D- and N-Methyl Amino Acids for Modulating the Therapeutic Properties of Antimicrobial Peptides and Lipopeptides
Abstract
1. Introduction
2. Results and Discussion
2.1. Peptide Synthesis and Physicochemical Characterization
2.2. Antimicrobial Activity
2.3. Toxicity Characterization and Therapeutic Index (TI)
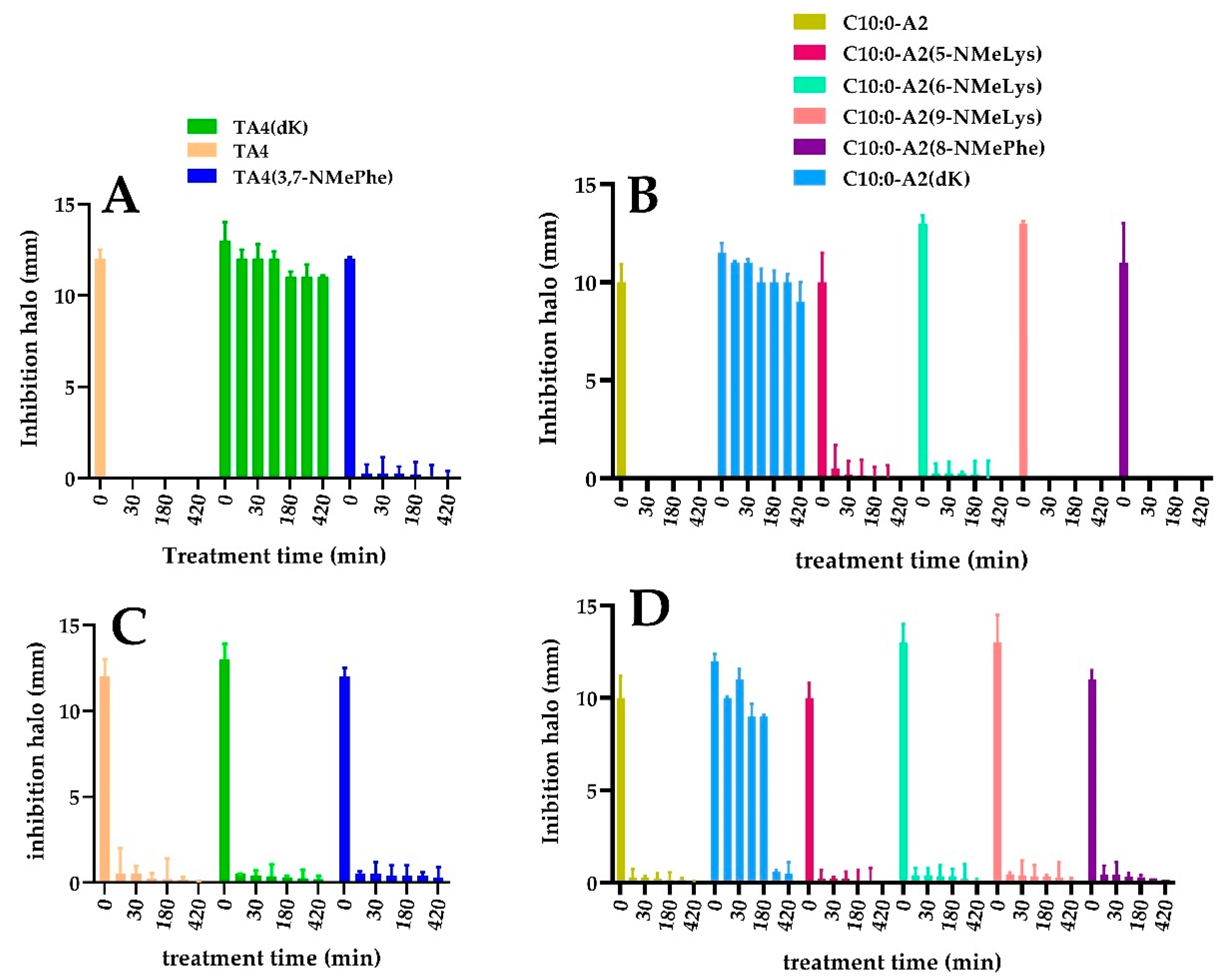
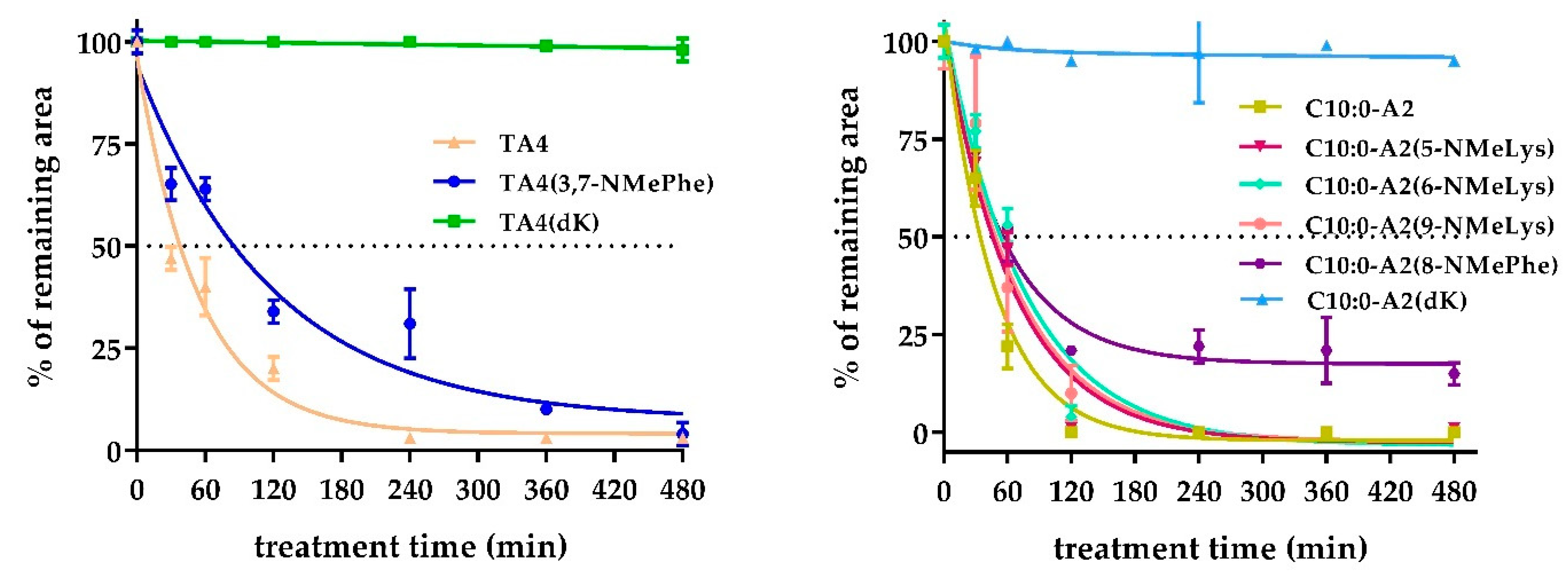
2.4. Enzymatic Stability Characterization in the Presence of Digestive Enzymes and Serum Proteases
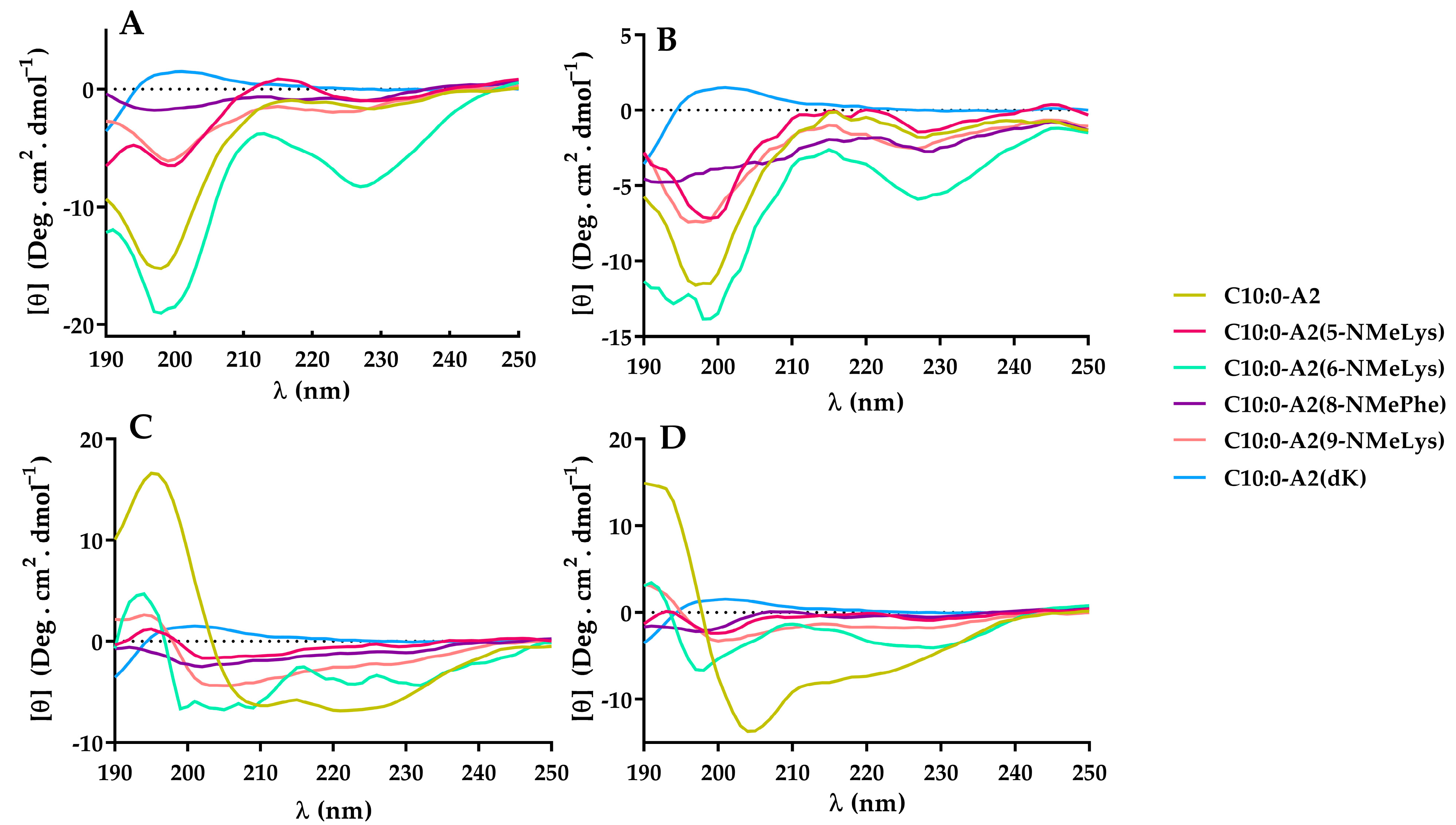
2.5. Secondary Structure Determination by Circular Dichroism (CD)
3. Material and Methods
3.1. Peptide Synthesis
3.2. Antimicrobial Activity
3.2.1. Minimal Inhibitory Concentration (MIC) against Bacteria
3.2.2. Minimal Inhibitory Concentration (MIC) against Yeast
3.3. Toxicity Characterization
3.3.1. Hemolysis Assay
3.3.2. Cytotoxicity Assay
3.4. Calculation of the Therapeutic Index (TI)
3.5. Characterization of Enzymatic Stability
3.5.1. Peptide Stability in the Presence of Digestive Enzymes
3.5.2. Peptide stability in Serum
3.6. Determination of Critical Micelle Concentration (CMC)
3.7. Secondary Structure Determination by Circular Dichroism (CD)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Loose, C.; Jensen, K.; Rigoutsos, I.; Stephanopoulos, G. A linguistic model for the rational design of antimicrobial peptides. Nature 2006, 443, 867–869. [Google Scholar] [CrossRef] [PubMed]
- Der Torossian Torres, M.; de la Fuente-Nunez, C. Reprogramming biological peptides to combat infectious diseases. Chem. Commun. 2019, 55, 15020–15032. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, A.; Torres, M.T.; de la Fuente-Nunez, C. Methods for the design and characterization of peptide antibiotics. Methods Enzymol. 2022, 663, 303–326. [Google Scholar] [CrossRef] [PubMed]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.; Harper, D.; et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Melo, M.C.R.; Flowers, L.; Crescenzi, O.; Notomista, E.; de la Fuente-Nunez, C. Mining for encrypted peptide antibiotics in the human proteome. Nat. Biomed. Eng. 2022, 6, 67–75. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Reffuveille, F.; Mansour, S.C.; Reckseidler-Zenteno, S.L.; Hernández, D.; Brackman, G.; Coenye, T.; Hancock, R.E. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 2015, 22, 196–205. [Google Scholar] [CrossRef]
- Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, Z.; Wang, X.; Wang, J. A recombinant fungal defensin-like peptide-P2 combats multidrug-resistant Staphylococcus aureus and biofilms. Appl. Microbiol. Biotechnol. 2019, 103, 5193–5213. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, N.; Teng, D.; Hao, Y.; Li, T.; Han, H.; Mao, R.; Wang, J. Resistance response to Arenicin derivatives in Escherichia coli. Appl. Microbiol. Biotechnol. 2022, 106, 211–226. [Google Scholar] [CrossRef]
- Liu, H.; Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Ma, X.; Wang, X.; Wang, J. Fatty acid modified-antimicrobial peptide analogues with potent antimicrobial activity and topical therapeutic efficacy against Staphylococcus hyicus. Appl. Microbiol. Biotechnol. 2021, 105, 5845–5859. [Google Scholar] [CrossRef]
- Ma, X.; Yang, N.; Mao, R.; Hao, Y.; Yan, X.; Teng, D.; Wang, J. The Pharmacodynamics Study of Insect Defensin DLP4 against Toxigenic Staphylococcus hyicus ACCC 61734 In Vitro and Vivo. Front. Cell. Infect. Microbiol. 2021, 11, 638598. [Google Scholar] [CrossRef] [PubMed]
- Molchanova, N.; Hansen, P.R.; Franzyk, H. Advances in Development of Antimicrobial Peptidomimetics as Potential Drugs. Molecules 2017, 22, 1430. [Google Scholar] [CrossRef] [PubMed]
- Böttger, R.; Hoffmann, R.; Knappe, D. Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum. PLoS ONE 2017, 12, e0178943. [Google Scholar] [CrossRef] [PubMed]
- Gentilucci, L.; De Marco, R.; Cerisoli, L. Chemical modifications designed to improve peptide stability: Incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization. Curr. Pharm. Des. 2010, 16, 3185–3203. [Google Scholar] [CrossRef]
- Nestor, J.J., Jr. The medicinal chemistry of peptides. Curr. Med. Chem. 2009, 16, 4399–4418. [Google Scholar] [CrossRef]
- Werle, M.; Bernkop-Schnürch, A. Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids 2006, 30, 351–367. [Google Scholar] [CrossRef]
- Hunter, H.N.; Jing, W.; Schibli, D.J.; Trinh, T.; Park, I.Y.; Kim, S.C.; Vogel, H.J. The interactions of antimicrobial peptides derived from lysozyme with model membrane systems. Biochim. Biophys. Acta 2005, 1668, 175–189. [Google Scholar] [CrossRef]
- Seebach, D.; Beck, A.K.; Bierbaum, D.J. The world of beta- and gamma-peptides comprised of homologated proteinogenic amino acids and other components. Chem. Biodivers. 2004, 1, 1111–1239. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, M.; Qiu, S.; Wang, J.; Peng, J.; Zhao, P.; Zhu, R.; Wang, H.; Li, Y.; Wang, K.; et al. Antimicrobial activity and stability of the D-amino acid substituted derivatives of antimicrobial peptide polybia-MPI. AMB Express 2016, 6, 122. [Google Scholar] [CrossRef]
- Húmpola, M.V.; Rey, M.C.; Carballeira, N.M.; Simonetta, A.C.; Tonarelli, G.G. Biological and structural effects of the conjugation of an antimicrobial decapeptide with saturated, unsaturated, methoxylated and branched fatty acids. J. Pept. Sci. 2017, 23, 45–55. [Google Scholar] [CrossRef]
- Húmpola, M.V.; Rey, M.C.; Spontón, P.G.; Simonetta, A.C.; Tonarelli, G.G. A Comparative Study of the Antimicrobial and Structural Properties of Short Peptides and Lipopeptides Containing a Repetitive Motif KLFK. Protein Pept. Lett. 2019, 26, 192–203. [Google Scholar] [CrossRef]
- Krause, E.; Bienert, M.; Schmieder, P.; Wenschuh, H. The Helix-Destabilizing Propensity Scale of d-Amino Acids: The Influence of Side Chain Steric Effects. J. Am. Chem. Soc. 2000, 122, 4865–4870. [Google Scholar] [CrossRef]
- De Vleeschouwer, M.; Sinnaeve, D.; Matthijs, N.; Coenye, T.; Madder, A.; Martins, J.C. Synthesis of N-Methylated Pseudodesmin A Analogues: On the Structural Importance of N-H Hydrogen Bonds. ChemistrySelect 2017, 2, 640–644. [Google Scholar] [CrossRef]
- Bach, A.C., II; Eyermann, C.J.; Gross, J.D.; Bower, M.J.; Harlow, R.L.; Weber, P.C.; DeGrado, W.F. Structural Studies of a Family of High Affinity Ligands for GPIIb/IIIa. J. Am. Chem. Soc. 1994, 116, 3207–3219. [Google Scholar] [CrossRef]
- Sikorska, E.; Dawgul, M.; Greber, K.; Iłowska, E.; Pogorzelska, A.; Kamysz, W. Self-assembly and interactions of short antimicrobial cationic lipopeptides with membrane lipids: ITC, FTIR and molecular dynamics studies. Biochim. Biophys. Acta (BBA)-Biomembr. 2014, 1838, 2625–2634. [Google Scholar] [CrossRef] [PubMed]
- Sikorska, E.; Stachurski, O.; Neubauer, D.; Małuch, I.; Wyrzykowski, D.; Bauer, M.; Brzozowski, K.; Kamysz, W. Short arginine-rich lipopeptides: From self-assembly to antimicrobial activity. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2242–2251. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Swarbrick, J.D.; Hussein, M.H.; Schneider-Futschik, E.K.; Hoyer, D.; Li, J.; Karas, J.A. The impact of backbone N-methylation on the structure-activity relationship of Leu10-teixobactin. J. Pept. Sci. 2019, 25, e3206. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; MacDonald, D.L.; Holroyd, K.J.; Thornsberry, C.; Wexler, H.; Zasloff, M. In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrob. Agents Chemother. 1999, 43, 782–788. [Google Scholar] [CrossRef]
- Fritsche, T.R.; Rhomberg, P.R.; Sader, H.S.; Jones, R.N. Antimicrobial Activity of Omiganan Pentahydrochloride against Contemporary Fungal Pathogens Responsible for Catheter-Associated Infections. Antimicrob. Agents Chemother. 2008, 52, 1187–1189. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Fedler, K.A.; Rennie, R.P.; Stevens, S.; Jones, R.N. Omiganan Pentahydrochloride (MBI 226), a Topical 12-Amino-Acid Cationic Peptide: Spectrum of Antimicrobial Activity and Measurements of Bactericidal Activity. Antimicrob. Agents Chemother. 2004, 48, 3112–3118. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bionda, N.; Yongye, A.; Geer, P.; Stawikowski, M.; Cudic, P.; Martinez, K.; Houghten, R.A. Dissociation of antimicrobial and hemolytic activities of gramicidin S through N-methylation modification. ChemMedChem 2013, 8, 1865–1872. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.M.; Edwards, M.A.; Li, J.; Yip, C.M.; Deber, C.M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 2012, 287, 7738–7745. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.L.; Hodges, R.S. Structure-activity relationships of de novo designed cyclic antimicrobial peptides based on gramicidin S. Biopolymers 2003, 71, 28–48. [Google Scholar] [CrossRef]
- Dong, Q.G.; Zhang, Y.; Wang, M.S.; Feng, J.; Zhang, H.H.; Wu, Y.G.; Gu, T.J.; Yu, X.H.; Jiang, C.L.; Chen, Y.; et al. Improvement of enzymatic stability and intestinal permeability of deuterohemin-peptide conjugates by specific multi-site N-methylation. Amino Acids 2012, 43, 2431–2441. [Google Scholar] [CrossRef]
- Hong, S.Y.; Oh, J.E.; Lee, K.H. Effect of D-amino acid substitution on the stability, the secondary structure, and the activity of membrane-active peptide. Biochem. Pharmacol. 1999, 58, 1775–1780. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, D.; Xi, X.; Wu, Y.; Ma, C.; Zhou, M.; Wang, L.; Yang, M.; Chen, T.; Shaw, C. Identification and Characterisation of the Antimicrobial Peptide, Phylloseptin-PT, from the Skin Secretion of Phyllomedusa tarsius, and Comparison of Activity with Designed, Cationicity-Enhanced Analogues and Diastereomers. Molecules 2016, 21, 1667. [Google Scholar] [CrossRef]
- Hancock, R.E.W. Recently Modified Methods Used by Hancock Laboratory. Available online: http://cmdr.ubc.ca/bobh/methods/MODIFIEDMIC.html (accessed on 19 March 2022).
- Siano, A.; Húmpola, M.V.; Rey, M.C.; Simonetta, A.; Tonarelli, G.G. Interaction of acylated and substituted antimicrobial peptide analogs with phospholipid-polydiacetylene vesicles. Correlation with their biological properties. Chem. Biol. Drug Des. 2011, 78, 85–93. [Google Scholar] [CrossRef]
- Tagg, J.R.; McGiven, A.R. Assay system for bacteriocins. Appl. Microbiol. 1971, 21, 943. [Google Scholar] [CrossRef] [PubMed]
- Fuguet, E.; Rafols, C.; Roses, M.; Bosch, E. Critical micelle concentration of surfactants in aqueous buffered and unbuffered systems. Anal. Chim. Acta 2005, 548, 95–100. [Google Scholar] [CrossRef]
- Ladokhin, A.S.; Fernández-Vidal, M.; White, S.H. CD spectroscopy of peptides and proteins bound to large unilamellar vesicles. J. Membr. Biol. 2010, 236, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]



| Identification | Sequence | Net Charge pH = 7 | Experi Mental MM | rt | CMC (mM) |
|---|---|---|---|---|---|
| TA4 | KLFKKLFKKLFK-NH2 | +7 | 1567.16 | 17.2 | ND |
| TA4(3,7-NMePhe) | KL(NMeF)KKL(NMeF)KKLFK-NH2 | +7 | 1595.20 | 16.7 | ND |
| TA4(dK) | dKdKLFdKdKLFdKdKLFdK-NH2 | +7 | 1567.04 | 15.4 | ND |
| C10:0-A2 | C10:0-IKQVKKLFKK-NH2 | +5 | 1413.08 | 17.8 | 5.74 |
| C10:0-A2(5-NMeLys) | C10:0-IKQV(NMe-K)KLFKK-NH2 | +5 | 1426.87 | 16.3 | 3.73 |
| C10:0-A2(6-NMeLys) | C10:0-IKQVK(NMe-K)LFKK-NH2 | +5 | 1426.99 | 16.1 | 3.63 |
| C10:0-A2(9-NMeLys) | C10:0-IKQVKKLF(NMe-K)K-NH2 | +5 | 1427.08 | 17.7 | 3.90 |
| C10:0-A2(8-NMePhe) | C10:0-IKQVKKL(NMe-F)KK-NH2 | +5 | 1427.03 | 15.3 | 2.01 |
| C10:0-A2(dK) | C10:0-IdKQVdKdKLFdKdK-NH2 | +5 | 1413.10 | 17.3 | 5.68 |
| Microorganism | TA4 | TA4(3,7-NMePhe) | TA4(dK) | C10:0-A2 | C10:0-A2(5-NMeLys) | C10:0-A2(6-NMeLys) | C10:0-A2(9-NMeLys) | C10:0-A2(8-NMePhe) | C10:0-A2(dK) | |
|---|---|---|---|---|---|---|---|---|---|---|
| MIC (µM) | ||||||||||
| Gram negative | E. coli ATCC 35218 | 5.1 | 5.0 | 5.1 | 1.4 | 2.8 | 1.4 | 2.8 | 11.2 | 22.6 |
| P. aeruginosa ATCC 27853 | 2.6 | 1.2 | 10.2 | 1.4 | 5.6 | 0.7 | 0.7 | 5.6 | 1.6 | |
| Gram positive | S. aureus ATCC 25923 | 2.6 | 5.0 | 10.2 | 1.4 | 2.8 | 1.4 | 2.8 | 2.8 | >90.6 |
| S. aureus RM * SA1 | 10.2 | 10.0 | 20.4 | 2.8 | 5.6 | 5.6 | 2.8 | 5.6 | >90.6 | |
| E. faecalis ATCC 29212 | 5.1 | 40.0 | 20.4 | 1.4 | 11.2 | 5.6 | 5.6 | 11.2 | >90.6 | |
| Yeast | C. albicans PEEC 2 | 40.8 | 80.2 | >163.3 | 90.6 | 179.5 | 44.9 | 89.7 | 179.5 | 90.6 |
| C. tropicalis DBFIQ 3 | 40.8 | 80.2 | 163.3 | 90.6 | 179.5 | 89.7 | 179.5 | 179.5 | 181.2 | |
| Identification | MIC Average (µM) | HC50 ± SD (µM) | IC50 ± SD (µM) | TI | ||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Yeast | Bacteria | Yeast | |||||
| Gram (−) | Gram (+) | Gram (−) | Gram (+) | |||||
| TA4 | 3.9 | 6.0 | 40.8 | >400 | 46.35 ± 10.13 | 102.6/11.9 | 66.7/7.7 | 9.8/1.1 |
| TA4(3,7-NMePhe) | 3.1 | 28.3 | 80.2 | >400 | 86.56 ± 7.35 | 129.0/27.9 | 14.1/3.1 | 5.0/1.1 |
| TA4(dK) | 7.7 | 17.0 | 163.3 | >400 | >400 | 51.9 | 23.5 | 2.4 |
| C10:0-A2 | 1.4 | 1.9 | 90.6 | 202.9 ± 17.5 | 32.62 ± 2.49 | 144.9/23.3 | 106.8/17.2 | 2.2/0.4 |
| C10:0-A2(5-NMeLys) | 4.2 | 6.5 | 179.5 | >400 | >400 | 95.2 | 61.5 | 2.2 |
| C10:0-A2(6-NMeLys) | 1.1 | 4.2 | 67.3 | >400 | 399.90 ± 28.96 | 363.6 | 95.2 | 5.9 |
| C10:0-A2(9-NMeLys) | 1.8 | 3.7 | 134.6 | 298.5 ± 12.0 | >400 | 165.8/222.2 | 80.7/108.1 | 2.2/3.0 |
| C10:0-A2(8-NMePhe) | 8.4 | 6.5 | 179.5 | >400 | >400 | 47.6 | 61.5 | 2.2 |
| C10:0-A2(dK) | 12.1 | >90.6 | 135.9 | >400 | >400 | 33.1 | <4.4 | 2.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Humpola, M.V.; Spinelli, R.; Erben, M.; Perdomo, V.; Tonarelli, G.G.; Albericio, F.; Siano, A.S. D- and N-Methyl Amino Acids for Modulating the Therapeutic Properties of Antimicrobial Peptides and Lipopeptides. Antibiotics 2023, 12, 821. https://doi.org/10.3390/antibiotics12050821
Humpola MV, Spinelli R, Erben M, Perdomo V, Tonarelli GG, Albericio F, Siano AS. D- and N-Methyl Amino Acids for Modulating the Therapeutic Properties of Antimicrobial Peptides and Lipopeptides. Antibiotics. 2023; 12(5):821. https://doi.org/10.3390/antibiotics12050821
Chicago/Turabian StyleHumpola, Maria Veronica, Roque Spinelli, Melina Erben, Virginia Perdomo, Georgina Guadalupe Tonarelli, Fernando Albericio, and Alvaro Sebastian Siano. 2023. "D- and N-Methyl Amino Acids for Modulating the Therapeutic Properties of Antimicrobial Peptides and Lipopeptides" Antibiotics 12, no. 5: 821. https://doi.org/10.3390/antibiotics12050821
APA StyleHumpola, M. V., Spinelli, R., Erben, M., Perdomo, V., Tonarelli, G. G., Albericio, F., & Siano, A. S. (2023). D- and N-Methyl Amino Acids for Modulating the Therapeutic Properties of Antimicrobial Peptides and Lipopeptides. Antibiotics, 12(5), 821. https://doi.org/10.3390/antibiotics12050821







