chondrotoxic). |
| Microorganism | Sample Source | Antibiotic Resistance | Prevalence (%) | Source |
|---|---|---|---|---|
| Escherichia coli | Bovine milk sample | Azithromycin | 53 | [53] |
| Chloramphenicol | 15 | |||
| Ceftriaxone | 17 | |||
| Penicillin | 69 | |||
| Gentamicin | 6 | |||
| Amoxicillin | 55 | |||
| Tetracycline | 20 | |||
| Cephalexin | 64 | |||
| Listeria monocytogenes | Bovine milk sample | Azithromycin | 12 | |
| Chloramphenicol | 22 | |||
| Ceftriaxone | 17 | |||
| Penicillin | 46 | |||
| Gentamicin | 24 | |||
| Amoxicillin | 46 | |||
| Tetracycline | 23 | |||
| Cephalexin | 46 | |||
| Salmonella spp. | Bovine milk sample | Azithromycin | 8 | |
| Chloramphenicol | 6 | |||
| Ceftriaxone | 5 | |||
| Penicillin | 21 | |||
| Amoxicillin | 15 | |||
| Tetracycline | 5 | |||
| Cephalexin | 21 | |||
| Staphylococcus aureus | Bovine milk sample | Azithromycin | 8 | |
| Chloramphenicol | 6 | |||
| Ceftriaxone | 6 | |||
| Penicillin | 21 | |||
| Gentamicin | 3 | |||
| Amoxicillin | 25 | |||
| Tetracycline | 7 | |||
| Cephalexin | 25 | |||
| E. coli | Healthy farm workers | β-lactams | 77.3 | [54] |
| Pigs | 76.7 | |||
| Poultry broilers | 40 | |||
| S. aureus | Pigs | Methicillin | 30 | [55] |
| Campylobacter jejuni | Chicken | Ampicillin | 5 | [56] |
| Tetracycline | 31.7 | |||
| Ciprofloxacin | 23.3 | |||
| C. coli | Pork | Ampicillin | 33.3 | |
| Erythromycin | 73.3 | |||
| Tetracycline | 73.3 | |||
| Chloramphenicol | 6.7 | |||
| Ciprofloxacin | 46.7 |
| Bacteriocin | Source | Food Use | Reference |
|---|---|---|---|
| Nisin and Nisin Z | Lactococcus lactis | Prevents food spoilage caused by Lactobacillus spp., L. monocytogenes, S. aureus, and Clostridium spp. | [79] |
| lactococcin-G β | Lactococcus lactis | Activity against L. monocytogenes in yogurt, cheese, and sauerkraut | [84] |
| Leucocin A | Leuconostoc gelidum | Activity against E. coli and L. monocytogenes in meat and fish products. | [84] |
| Carnobacteriocin B2 | Carnobacterium maltaromaticum | Activity against L. monocytogenes in dairy, meat, or fish food and feed products | [85] |
| Curvacin A | Latilactobacillus curvatus | Activity against Listeria monocytogenes | [86] |
| Enterocin 7A | Enterococcus faecalis | Activity against L. monocytogenes in meat and meat-based products | [87] |
| Name | Source Organism | Molecular Weight (Da) | Net Charge pH 7 | Isoelectric Point | Hydrophobicity | Hydrophobic Moment |
|---|---|---|---|---|---|---|
| Nisin | L. lactis | 3456.62 | 3 | 8.52 | −0.29 | 0.48 |
| lactococcin-G β | L. lactis | 4107.19 | 4 | 10.42 | 0.25 | 0.71 |
| Leucocin A | L. gelidum | 3929.80 | 2 | 8.77 | 0.26 | 1.58 |
| Carnobacteriocin B2 | C. maltaromaticum | 4966.40 | 4 | 9.96 | 0.00 | 1.60 |
| Curvacin A | L. curvatus | 4306.03 | 3 | 9.37 | 0.11 | 1.69 |
| Enterocin 7A | E. faecalis | 5172.91 | 6 | 10.68 | 0.20 | 2.12 |
| Average | 4815.12 | 4 | 10.00 | 0.10 | 1.81 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu-Wu, J.W.F.; Guadamuz-Mayorga, C.; Oviedo-Cerdas, D.; Zamora, W.J. Antibiotic Resistance and Food Safety: Perspectives on New Technologies and Molecules for Microbial Control in the Food Industry. Antibiotics 2023, 12, 550. https://doi.org/10.3390/antibiotics12030550
Wu-Wu JWF, Guadamuz-Mayorga C, Oviedo-Cerdas D, Zamora WJ. Antibiotic Resistance and Food Safety: Perspectives on New Technologies and Molecules for Microbial Control in the Food Industry. Antibiotics. 2023; 12(3):550. https://doi.org/10.3390/antibiotics12030550
Chicago/Turabian StyleWu-Wu, Jannette Wen Fang, Carolina Guadamuz-Mayorga, Douglas Oviedo-Cerdas, and William J. Zamora. 2023. "Antibiotic Resistance and Food Safety: Perspectives on New Technologies and Molecules for Microbial Control in the Food Industry" Antibiotics 12, no. 3: 550. https://doi.org/10.3390/antibiotics12030550
APA StyleWu-Wu, J. W. F., Guadamuz-Mayorga, C., Oviedo-Cerdas, D., & Zamora, W. J. (2023). Antibiotic Resistance and Food Safety: Perspectives on New Technologies and Molecules for Microbial Control in the Food Industry. Antibiotics, 12(3), 550. https://doi.org/10.3390/antibiotics12030550