Direct Detection of KPC Peak from Positive Blood Cultures Using MALDI-TOF MS: Are We There Yet?
Abstract
1. Introduction
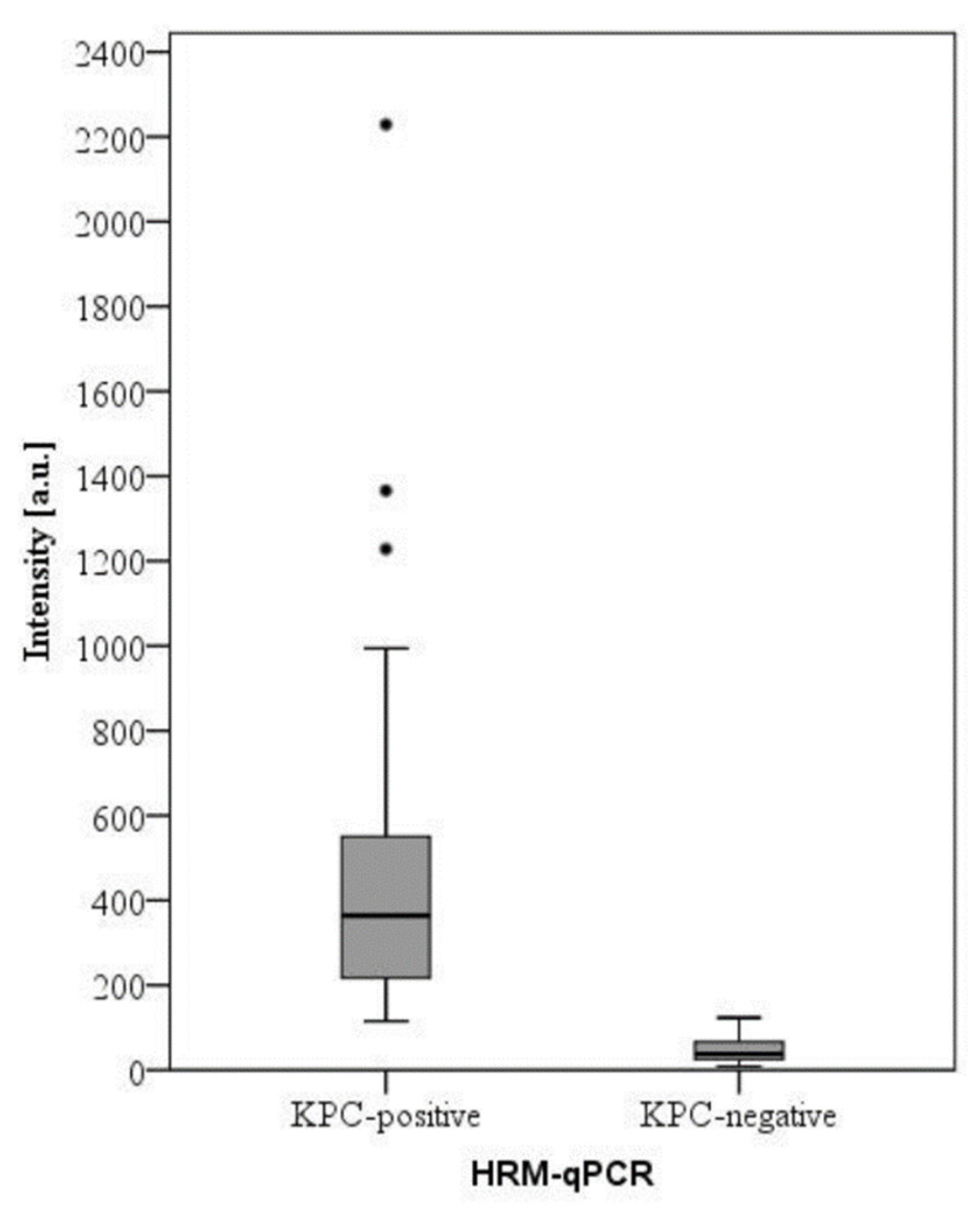
2. Results and Discussion
3. Materials and Methods
3.1. Bacterial Isolates and Carbapenemase Characterization
3.2. Blood Culture Samples
3.3. Protein Extraction
3.4. Target Spot Loading
3.5. Spectra Acquisition
3.6. Data Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO 10 Global Health Issues to Track in 2021. Available online: https://www.who.int/news-room/spotlight/10-global-health-issues-to-track-in-2021 (accessed on 20 March 2022).
- Lee, C.-R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global Dissemination of Carbapenemase-Producing Klebsiella Pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef] [PubMed]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World Health Organization Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. J. Med. Soc. 2018, 32, 76. [Google Scholar] [CrossRef]
- Perez, F.; Bonomo, R.A. Carbapenem-Resistant Enterobacteriaceae: Global Action Required. Lancet Infect. Dis. 2019, 19, 561–562. [Google Scholar] [CrossRef]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of Hypotension before Initiation of Effective Antimicrobial Therapy Is the Critical Determinant of Survival in Human Septic Shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef]
- Kumar, A.; Ellis, P.; Arabi, Y.; Roberts, D.; Light, B.; Parrillo, J.E.; Dodek, P.; Wood, G.; Kumar, A.; Simon, D.; et al. Initiation of Inappropriate Antimicrobial Therapy Results in a Fivefold Reduction of Survival in Human Septic Shock. Chest 2009, 136, 1237–1248. [Google Scholar] [CrossRef]
- Tariq, T.M.; Rasool, E. Emerging Trends of Bloodstream Infections: A Six-Year Study at a Paediatric Tertiary Care Hospital in Kabul. J. Coll. Physicians Surg. Pak. 2016, 26, 887–891. [Google Scholar]
- Rice, L.B. Antimicrobial Stewardship and Antimicrobial Resistance. Med. Clin. N. Am. 2018, 102, 805–818. [Google Scholar] [CrossRef]
- Nauclér, P.; Huttner, A.; van Werkhoven, C.H.; Singer, M.; Tattevin, P.; Einav, S.; Tängdén, T. Impact of Time to Antibiotic Therapy on Clinical Outcome in Patients with Bacterial Infections in the Emergency Department: Implications for Antimicrobial Stewardship. Clin. Microbiol. Infect. 2021, 27, 175–181. [Google Scholar] [CrossRef]
- Noval, M.; Banoub, M.; Claeys, K.C.; Heil, E. The Battle Is on: New Beta-Lactams for the Treatment of Multidrug-Resistant Gram-Negative Organisms. Curr. Infect. Dis. Rep. 2020, 22, 1. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef]
- AlTamimi, M.; AlSalamah, A.; AlKhulaifi, M.; AlAjlan, H. Comparison of Phenotypic and PCR Methods for Detection of Carbapenemases Production by Enterobacteriaceae. Saudi J. Biol. Sci. 2017, 24, 155–161. [Google Scholar] [CrossRef]
- Hrabák, J. Detection of Carbapenemases Using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) Meropenem Hydrolysis Assay. Sepsis 2015, 1237, 91–96. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Dortet, L. Rapid Detection of Carbapenemase-Producing Enterobacteriaceae. Emerg. Infect. Dis. 2012, 18, 1503–1507. [Google Scholar] [CrossRef]
- Tamma, P.D.; Opene, B.N.A.; Gluck, A.; Chambers, K.K.; Carroll, K.C.; Simner, P.J. Comparison of 11 Phenotypic Assays for Accurate Detection of Carbapenemase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Simner, P.J. Phenotypic Detection of Carbapenemase-Producing Organisms from Clinical Isolates. J. Clin. Microbiol. 2018, 56, e01140-18. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-Negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef]
- Perez, K.K.; Olsen, R.J.; Musick, W.L.; Cernoch, P.L.; Davis, J.R.; Land, G.A.; Peterson, L.E.; Musser, J.M. Integrating Rapid Pathogen Identification and Antimicrobial Stewardship Significantly Decreases Hospital Costs. Arch. Pathol. Lab. Med. 2013, 137, 1247–1254. [Google Scholar] [CrossRef]
- Byun, J.-H.; Kim, Y.A.; Kim, M.; Kim, B.; Choi, J.Y.; Park, Y.S.; Yong, D. Evaluation of Xpert Carba-R Assay v.2 to Detect Carbapenemase Genes in Two Hospitals in Korea. Ann. Lab. Med. 2020, 40, 209–215. [Google Scholar] [CrossRef]
- Lutgring, J.D.; Limbago, B.M. The Problem of Carbapenemase-Producing-Carbapenem-Resistant-Enterobacteriaceae Detection. J. Clin. Microbiol. 2016, 54, 529–534. [Google Scholar] [CrossRef]
- Hamprecht, A.; Vehreschild, J.J.; Seifert, H.; Saleh, A. Rapid Detection of NDM, KPC and OXA-48 Carbapenemases Directly from Positive Blood Cultures Using a New Multiplex Immunochromatographic Assay. PLoS ONE 2018, 13, e0204157. [Google Scholar] [CrossRef]
- Wink, P.L.; Martins, A.S.; Inamine, E.; Dalmolin, T.V.; Barth, A.L. Rapid Detection of the Main Carbapenemases in Brazil Directly from Spiked Blood Culture Using the RESIST-3 O.K.N. Immunoassay. Braz. J. Microbiol. 2019, 50, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Baeza, L.L.; Pfennigwerth, N.; Greissl, C.; Göttig, S.; Saleh, A.; Stelzer, Y.; Gatermann, S.G.; Hamprecht, A. Comparison of Five Methods for Detection of Carbapenemases in Enterobacterales with Proposal of a New Algorithm. Clin. Microbiol. Infect. 2019, 25, 1286.e9–1286.e15. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, I.; Zimmermann, S. Susceptibility Testing of Bacteria Using Maldi-Tof Mass Spectrometry. Front. Microbiol. 2018, 9, 1744. [Google Scholar] [CrossRef]
- Florio, W.; Baldeschi, L.; Rizzato, C.; Tavanti, A.; Ghelardi, E.; Lupetti, A. Detection of Antibiotic-Resistance by MALDI-TOF Mass Spectrometry: An Expanding Area. Front. Cell. Infect. Microbiol. 2020, 10, 572909. [Google Scholar] [CrossRef] [PubMed]
- Akyar, I.; Kaya Ayas, M.; Karatuna, O. Performance Evaluation of MALDI-TOF MS MBT STAR-BL Versus In-House Carba NP Testing for the Rapid Detection of Carbapenemase Activity in Escherichia Coli and Klebsiella Pneumoniae Strains. Microb. Drug Resist. 2019, 25, 985–990. [Google Scholar] [CrossRef]
- Papagiannitsis, C.C.; Študentová, V.; Izdebski, R.; Oikonomou, O.; Pfeifer, Y.; Petinaki, E.; Hrabák, J. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Meropenem Hydrolysis Assay with NH4 HCO3, a Reliable Tool for Direct Detection of Carbapenemase Activity. J. Clin. Microbiol. 2015, 53, 1731–1735. [Google Scholar] [CrossRef]
- Oviaño, M.; Bou, G. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry for the Rapid Detection of Antimicrobial Resistance Mechanisms and Beyond. Clin. Microbiol. Rev. 2018, 32, e00037-18. [Google Scholar] [CrossRef]
- Oviaño, M.; Sparbier, K.; Barba, M.J.; Kostrzewa, M.; Bou, G. Universal Protocol for the Rapid Automated Detection of Carbapenem-Resistant Gram-Negative Bacilli Directly from Blood Cultures by Matrix-Assisted Laser Desorption/Ionisation Time-of-Flight Mass Spectrometry (MALDI-TOF/MS). Int. J. Antimicrob. Agents 2016, 48, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Oueslati, S.; Nordmann, P.; Poirel, L. Heterogeneous Hydrolytic Features for OXA-48-like β-Lactamases. J. Antimicrob. Chemother. 2015, 70, 1059–1063. [Google Scholar] [CrossRef]
- Bakthavatchalam, Y.; Anandan, S.; Veeraraghavan, B. Laboratory Detection and Clinical Implication of Oxacillinase-48 like Carbapenemase: The Hidden Threat. J. Glob. Infect. Dis. 2016, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-H.; Drake, S.K.; Weingarten, R.A.; Frank, K.M.; Dekker, J.P.; Lau, A.F. Clinical Performance of a Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Method for Detection of Certain Bla KPC -Containing Plasmids. J. Clin. Microbiol. 2016, 54, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; Galea, A.; Fagioni, M.; Ambretti, S.; Sambri, V.; Landini, M.P. Evaluation of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry for Identification of KPC-Producing Klebsiella Pneumoniae. J. Clin. Microbiol. 2016, 54, 2609–2613. [Google Scholar] [CrossRef]
- Lau, A.F.; Wang, H.; Weingarten, R.A.; Drake, S.K.; Suffredini, A.F.; Garfield, M.K.; Chen, Y.; Gucek, M.; Youn, J.-H.; Stock, F.; et al. A Rapid Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry-Based Method for Single-Plasmid Tracking in an Outbreak of Carbapenem-Resistant Enterobacteriaceae. J. Clin. Microbiol. 2014, 52, 2804–2812. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, R.F.; Rumi, V.; Marchisio, M.; Cejas, D.; Radice, M.; Vay, C.; Barrios, R.; Gutkind, G.; Di Conza, J. Fast and Easy Detection of CMY-2 in Escherichia Coli by Direct MALDI-TOF Mass Spectrometry. J. Microbiol. Methods 2018, 148, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Espinosa, R.; Costa, A.; Cejas, D.; Barrios, R.; Vay, C.; Radice, M.; Gutkind, G.; Di Conza, J. MALDI-TOF MS Based Procedure to Detect KPC-2 Directly from Positive Blood Culture Bottles and Colonies. J. Microbiol. Methods 2019, 159, 120–127. [Google Scholar] [CrossRef]
- Yoon, E.-J.; Lee, E.H.; Hwang, D.H.; Lee, H.; Baek, J.-H.; Jeong, S.H. Direct Detection of Intact Klebsiella Pneumoniae Carbapenemases Produced by Enterobacterales Using MALDI-TOF MS. J. Antimicrob. Chemother. 2020, 75, 1174–1181. [Google Scholar] [CrossRef]
- Moreira, N.K.; Wilhelm, C.M.; Wink, P.L.; Barth, A.L.; Caierão, J. MALDI-TOF Mass Spectrometry for Direct KPC Detection among Enterobacterales. Braz. J. Microbiol. 2022, 53, 1907–1913. [Google Scholar] [CrossRef]
- Monteiro, J.; Widen, R.H.; Pignatari, A.C.C.; Kubasek, C.; Silbert, S. Rapid Detection of Carbapenemase Genes by Multiplex Real-Time PCR. J. Antimicrob. Chemother. 2012, 67, 906–909. [Google Scholar] [CrossRef]
- Jung, J.S.; Popp, C.; Sparbier, K.; Lange, C.; Kostrzewa, M.; Schubert, S. Evaluation of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Rapid Detection of -Lactam Resistance in Enterobacteriaceae Derived from Blood Cultures. J. Clin. Microbiol. 2014, 52, 924–930. [Google Scholar] [CrossRef]
- Meier, M.; Hamprecht, A. Systematic Comparison of Four Methods for Detection of Carbapenemase-Producing Enterobacterales Directly from Blood Cultures. J. Clin. Microbiol. 2019, 57, e00709-19. [Google Scholar] [CrossRef] [PubMed]
- ISO Clinical Laboratory Testing and in Vitro Diagnostic Test Systems—Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Broth Micro-Dilution Reference Method for Testing the in Vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. 2019. Available online: https://www.iso.org/standard/70464.html (accessed on 12 October 2022).
- EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: Http://Www.Eucast.Org (accessed on 19 August 2022).
- EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance 2017. Version 2.01. 2017. Available online: http://www.eucast.org/ (accessed on 19 August 2022).
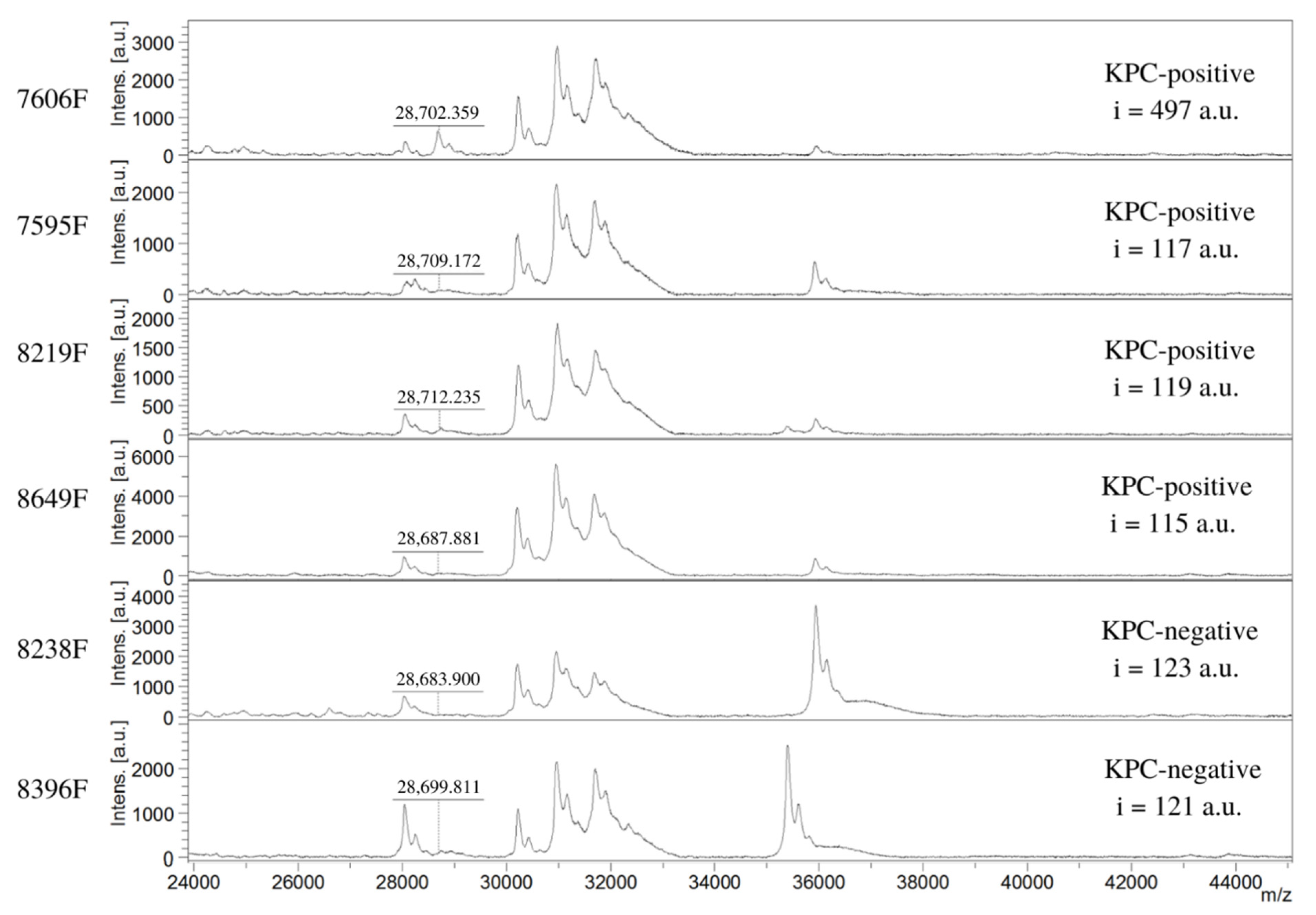

| ID | Species | KPC Peak m/z | i (a.u.) | Meropenem MIC (µg/mL) |
|---|---|---|---|---|
| 7023F | E. coli | 28,713 | 563 | 64 |
| 7431F | K. pneumoniae | 28,727 | 351 | 256 |
| 7434F | K. pneumoniae | 28,663 | 663 | 64 |
| 7437F | E. cloacae complex | 28,671 | 951 | 64 |
| 7440F | K. pneumoniae | 28,726 | 1366 | 256 |
| 7442F | K. pneumoniae | 28,726 | 994 | 128 |
| 7462F | K. pneumoniae | 28,717 | 530 | 64 |
| 7464F | K. pneumoniae | 28,688 | 883 | >256 |
| 7467F | K. pneumoniae | 28,724 | 683 | 128 |
| 7480F | K. pneumoniae | 28,721 | 434 | 128 |
| 7481F | K. pneumoniae | 28,729 | 298 | 256 |
| 7502F | K. pneumoniae | 28,710 | 360 | >256 |
| 7514F | K. pneumoniae | 28,720 | 200 | 128 |
| 7590F | K. pneumoniae | 28,684 | 584 | >256 |
| 7592F | K. pneumoniae | 28,663 | 2229 | 128 |
| 7593F | K. pneumoniae | 28,721 | 149 | 128 |
| 7594F | K. pneumoniae | 28,665 | 450 | 128 |
| 7595F | K. pneumoniae | Absent | 117 | >256 |
| 7599F | K. pneumoniae | 28,715 | 202 | 128 |
| 7605F | K. pneumoniae | 28,713 | 224 | 64 |
| 7606F | K. pneumoniae | 28,679 | 497 | 64 |
| 8156F | K. pneumoniae | 28,736 | 139 | 16 |
| 8219F | K. pneumoniae | Absent | 119 | 128 |
| 8285F * | K. pneumoniae | 28,687 | 364 | 64 |
| 8467F | K. pneumoniae | 28,721 | 502 | >256 |
| 8649F | K. pneumoniae | Absent | 115 | >256 |
| 8799F | S. marcescens | 28,677 | 1228 | >256 |
| 8817F | K. pneumoniae | 28,715 | 668 | 64 |
| 8829F | S. marcescens | 28,688 | 134 | 256 |
| 8848F | S. marcescens | 28,717 | 125 | >256 |
| 8871F | S. marcescens | 28,674 | 173 | >256 |
| 8882F | K. pneumoniae | 28,731 | 520 | 32 |
| 8884F | E. coli | 28,704 | 264 | 256 |
| 8910F | S. marcescens | 28,672 | 245 | 64 |
| 8946F | S. marcescens | 28,674 | 210 | 256 |
| 8974F | E. coli | 28,668 | 394 | 256 |
| 8982F | S. marcescens | 28,666 | 746 | 128 |
| 9077F | K. pneumoniae | 28,690 | 237 | 16 |
| 9079F | S. marcescens | 28,709 | 131 | 16 |
| 9100F | K. pneumoniae | 28,677 | 298 | >256 |
| 9169F | E. cloacae complex | 28,715 | 298 | >256 |
| 9173F | K. pneumoniae | 28,722 | 375 | 32 |
| 9176F | S. marcescens | 28,699 | 331 | 128 |
| 9221F | S. marcescens | 28,677 | 121 | 32 |
| 9236F | K. pneumoniae | 28,677 | 593 | 128 |
| 9274F | K. pneumoniae | 28,661 | 414 | >256 |
| 9279F | K. pneumoniae | 28,702 | 310 | >256 |
| 9283F | K. pneumoniae | 28,702 | 796 | 256 |
| 9288F | K. pneumoniae | 28,661 | 460 | 64 |
| 9289F | K. pneumoniae | 28,704 | 393 | 128 |
| 9294F | K. pneumoniae | 28,725 | 855 | 16 |
| 9295F | K. pneumoniae | 28,670 | 538 | 32 |
| 9297F | K. pneumoniae | 28,720 | 322 | 256 |
| 9299F | K. pneumoniae | 28,670 | 173 | 128 |
| 9300F | S. marcescens | 28,725 | 270 | 128 |
| 9301F | K. pneumoniae | 28,664 | 372 | 256 |
| 9304F | K. pneumoniae | 28,676 | 442 | 256 |
| 9305F | K. pneumoniae | 28,706 | 336 | 32 |
| 9310F | K. pneumoniae | 28,707 | 202 | 128 |
| ID | Species | HRM-qPCR | KPC Peak m/z | i [a.u.] | Meropenem MIC (µg/mL) |
|---|---|---|---|---|---|
| 7282F | K. pneumoniae | Negative | Absent | 96 | 4 |
| 7452F | K. pneumoniae | Negative | Absent | 42 | 64 |
| 7523F | K. pneumoniae | blaNDM-1 | Absent | 68 | 8 |
| 8113F | K. oxytoca | Negative | Absent | 65 | 16 |
| 8143F | K. pneumoniae | Negative | Absent | 35 | 32 |
| 8144F | K. pneumoniae | Negative | Absent | 36 | 8 |
| 8152F | K. pneumoniae | Negative | Absent | 20 | 4 |
| 8155F | K. pneumoniae | Negative | Absent | 44 | 16 |
| 8158F | K. pneumoniae | Negative | Absent | 54 | ≤0.5 |
| 8165F | K. aerogenes | Negative | Absent | 80 | 4 |
| 8215F | E. cloacae complex | Negative | Absent | 34 | 4 |
| 8238F | K. pneumoniae | Negative | Absent | 123 | 1 |
| 8300F | K. oxytoca | Negative | Absent | 28 | 4 |
| 8311F | K. pneumoniae | Negative | Absent | 54 | ≤0.5 |
| 8314F | K. oxytoca | Negative | Absent | 16 | 4 |
| 8333F | K. pneumoniae | blaNDM-1 | Absent | 112 | 64 |
| 8348F | K. pneumoniae | blaOXA-48like | Absent | 31 | 8 |
| 8355F | E. cloacae complex | blaNDM-1 | Absent | 20 | 8 |
| 8378F | E. cloacae complex | blaNDM-1 | Absent | 24 | 8 |
| 8382F | K. pneumoniae | blaNDM-1 | Absent | 9 | 4 |
| 8387F | E. coli | blaNDM-1 | Absent | 23 | ≤0.5 |
| 8389F | E. cloacae complex | blaNDM-1 | Absent | 31 | 16 |
| 8396F | E. cloacae complex | blaNDM-1 | Absent | 121 | 4 |
| 8400F | K. pneumoniae | Negative | Absent | 95 | 32 |
| 8411F | K. pneumoniae | blaNDM-1 | Absent | 59 | 128 |
| 8412F | K. pneumoniae | blaNDM-1 | Absent | 34 | 64 |
| 8414F | E. cloacae complex | blaNDM-1 | Absent | 28 | 4 |
| 8420F | E. cloacae complex | Negative | Absent | 101 | 2 |
| 8424F | E. cloacae complex | blaNDM-1 | Absent | 14 | 8 |
| 8432F | K. pneumoniae | blaNDM-1 | Absent | 83 | 8 |
| 8435F | K. aerogenes | Negative | Absent | 38 | 32 |
| 8449F | K. pneumoniae | blaNDM-1 | Absent | 32 | 8 |
| 8471F | K. pneumoniae | blaNDM-1 | Absent | 19 | 1 |
| 8474F | K. pneumoniae | blaOXA-48like | Absent | 107 | 16 |
| 8478F | E. cloacae complex | Negative | Absent | 61 | 8 |
| 8481F | K. pneumoniae | blaOXA-48like | Absent | 50 | 16 |
| 8485F | K. oxytoca | blaNDM-1 | Absent | 11 | ≤0.5 |
| 8486F | E. cloacae complex | blaNDM-1 | Absent | 27 | 32 |
| 8889F | E. cloacae complex | blaNDM-1 | Absent | 47 | 64 |
| 8890F | E. cloacae complex | blaOXA-48like | Absent | 12 | 8 |
| 9110F | K. pneumoniae | Negative | Absent | 39 | 64 |
| 9112F | K. pneumoniae | blaNDM-1 + blaOXA-48like | Absent | 97 | 256 |
| 9291F | K. pneumoniae | blaNDM-1 | Absent | 16 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira, N.K.; Wilhelm, C.M.; Echevarria, A.D.; Volpato, F.C.Z.; Wink, P.L.; Barth, A.L.; Caierão, J. Direct Detection of KPC Peak from Positive Blood Cultures Using MALDI-TOF MS: Are We There Yet? Antibiotics 2023, 12, 601. https://doi.org/10.3390/antibiotics12030601
Moreira NK, Wilhelm CM, Echevarria AD, Volpato FCZ, Wink PL, Barth AL, Caierão J. Direct Detection of KPC Peak from Positive Blood Cultures Using MALDI-TOF MS: Are We There Yet? Antibiotics. 2023; 12(3):601. https://doi.org/10.3390/antibiotics12030601
Chicago/Turabian StyleMoreira, Natália Kehl, Camila Mörschbächer Wilhelm, Aymê Duarte Echevarria, Fabiana Caroline Zempulski Volpato, Priscila Lamb Wink, Afonso Luís Barth, and Juliana Caierão. 2023. "Direct Detection of KPC Peak from Positive Blood Cultures Using MALDI-TOF MS: Are We There Yet?" Antibiotics 12, no. 3: 601. https://doi.org/10.3390/antibiotics12030601
APA StyleMoreira, N. K., Wilhelm, C. M., Echevarria, A. D., Volpato, F. C. Z., Wink, P. L., Barth, A. L., & Caierão, J. (2023). Direct Detection of KPC Peak from Positive Blood Cultures Using MALDI-TOF MS: Are We There Yet? Antibiotics, 12(3), 601. https://doi.org/10.3390/antibiotics12030601






