Abstract
The presence of antimicrobial-resistant Enterococci in poultry is a growing public health concern worldwide due to its potential for transmission to humans. The aim of this study was to determine the prevalence and patterns of antimicrobial resistance and to detect drug-resistant genes in Enterococcus faecalis and E. faecium in poultry from four districts in Zambia. Identification of Enterococci was conducted using phenotypic methods. Antimicrobial resistance was determined using the disc diffusion method and antimicrobial resistance genes were detected using polymerase chain reaction and gene-specific primers. The overall prevalence of Enterococci was 31.1% (153/492, 95% CI: 27.1–35.4). Enterococcus faecalis had a significantly higher prevalence at 37.9% (58/153, 95% CI: 30.3–46.1) compared with E. faecium, which had a prevalence of 10.5% (16/153, 95% CI: 6.3–16.7). Most of the E. faecalis and E. faecium isolates were resistant to tetracycline (66/74, 89.2%) and ampicillin and erythromycin (51/74, 68.9%). The majority of isolates were susceptible to vancomycin (72/74, 97.3%). The results show that poultry are a potential source of multidrug-resistant E. faecalis and E. faecium strains, which can be transmitted to humans. Resistance genes in the Enterococcus species can also be transmitted to pathogenic bacteria if they colonize the same poultry, thus threatening the safety of poultry production, leading to significant public health concerns.
1. Introduction
Enterococcus is a genus of Gram-positive bacteria in the family Enterococcaceae, the order Lactobacillales and the phylum Firmicutes [1]. Enterococcus is part of the normal flora in the gastrointestinal tract (GIT) of mammals, fish, reptiles, insects, and birds [2,3]. Being ubiquitous in nature, it is also found in soil, plants, sewage and fresh and salt water [4,5]. Species in the genus Enterococcus (E) have emerged as pathogens of medical and public health importance [6]. This is partly due to their adaptability to the selective pressures of antimicrobials. They also have the ability to acquire, express, and transmit mobile genetic elements (MGEs) from/to pathogenic as well as non-pathogenic species in the same or different genus [7,8], leading to the development of antimicrobial resistance. MGEs play an important role in facilitating horizontal genetic exchange and promoting the acquisition and transmission of resistance genes [9]. These properties have made Enterococcus an important human pathogen responsible for a number of clinical conditions, including urinary tract infections (UTI), endocarditis, bacteremia and mastitis in humans and animals [10,11]. Enterococcus species also cause locomotive disorders and septicemia in broilers [12]. Enterococci is ranked among the major causes of nosocomial infections worldwide [13]. This is especially true for Enterococcus (E) faecalis and E. faecium. The emergence of multidrug-resistant (MDR) Enterococci such as vancomycin-resistant Enterococci (VRE) and drug-resistant Enterococci in poultry are of major public health concern because of the limited treatment options available for infections caused by such species, as well as the possibility of dispersion between poultry and humans [3,14,15,16] and the transfer of resistance genes to other bacteria (9). This has led to an increase in infections caused by multidrug-resistant Enterococci, which can not only be very difficult to treat but can also lead to increased mortality rates [17].
Enterococcal infections can be serious and are associated with increased healthcare costs, including the cost of hospitalization, laboratory testing and antibiotic treatment [18]. Enterococcal infections can also lead to lost productivity due to missed work or school.
Although Enterococcus faecalis and Enterococcus faecium are commonly found in the guts of poultry, they can cause infections in poultry that can lead to significant economic losses for the industry. Enterococcal infections in poultry can result in decreased growth rates, reduced feed efficiency and increased mortality rates [19]. Poultry and food products of poultry origin are the most consumed worldwide [20]. Enterococci can contaminate poultry products and pose a risk to human health if consumed [21]. Antibiotic resistance in Enterococci is also a concern for the poultry industry, as the use of antibiotics in poultry production can contribute to the development and spread of antibiotic-resistant strains [22]. Therefore, the presence of antimicrobial-resistant Enterococci, especially multidrug resistance Enterococcus species, in poultry is of public health concern as it may serve as a pool from which antimicrobial resistance genes are disseminated. VRE is a nosocomial pathogen that exhibits multidrug resistance (MDR) and virulence.
Enterococcus faecium has transitioned from a commensal organism to an ESKAPE (E. faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogen. ESKAPE is an acronym for a group of life-threatening nosocomial pathogens that successfully evade the effect of antimicrobial drugs and represent a model for pathogenesis, transmission, and resistance [23]. VRE cause a greater number of infections than other nosocomial pathogens in hospitals in the United States [23].
Vancomycin-resistant Enterococci have been reported worldwide [24], including in Zambia [25]. However, they have not been given the same attention as other commensals of the GIT such as Staphylococci, Salmonella, Shigella, Campylobacter and Escherichia coli. Zambia developed a multi-sectoral national action plan in recognition of the public health threat of morbidity, mortality, and economic outcomes of antimicrobial resistance. However, minimal surveillance and research have been conducted on MDR Enterococci in Zambia. This study aimed to determine the prevalence of antimicrobial resistance and the presence of antimicrobial-resistant genes in Enterococcus faecalis and Enterococcus faecium isolates from poultry in four districts in Zambia.
2. Results
2.1. Identification
2.1.1. Identification of Enterococci Using Analytical Profile Index (API)
Of the 37 poultry isolates subjected to API identification using BioMérieux’s Analytical Profile Index (API) 20 Strep test kits, 19 were identified as Enterococcus faecalis, 15 as Enterococcus faecium and one as Enterococcus durans. Two were not identified. The reason for performing API tests on only 37 isolates was due to insufficient reagents. Particularly, the NIN, VP 1 + VP 2, ZYM A and ZYM B were enough for only 38 samples (one control Enterococcus faecalis ATCC 29212 strain and the 37 isolates).
2.1.2. Identification of Enterococci Using Polymerase Chain Reaction (PCR)
PCR was run on 343 suspect Enterococcus DNA samples extracted from poultry droppings using genus-specific primers for elongation factor (tuf) and d-alanine-d-alanine ligase (ddl) genes. PCR was subsequently run on 153 positive DNA samples using species-specific primers for Enterococcus faecalis and Enterococcus faecium. The most common Enterococcus species was E. faecalis (37.9%), followed by E. faecium (10.5%). Remarkably, 38.6% of the isolates contained more than one species, with 34.6% of the total enterococcus isolates containing both E. faecalis and E. faecium. Adding the latter to E. faecalis and E. faecium, E. faecalis would still be the most predominant species, followed by E. faecium (Figure 1). The word “Other” represents Enterococcus species—identified by PCR using genus-specific ddl and tuf gene primers—which could not be identified through PCR due to lack of additional species-specific primers, or DNA sequencing due to the unavailability of reagents. Figure 1 shows the species identified using E. faecalis and E. faecium species-specific primers.
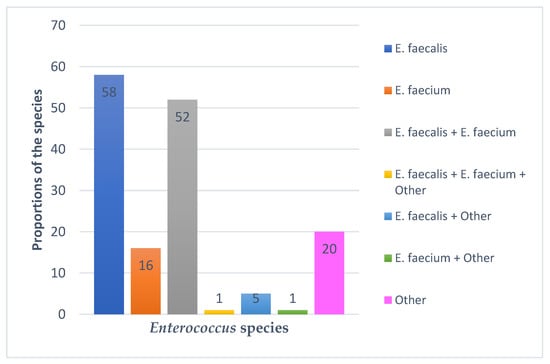
Figure 1.
Enterococcus species identified using species-specific E. faecalis and E. faecium primers.
2.1.3. Comparing API and PCR Identification
API and PCR results were compared to ascertain the agreement between the two methods. API correctly identified 17 (45.9%) of the 37 isolates. API could not identify isolates with more than one species and only picked one of the species in samples with two or more species (16, 43.2%). It also misidentified an isolate that contained E. faecalis and another species as E. faecium, and it was not able to identify two isolates. Additionally, API identified one isolate as E. durans1, but this could not be confirmed as the corresponding species-specific primers were not available (Table 1).

Table 1.
API and PCR Identities.
2.2. Prevalence of Enterococci
2.2.1. Overall Prevalence
The overall prevalence of Enterococci was 31.1% (153/492, CI: 27.1–35.4), while the prevalence in Lusaka Province was 30.8% (33/107, CI: 22.5–40.6) and the prevalence in Copperbelt Province was 31.2% (120/385, 26.6–36.1). Table 2 contains summaries of the prevalence of E. faecalis and E. faecium (combined and separate) in poultry from districts in the Copperbelt and Lusaka Provinces.

Table 2.
Prevalence of Enterococcus in Poultry from the four Districts.
2.2.2. Species-Specific Prevalence of Isolates
The prevalence of Enterococci varied significantly across the districts. Lusaka district had the highest prevalence at 44.0% (22/50, CI: 30.3–58.7) compared with the other three districts of Kitwe, Ndola and Chongwe (p = 0.038). The prevalence of E. faecalis was higher than that of E. faecium in all districts (p = 0.012) except Kitwe district (p = 0.044) (Table 3).

Table 3.
Prevalence of specific species across the study area.
2.3. Antimicrobial Susceptibility Test Results
2.3.1. Antimicrobial Susceptibility of Enterococci
All intermediate test results were considered resistant. Both Enterococcus species showed very high (97.3%) resistance to tetracycline, while 94.6% were resistant to erythromycin and 77.0% were resistant to ciprofloxacin. Remarkably, 64.9% of both Enterococcus species were susceptible to vancomycin. More than 90.0% of E. faecalis isolates were resistant to erythromycin and tetracycline and more than 50.0% were resistant to ampicillin, chloramphenicol and ciprofloxacin. Less than 20.0% of the E. faecalis isolates were resistant to vancomycin. All E. faecium isolates in this study were resistant to erythromycin. More than 80.0% of E. faecium isolates exhibited phenotypic resistance to ampicillin, ciprofloxacin and tetracycline, while less than 40.0% showed resistance to chloramphenicol and vancomycin. Susceptibility profiles of E. faecalis and E. faecium to the eight antimicrobials tested are provided in Table 4.

Table 4.
Antimicrobial Susceptibility Profiles of Enterococcus faecalis and Enterococcus faecium.
2.3.2. Number of Enterococcus Isolates Resistant to One, Two, Three or More Antimicrobial Classes
Multidrug resistance (MDR) is defined as resistance to three or more classes of antimicrobials. The results of our study show that none of the isolates were susceptible to all antimicrobial classes tested. Of the 74 E. faecalis and E. faecium isolates tested against eight antimicrobials, only two (2.7%) were resistant to one class of antimicrobials. A total of 5 (6.8%) isolates were resistant to two classes of antimicrobials. The majority of E. faecalis and E. faecium isolates (67, 90.5%) were MDR (Table 5).

Table 5.
Number of Isolates Resistant against one, two, three or more Antimicrobial Classes.
2.4. Detected Antimicrobial Resistance Genes
2.4.1. Presence of Antimicrobial Resistance Genes in E. faecalis and E. faecium
The aac(6′)-Ie-aph(2″)-LA resistance gene encoding resistance to gentamycin was detected in 33 and 12 E. faecalis and E. faecium isolates, respectively, representing 60.8% of the isolates. The ermB resistance gene was more common in both E. faecalis and E. faecium compared with the ermA gene. The vanA resistance gene was detected in only two E. faecalis isolates and in none of the E. faecium isolates. Table 6 shows the number of different resistance genes detected in the E. faecalis and E. faecium isolates.

Table 6.
The number of different resistance genes detected in E. faecalis and E. faecium isolates.
2.4.2. Resistance Genes in E. faecalis and E. faecium isolates across the Study Area
The most common resistance genes in E. faecalis isolates from Chongwe district in Lusaka Province were tetL and tetM, as they were found in all five isolates. These were followed by aac and tetK, which were detected in four of the five isolates, and ermB, which was detected in three isolates. The most common resistance genes in isolates from Lusaka district were tetK and tetM, which were found in all 8 E. faecalis isolates. These were followed by tetL (7/8), aac (6/8) and ermB (5/8). Resistant genes commonly detected in E. Faecalis from Ndola district were tetM (16/20), tetL (15/20), aac (13/20), ermB (12/20) and tetK (11/20). In E. faecalis isolates from Kitwe, tetK (17/25) was the most prevalent resistance gene, followed by tetM (16/25), ermB (15/25), tetL (14/25) and aac (10/25). The most commonly detected resistance genes in E. faecium isolates from Kitwe district were aac (5/8) and ermB (5/8), followed by tetM (4/8) and tetK (3/8). In Lusaka district, the most common genes were aac (5/5) followed by ermB (4/5), tetL (4/5), tetK (3/5) and tetM (3/5). Table 7 shows all resistance genes detected in E. faecalis and E. faecium isolates from the four districts in Zambia.

Table 7.
Resistance genes detected in E. faecalis and E. faecium isolates from poultry in Copperbelt and Lusaka Provinces.
2.5. Association between Antimicrobials and Resistance Genes
Differences in antimicrobial resistance patterns and resistance genes in both enterococcus species were analyzed to assess possible associations between resistance phenotypes and their corresponding genotypes. A positive association between phenotype and genotype was found for tetracycline (p = 0.047) and erythromycin (p = 0.008), but there was no association between genotype and the vancomycin resistance phenotype (p = 0.051) (Table 8).

Table 8.
Association between antimicrobial results and their corresponding resistance genes.
3. Discussion
The prevalence, antimicrobial susceptibility patterns and presence of resistance genes in poultry droppings from four districts in Zambia were determined. The overall prevalence of Enterococci was 31.1%. This is in agreement with other studies that reported similar results in Poland [26], Malaysia [27] and Nigeria [28]. However it was lower than that reported in a similar study conducted in Zambia, where the prevalence was 88.4% in laying hens [29]. This could be due to differences in sampling methods, farms sampled and the number of farms sampled. Another previous study [30] also reported a higher prevalence than that reported in the present study. Conversely, the prevalence rate in our study was higher than the rates reported in Ethiopia [31], Pakistan [32] and Thailand [33]. The differences in the isolation rates of Enterococci can be attributed to several factors, including antibiotic use, environmental factors and methodology. The widespread use of antibiotics has led to the selection and dissemination of antibiotic-resistant Enterococci. Enterococci are found in soil and water and can persist in the environment for long periods of time, making them more difficult to control and leading to increased isolation rates. The isolation rate can also be influenced by the type of culture method used and the presence of selective media that may preferentially isolate Enterococci [34].
Among the Enterococcus species isolated in this study, E. faecalis was the most prevalent (37.9%), followed by E. faecium (10.5%). This was in agreement with other studies [35,36,37,38] which found E. faecalis to be the most prevalent species in poultry. However, our study differed slightly from some studies that found species other than E. faecalis to be the most predominant [16,39,40]. The variations in species levels between studies might be due to differences in the type of poultry, source of chicks, sampling methods, geographical disparities, the time of study and isolation and identification procedures [40].
Although API 20 strep is considered the best identification system for bacteria [41], it does not accurately identify some species of Enterococci [42]. In the present study, we validated API 20 strep results using PCR. PCR conducted using species-specific primers identified 91.9% of samples containing both single and multiple species. API 20 strep accurately identified 45.3% of Enterococcus species, but identified only one species in isolates containing more than one species. It also misidentified 2.7% of the Enterococcus species. Our findings were in agreement with the results of previous studies [42,43,44,45].
In the present study, phenotypic resistance to critically important antimicrobials, as defined by the WHO [46], was observed and 90.5% of E. faecalis and E. faecium isolates were multidrug resistant (MDR) (Table 4). Notably, all E. faecalis and E. faecium isolates were resistant to one or more of the tested antimicrobials (Table 5). These findings were similar to those of a study done earlier [47] in which the majority of E. faecalis and E. faecium isolates were resistant to one or more of the tested antimicrobials. Resistance to all tested antimicrobials was also observed in both E. faecalis and E. faecium isolates.
More than 50.0% of E. faecalis isolates were resistant to all tested antimicrobials, while 100.0% of the isolates were resistant to tetracycline. On average, E. faecium exhibited increased resistance to antimicrobials in comparison with E. faecalis. Our findings are in agreement with a recent study conducted in Zambia [29]. Our study also has some similarities with a study conducted in the Czech Republic [48], in which increased resistance of Enterococci to tetracycline, erythromycin and nitrofurantoin were observed, as well as a study from USA [49], in which Enterococci resistance to tetracycline, penicillin and ciprofloxacin was documented. Furthermore, our results are consistent with findings from previous studies [50,51,52,53,54,55,56] where high tetracycline resistance was reported. Our results were also comparable with those of the study by Fracalanzza et al. [57], which recorded the resistance of Enterococci to erythromycin to be at 82.0% when intermediate results were included. Nevertheless, that study noted reduced resistance to tetracycline (38.3%) and chloramphenicol (5.7%) compared with our study. The observed increase in resistance to all the antimicrobials tested indicate that poultry from these four districts in Zambia can be a source of MDR Enterococci. However, our study contrasted with other studies [58,59], which reported lower levels of resistance to antimicrobials.
Although the gene aac(6′)-Ie-aph(2″)-LA, which encodes resistance to gentamicin, was detected in 60.8% of both Enterococci species tested, an association with susceptibility could not be determined, as discs containing high concentrations of gentamicin (for example 120 μg or 500 μg), which are used to detect high-level aminoglycoside resistance, were not available.
The associations between antimicrobial resistance phenotypes and genotypes in E. faecalis and E. faecium isolates were analyzed. Associations were found between genotypes and tetracycline and erythromycin resistant phenotypes. However, genotypes showed no relationship with vancomycin resistant phenotypes. The disparity observed between the phenotypes and genotypes in the case of vancomycin could be due to the fact that vancomycin resistance in Enterococci can be conferred by different gene clusters [60,61,62].
4. Materials and Methods
4.1. Study Design and Sites
A cross-sectional study was conducted in selected farms in Chongwe and Lusaka (Lusaka Province) and Ndola and Kitwe (Copperbelt Province) districts in Zambia (Figure 2). The two provinces are among those that harbor most of the commercial poultry farms in Zambia.
Figure 2.
Map of the study areas.
4.2. Sample Collection
A total of 492 freshly voided poultry droppings were collected from layers, broilers and village chickens. Five different visits were made to selected poultry farms in four districts in the Copperbelt and Lusaka Provinces in Zambia (Figure 2). Of the total samples collected, 57 were from farms in Chongwe, while 50 were from Lusaka district in Lusaka Province. Of the 385 samples from Copperbelt Province, 140 and 245 came from Ndola and Kitwe districts, respectively.
4.3. Laboratory Investigations
4.3.1. Isolation of Enterococci
Conventional microbiological assays were performed to detect and identify Enterococcus species as described by Facklam and Collins [63]. Briefly, 1 g of poultry droppings was suspended in 9 mL buffered peptone water (BPW) (HIMEDIA, India), mixed and incubated at 37 °C for 24 h. A loopful of the BPW suspension was streaked on Bile Esculin Agar (BEA) (HIMEDIA, India) and incubated at 37 °C for 24 h. Following this, colonial traits were noted and smears of suspect colonies (small black shiny colonies on BEA) were made and stained using Central Drug House’s (CDH) Gram’s color staining kit from India. Gram-positive cocci appearing in chains, doubles or singles were characteristic of enterococci. A total of 343 suspected Enterococcus isolates were recovered from the 492 samples tested. These were stored in 20% glycerol at −20 °C for subsequent experiments.
4.3.2. Identification of Enterococci Using Analytical Profile Index (API)
Species identification based on phenotypic characteristics and biochemical tests was conducted using BioMérieux’s Analytical Profile Index (API) 20 Strep test kits. A total of 37 isolates were identified using the API 20 Strep test kits. The reasons for this are stated in Section 2.1.1.
4.3.3. DNA Extraction
Colonies grown overnight on a blood agar plate were placed in a test tube containing 0.5 mL of molecular grade water, vortexed and boiled at 95 °C for 10 min and then centrifuged for 5 min at 1500× g. The supernatant was pipetted into cryo-vials and stored at −20 °C for further analysis.
4.3.4. Molecular Identification of Enterococci
Molecular identification of the Enterococcus species was conducted using single PCR and the genus-specific and species-specific primers shown in Table 9, following the procedure described by Li et al. (2012) [64]. PCR amplification of elongation factor (tuf) and D -Ala- D -Ala ligase (ddl) in the extracted DNA was conducted using Phusion Flash High-Fidelity PCR Master Mix (Thermofisher Scientific, USA) in a thermal cycler (Applied Biosystems, Chiba, Japan) under the following PCR conditions: initial denaturation at 98 °C for 2 min followed by 30 cycles of denaturation at 98 °C for 5 s, annealing at 56 °C for 5 s, and extension at 72 °C for 30 s. A final extension was performed at 72 °C for 1 min. PCR amplicons were run on 1.5% agarose gels. The expected bandwidths for tuf and ddl PCR products were 112 bp and 475 bp, respectively. For species identification, species-specific primers (Table 1) targeting the superoxide dismutase (sodA) gene in E. faecalis and E. faecium were used. No primers were available for other species. The PCR conditions were similar to those used for genus amplification, except for the annealing temperature, which was set to 52 °C for both species.

Table 9.
Primers for Enterococcus Genus and Species identification.
4.3.5. Determination of Antimicrobial Resistance Levels
Susceptibility to vancomycin (30 μg), erythromycin (15 μg), ampicillin (10 μg), penicillin (10 U), tetracycline (30 μg), nitrofurantoin (300 μg), ciprofloxacin (5 μg) and chloramphenicol (30 μg) was determined using the disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines [68]. The disks used for susceptibility testing were manufactured by HIMEDIA, India. Diameters of the zones of inhibition were recorded in millimeters (mm) and interpreted as susceptible, intermediate or resistant. In this study, intermediate results were taken as resistant. A reference strain, Enterococcus faecalis 29,212, was used as a control strain.
4.3.6. Detection of Antimicrobial Resistant Genes (ARG)
Genes conferring resistance to aminoglycosides [aac(6′)-le-aph(2″)-LA], which in this study was abbreviated as “aac”, macrolides (ermA and ermB), tetracyclines (tetM, tetL, tetK, and tetX) and glycopeptides (vanA) were detected in a single PCR using the gene-specific primers shown in Table 10. One Taq Quick-load 2X Master Mix (Biolabs, Durham, NC, USA) was used for amplification in a thermal cycler (Applied Biosystems, Chiba, Japan). The following PCR conditions were employed: initial denaturation at 93 °C for 3 min followed by 35 cycles of denaturation at 93 °C for 60 s, annealing at 52 °C for 60 s and elongation at 72 °C for 60 s. The final elongation step was performed at 72 °C for 5 min. PCR amplicons were run on 1.5% agarose gels. The expected sizes of the PCR products differed for each gene (Table 10).

Table 10.
Primers used for the Detection of Resistance Genes.
4.4. Data Analysis
Data were entered, cleaned and validated in a Microsoft™ excel spreadsheet (MS Office Excel® 2016). The data were then exported to SPSS software ver. 21 (IBM Corp., Armonk, NY, USA). PCR results (positive or negative) were reference variables for descriptive analyses. Univariate analyses were conducted for descriptive statistics and data are presented as frequencies, percentages and prevalence.
Author Contributions
Conceptualization, G.M. and B.M.H.; methodology, G.M.; software, G.M. and H.K.; validation, G.M.; formal analysis, G.M., H.K. and S.A.K.; investigation, G.M.; resources, G.M., C.N. and Y.S.; data curation, G.M. and H.K.; writing—original draft preparation, G.M.; writing—review and editing, G.M., B.M.H., H.K. and S.A.K.; visualization, G.M., B.M.H., H.K. and S.A.K.; supervision, B.M.H., C.N. and Y.S.; project administration, G.M.; funding acquisition, G.M., C.N. and Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Africa Center of Excellence for Infectious Diseases of Humans and Animals (ACEIDHA), grant number P151847 funded by the World Bank and “the APC was funded by ACEIDHA”. This work was partially supported by the Japan Agency for Medical Research and Development with grant numbers JP223fa627005 and JP21wm0125008.
Institutional Review Board Statement
Ethics approval was obtained from The University of Zambia Biomedical Research Ethics Committee (UNZABREC), (Protocol code 797-2020, 16 July 2020). Final study clearance and the authority to conduct research were obtained from the National Health Research Authority.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data supporting the reported results have been provided in this study. Any questions regarding data in this study or any supplementary data that may be required may be provided by the corresponding author upon request.
Acknowledgments
The authors would like to thank the University of Zambia, School of Medicine, Departments of Disease control and Clinical Studies and their technical staff for their support. Gratitude is also extended to Kitwe Teaching Hospital laboratory staff and the Head of Department, Francis Musonda, for his unwavering support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ludwig, W.; Schleifer, K.H.; Whitman, W.B. Revised road map to the phylum Firmicutes. In Bergey’s Manual® of Systematic Bacteriology; Springer: New York, NY, USA, 2009; pp. 1–13. [Google Scholar]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 21–61. [Google Scholar]
- Soodmand, J.; Zeinali, T.; Kalidari, G.; Hashemitabar, G.; Razmyar, J. Antimicrobial Susceptibility Profile of Enterococcus Species Isolated from Companion Birds and Poultry in the Northeast of Iran. Arch. Razi. Inst. 2018, 73, 207–213. [Google Scholar]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef]
- Boehm, A.B.; Sassoubre, L.M. Enterococci as Indicators of Environmental Fecal Contamination. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef]
- Eaton, T.J.; Gasson, M.J. Molecular screening of enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef]
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2002, 26, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Agudelo Higuita, N.I.; Huycke, M.M. Enterococcal Disease, Epidemiology, and Implications for Treatment. Curr. Infect. Dis. Rep. 2014, 16, 385. [Google Scholar]
- Abat, C.; Huart, M.; Garcia, V.; Dubourg, G.; Raoult, D. Enterococcus faecalis urinary-tract infections: Do they have a zoonotic origin? J. Infect. 2016, 73, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Souillard, R.; Laurentie, J.; Kempf, I.; Le Caër, V.; Le Bouquin, S.; Serror, P.; Allain, V. Increasing incidence of Enterococcus-associated diseases in poultry in France over the past 15 years. Vet. Microbiol. 2022, 269, 109426. [Google Scholar] [CrossRef]
- Pillay, S.; Zishiri, O.T.; Adeleke, M.A. Prevalence of virulence genes in Enterococcus species isolated from companion animals and livestock. Onderstepoort J. Vet. Res. 2018, 85, e1–e8. [Google Scholar] [CrossRef]
- Daniel, D.S.; Lee, S.M.; Dykes, G.A.; Rahman, S. Public health risks of multiple-drug-resistant Enterococcus spp. in Southeast Asia. Appl. Environ. Microbiol. 2015, 81, 6090–6097. [Google Scholar] [CrossRef]
- Borst, L.B.; Suyemoto, M.M.; Sarsour, A.H.; Harris, M.C.; Martin, M.P.; Strickland, J.D.; Oviedo, E.O.; Barnes, H.J. Pathogenesis of Enterococcal Spondylitis Caused by Enterococcus cecorum in Broiler Chickens. Vet. Pathol. 2017, 54, 61–73. [Google Scholar] [CrossRef]
- Rehman, M.A.; Yin, X.; Zaheer, R.; Goji, N.; Amoako, K.K.; McAllister, T.; Pritchard, J.; Topp, E.; Diarra, M.S. Genotypes and phenotypes of Enterococci isolated from broiler chickens. Front. Sustain. Food Syst. 2018, 2, 83. [Google Scholar] [CrossRef]
- Adams, D.J.; Eberly, M.D.; Goudie, A.; Nylund, C.M. Rising vancomycin-resistant Enterococcus infections in hospitalized children in the United States. Hosp. Pediatr. 2016, 6, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.-Y.; Perencevich, E.N.; Nair, R.; Nelson, R.E.; Samore, M.; Khader, K.; Chorazy, M.L.; Herwaldt, L.A.; Blevins, A.; Ward, M.A.; et al. Incidence and Outcomes Associated with Infections Caused by Vancomycin-Resistant Enterococci in the United States: Systematic Literature Review and Meta-Analysis. Infection Control & Hospital Epidemiology. Camb. Univ. Press 2017, 38, 203–215. [Google Scholar]
- Jung, A.; Rautenschlein, S. Comprehensive report of an Enterococcus cecorum infection in a broiler flock in Northern Germany. BMC Vet. Res. 2014, 10, 311. [Google Scholar] [CrossRef]
- Shahbandeh, M. Meat Consumption Worldwide 1990–2021, by Type 2022. Available online: https://www.statista.com/statistics/274522/global-per-capita-consumption-of-meat/ (accessed on 19 January 2023).
- Chai, S.J.; Cole, D.; Nisler, A.; Mahon, B.E. Poultry: The most common food in outbreaks with known pathogens, United States, 1998–2012. Epidemiol. Infect. 2017, 145, 316–325. [Google Scholar] [CrossRef]
- Kousar, S.; Rehman, N.; Javed, A.; Hussain, A.; Naeem, M.; Masood, S.; Ali, H.A.; Manzoor, A.; Khan, A.A.; Akrem, A.; et al. Intensive Poultry Farming Practices Influence Antibiotic Resistance Profiles in Pseudomonas Aeruginosa Inhabiting Nearby Soils. Infect. Drug Resist. 2021, 29, 4511–4516. [Google Scholar] [CrossRef]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESCAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Lee, T.; Pang, S.; Abraham, S.; Coombs, G.W. Molecular characterization and evolution of the first outbreak of vancomycin-resistant enterococcus faecium in Western Australia. Int. J. Antimicrob. Agents 2019, 53, 814–819. [Google Scholar] [CrossRef]
- Zambia National Public Health Institute. Government of the Republic of Zambia Multisectoral National Action Plan on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2017; pp. 24–27. Available online: https://www.afro.who.int/publications/multi-sectoral-national-action-plan-antimicrobial-resistance-2017-2027 (accessed on 21 November 2022).
- Dolka, B.; Gołębiewska–Kosakowska, M.; Krajewski, K.; Kwieciński, P.; Nowak, T.; Szubstarski, J.; Wilczyński, J.; Szeleszczuk, P. Occurrence of Enterococcus spp. in poultry in Poland based on 2014–2015 data. Med. Water 2017, 73, 220–224. [Google Scholar] [CrossRef]
- Getachew, Y.M.; Hassan, L.; Zakaria, Z.; Saleha, A.A.; Kamaruddin, M.I.; Che Zalina, M.Z. Characterization of vancomycin-resistant Enterococcus isolates from broilers in Selangor, Malaysia. Trop Biomed. 2009, 26, 280–288. [Google Scholar] [PubMed]
- Ayeni, F.A.; Odumosu, B.T.; Oluseyi, A.E.; Ruppitsch, W. Identification and prevalence of tetracycline resistance in enterococci isolated from poultry in Ilishan, Ogun State, Nigeria. J. Pharm. Bioallied Sci. 2016, 8, 69–73. [Google Scholar] [PubMed]
- Mudenda, S.; Matafwali, S.K.; Malama, S.; Munyeme, M.; Yamba, K.; Katemangwe, P.; Siluchali, G.; Mainda, G.; Mukuma, M.; Bumbangi, F.N.; et al. Prevalence and antimicrobial resistance patterns of Enterococcus species isolated from laying hens in Lusaka and Copperbelt provinces of Zambia: A call for AMR surveillance in the poultry sector. JAC-Antimicrob. Resist. 2022, 4, 126. [Google Scholar] [CrossRef]
- Eldaly, E.A.; Rasha, M.; Elazam, R.A. Prevalence of Enterococcus species in chicken meat in Sharkia Governorate. Egypt. J. Appl. Sci. 2019, 34, 317–323. [Google Scholar] [CrossRef]
- Ferede, Z.T.; Tullu, K.D.; Derese, S.G.; Yeshanew, A.G. Prevalence and antimicrobial susceptibility pattern of Enterococcus species isolated from different clinical samples at Black Lion Specialized Teaching Hospital, Addis Ababa, Ethiopia. BMC Res. Notes 2018, 11, 793. [Google Scholar] [CrossRef]
- Ali, S.A.; Hasan, K.A.; Bin Asif, H.; Abbasi, A. Environmental enterococci: I. Prevalence of Virulence, Antibiotic Resistance and species distribution in poultry and its related environment in Karachi, Pakistan. Lett. Appl. Microbiol. 2014, 58, 423–432. [Google Scholar] [CrossRef]
- Noenchat, P.; Nhoonoi, C.; Srithong, T.; Lertpiriyasakulkit, S.; Sornplang, P. Prevalence and Multidrug Resistance of Enterococcus Species isolated from Chickens at Slaughterhouses in Nakhon Ratchasima Province, Thailand. Vet. World 2022, 15, 2535–2542. [Google Scholar] [CrossRef]
- Suyemoto, M.M.; Barnes, H.J.; Borst, L.B. Culture methods impact recovery of antibiotic-resistant Enterococci including Enterococcus cecorum from pre-and postharvest chicken. Lett. Appl. Microbiol. 2017, 64, 210–216. [Google Scholar] [CrossRef]
- Chingwaru, W.; Mpuchane, S.F.; Gashe, B.A. Enterococcus faecalis and Enterococcus faecium isolates from milk, beef, and chicken and their antibiotic resistance. J. Food Prot. 2003, 66, 931–936. [Google Scholar] [CrossRef]
- Çitak, S.; Yucel, N.; Orhan, S. Antibiotic resistance and incidence of Enterococcus species in Turkish white cheese. Int. J. Dairy Technol. 2004, 57, 27–31. [Google Scholar] [CrossRef]
- Stępień-Pyśniak, D.; Marek, A.; Banach, T.; Adaszek, Ł.; Pyzik, E.; Wilczyński, J.; Winiarczyk, S. Prevalence and antibiotic resistance of Enterococcus strains isolated from poultry. Acta Vet. Hung. 2016, 64, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Karunarathna, R.; Popowich, S.; Wawryk, M.; Chow-Lockerbie, B.; Ahmed, K.A.; Yu, C.; Liu, M.; Goonewardene, K.; Gunawardana, T.; Kurukulasuriya, S.; et al. Increased Incidence of enterococcal infection in nonviable broiler chicken embryos in Western Canadian hatcheries as detected by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Avian Dis. 2017, 61, 472–480. [Google Scholar] [CrossRef]
- Gong, J.; Forster, R.J.; Yu, H.; Chambers, J.R.; Wheatcroft, R.; Sabour, P.M.; Chen, S. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol. 2002, 41, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.R.; English, L.L.; Carter, P.J.; Proescholdt, T.; Lee, K.Y.; Wagner, D.D.; White, D.G. Prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Environ. Microbiol. 2003, 69, 7153–7160. [Google Scholar] [CrossRef] [PubMed]
- Rakotovao-Ravahatra, Z.D.; Antilahy, J.A.; Rakotovao-Ravahatra, J.N.; Rakotovao, A.L. Comparison of Bis-Plus-D and API 20 Strep for the identification of streptococci in the Laboratory of the University Hospital of Befelatanana Antananarivo Madagascar. J. Anal. Tech. Res. 2022, 4, 130–134. [Google Scholar] [CrossRef]
- Gomes, B.C.; Esteves, C.T.; Palazzo, I.C.; Darini AL, C.; Franco, B.D.; de Martinis, E.C. Correlation between API 20 STREP and multiplex PCR for identification of Enterococcus spp. isolated from Brazilian foods. Braz. J. Microbiol. 2007, 38, 617–619. [Google Scholar] [CrossRef]
- Facklam, R.; Teixeira, L.M. Enterococcus. In Manual of Clinical Microbiology, 8th ed.; Murray, P.R., Baron, E.J., Jorgensen, J.H., Pfaller, M.A., Yolken, R.H., Eds.; ASM Press: Washington, DC, USA, 2003; pp. 422–433. [Google Scholar]
- Velasco, D.; Perez, S.; Peña, F.; Dominguez, M.A.; Cartelle, M.; Molina, F.; Moure, R.; Villanueva, R.; Bou, G. Lack of correlation between phenotypic techniques and PCR-based genotypic methods for identification of Enterococcus spp. Diagn. Microbiol. Infect. Dis. 2004, 49, 151–156. [Google Scholar] [CrossRef]
- Winston, L.G.; Pang, S.; Haller, B.L.; Wong, M.; Chambers, H.F., III; Perdreau-Remington, F. API 20 STREP identification system may incorrectly speciate enterococci with low level resistance to vancomycin. Diagn. Microbiol. Infect. Dis. 2004, 48, 287–288. [Google Scholar] [CrossRef]
- World Health Organization. Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human Use. 2017. Available online: https://apps.who.int/iris/bitstream/handle/10665/255027/978?sequence=1 (accessed on 20 January 2023).
- Kim, M.H.; Moon, D.C.; Kim, S.J.; Mechesso, A.F.; Song, H.J.; Kang, H.Y.; Choi, J.H.; Yoon, S.S.; Lim, S.K. Nationwide surveillance on antimicrobial resistance profiles of Enterococcus faecium and Enterococcus faecalis isolated from healthy food animals in South Korea, 2010 to 2019. Microorganisms 2021, 9, 925. [Google Scholar] [CrossRef]
- Kolář, M.; Pantůček, R.; Bardoň, J.; Vágnerová, I.; Typovska, H.; Válka, I.; Doškař, J. Occurrence of antibiotic-resistant bacterial strains isolated in poultry. Veterinární Med. 2002, 47, 52–59. [Google Scholar] [CrossRef]
- Furtula, V.; Jackson, C.R.; Farrell, E.G.; Barrett, J.B.; Hiott, L.M.; Chambers, P.A. Antimicrobial resistance in Enterococcus spp. isolated from environmental samples in an area of intensive poultry production. Int. J. Environ. Res. Public Health 2013, 10, 1020–1036. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, C.L.; Letellier, A.; Quessy, S.; Boulianne, M.; Daignault, D.; Archambault, M. Multiple-antibiotic resistance of Enterococcus faecalis and Enterococcus faecium from cecal contents in broiler chicken and turkey flocks slaughtered in Canada and plasmid colocalization of tetO and ermB genes. J. Food Prot. 2011, 74, 1639–1648. [Google Scholar] [CrossRef]
- Šeputiene, V.; Bogdaite, A.; Ružauskas, M.; Sužiedeliene, E. Antibiotic resistance genes and virulence factors in Enterococcus faecium and Enterococcus faecalis from diseased farm animals: Pigs, cattle, and poultry. Pol. J. Vet. Sci. 2012, 15, 431–438. [Google Scholar] [PubMed]
- Liu, Y.; Liu, K.; Lai, J.; Wu, C.; Shen, J.; Wang, Y. Prevalence and antimicrobial resistance of Enterococcus species of food animal origin from Beijing and Shandong Province, China. J. Appl. Microbiol. 2013, 114, 555–563. [Google Scholar] [CrossRef]
- Sung, C.H.; Chon, J.W.; Kwak, H.S.; Kim, H.; Seo, K.H. Prevalence and antimicrobial resistance of Enterococcus faecalis and Enterococcus faecium isolated from beef, pork, chicken, and sashimi. Korean J. Food Sci. Anim. Resour. 2013, 33, 133–138. [Google Scholar] [CrossRef]
- Maasjost, J.; Mühldorfer, K.; De Jäckel, S.C.; Hafez, H.M. Antimicrobial susceptibility patterns of Enterococcus faecalis and Enterococcus faecium isolated from poultry flocks in Germany. Avian Dis. 2015, 59, 143–148. [Google Scholar] [CrossRef]
- Kim, Y.J.; Park, J.H.; Seo, K.H. Comparison of the loads and antibiotic-resistance profiles of Enterococcus species from conventional and organic chicken carcasses in South Korea. Poult. Sci. 2018, 97, 271–278. [Google Scholar] [CrossRef]
- Noh, E.B.; Kim, Y.B.; Seo, K.W.; Son, S.H.; Ha, J.S.; Lee, Y.J. Antimicrobial resistance monitoring of commensal Enterococcus faecalis in broiler breeders. Poult. Sci. 2020, 99, 2675–2683. [Google Scholar] [CrossRef]
- Fracalanzza, S.A.P.; Scheidegger, E.M.D.; Dos Santos, P.F.; Leite, P.C.; Teixeira, L.M. Antimicrobial resistance profiles of enterococci isolated from poultry meat and pasteurized milk in Rio de Janeiro, Brazil. Memórias do Inst. Oswaldo Cruz 2007, 102, 853–859. [Google Scholar] [CrossRef]
- Yoshimura, H.; Ishimaru, M.; Endoh, Y.S.; Kojima, A. Antimicrobial susceptibilities of enterococci isolated from faeces of broiler and layer chickens. Lett. Appl. Microbiol. 2000, 31, 427–432. [Google Scholar] [CrossRef]
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animals. Antimicrob. Agents Chemother. 2001, 45, 1374–1378. [Google Scholar] [CrossRef]
- Depardieu, F.; Perichon, B.; Courvalin, P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 2004, 42, 5857–5860. [Google Scholar] [CrossRef]
- Xu, X.; Lin, D.; Yan, G.; Ye, X.; Wu, S.; Guo, Y.; Zhu, D.; Hu, F.; Zhang, Y.; Wang, F.; et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 2010, 54, 4643–4647. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Depardieu, F.; Bourdon, N.; Fines-Guyon, M.; Berger, P.; Camiade, S.; Leclercq, R.; Courvalin, P.; Cattoir, V. D-Ala-D-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 2011, 55, 4606–4612. [Google Scholar] [CrossRef] [PubMed]
- Facklam, R.R.; Collins, M.D. Identification of Enterococcus species isolated from human infections by conventional test scheme. J. Clin. Microbiol. 1989, 27, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xing, J.; Li, B.; Wang, P.; Liu, J. Use of tuf as a target for sequence-based identification of Gram positive cocci of the genus Enterococcus, Streptococcus, coagulase-negative Staphylococcus, and Lactococcus. Annu. Clin. Microbiol. Antimicrob. 2012, 11, 31. [Google Scholar] [CrossRef]
- Ke, D.; Picard, F.; Martineau, F.; Menard, C.; Roy, P.; Ouellette, M.; Bergeron, M. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 1999, 37, 3497–3503. [Google Scholar] [CrossRef] [PubMed]
- Vilela, M.A.; Souz, S.L.; Palazzo, I.C.V.; Ferreira, J.C.; Morais, M.A., Jr.; Darini, A.L.C.; Morais, M.M.C. Identification and molecular characterization of Van A-type vancomycin-resistant Enterococcus faecalis in Northeast of Brazil. Memórias do Inst. Oswaldo Cruz 2006, 101, 716–719. [Google Scholar] [CrossRef]
- Bensalah, F.; Flores, M.J.A.; Mouats, A. Rapid PCR based method to distinguish between Enterococcus species by using degenerate and species-specific sodA gene primers. Afr. J. Biotechnol. 2006, 5, 697–702. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Tech. Rep. M100-S22; PA Publication: Wayne, PA, USA, 2012. [Google Scholar]
- Sabouni, F.; Movahedi, Z.; Mahmoudi, S.; Pourakbari, B.; Keshavarz, V.S.; Mamishi, S. High frequency of vancomycin resistant Enterococcus faecalis in children: An alarming concern. J. Prev. Med. Hyg. 2016, 57, E201–E204. [Google Scholar] [PubMed]
- Goudarzi, G.; Tahmasbi, F.; Anbari, K.; Ghafarzadeh, M. Distribution of genes encoding resistance to macrolides among Staphylococci isolated from the nasal cavity of hospital employees in Khorramabad, Iran. Iran. Red Crescent Med. J. 2016, 18, e25701. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.; Grebe, T.; Tait-Kamradt, A.; Wondrack, L. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 1996, 40, 2562–2566. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Agerso, Y.; Gerner–Smidt, P.; Madsen, M.; Jensen, L.B. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 2000, 37, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.K.; Martin, I.; Alfo, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistance genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Sting, R.; Richter, A.; Popp, C.; Hafez, H.M. Occurrence of vancomycin-resistant enterococci in turkey flocks. Poult. Sci. 2013, 92, 346–351. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).