In Vitro Bacterial Competition of Staphylococcus aureus, Streptococcus agalactiae, and Escherichia coli against Coagulase-Negative Staphylococci from Bovine Mastitis Milk
Abstract
1. Introduction
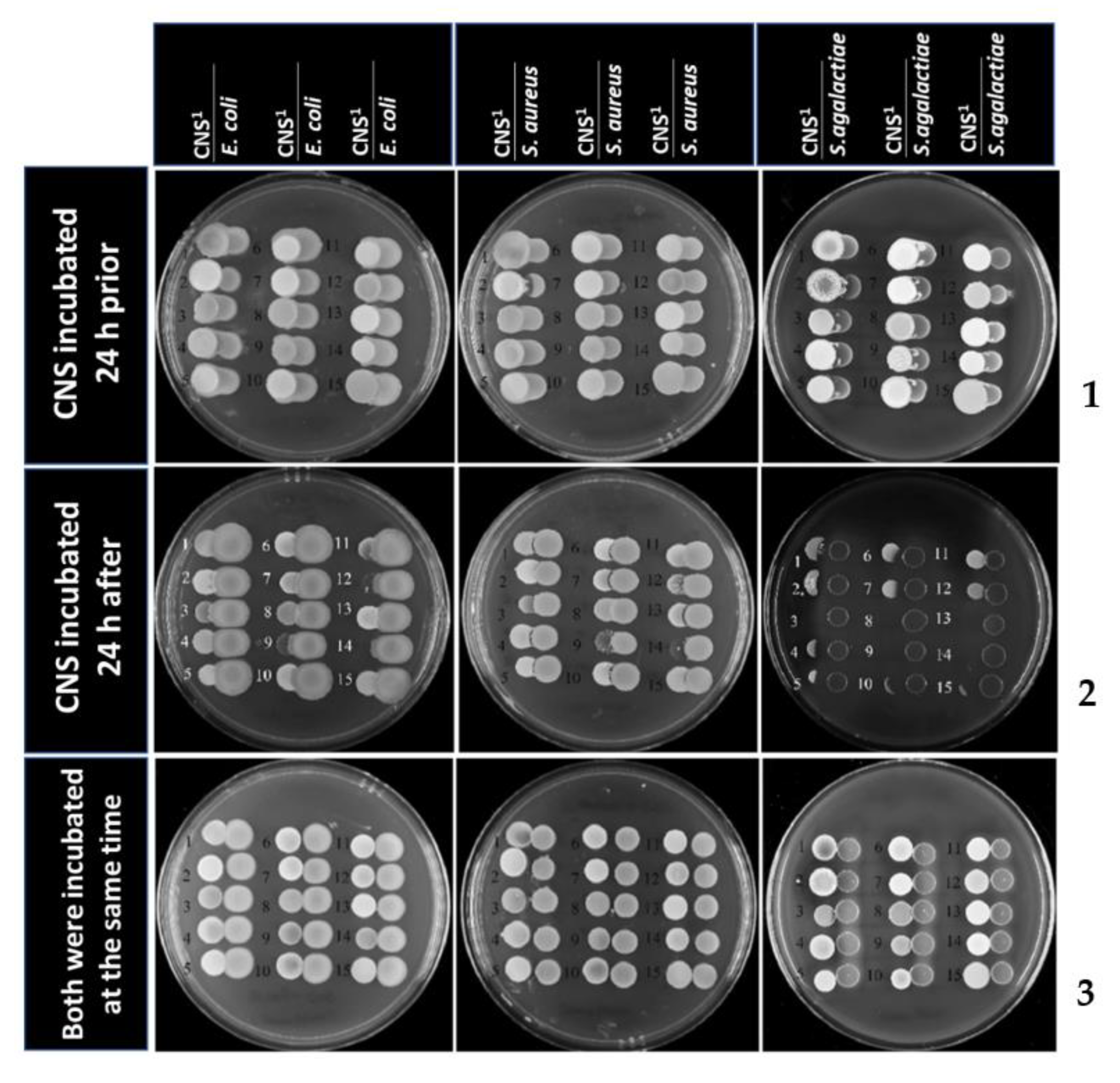
2. Results
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Study Design
5.2. Agar Spot Competition Assays
5.3. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 17 November 2021).
- Beyene, T.; Hayishe, H.; Gizaw, F.; Beyi, A.F.; Abunna, F.; Mammo, B.; Ayana, D.; Waktole, H.; Abdi, R.D. Prevalence and antimicrobial resistance profile of Staphylococcus in dairy farms, abattoir and humans in Addis Ababa, Ethiopia. BMC Res. Notes 2017, 10, 171. [Google Scholar] [CrossRef]
- Suriyasathaporn, W.; Chupia, V.; Sing-Lah, T.; Wongsawan, K.; Mektrirat, R.; Chaisri, W. Increases of Antibiotic Resistance in Excessive Use of Antibiotics in Smallholder Dairy Farms in Northern Thailand. Asian-Australas J. Anim. Sci. 2012, 25, 1322–1328. [Google Scholar] [CrossRef] [PubMed]
- Zigo, F.; Vasil, M.; Ondrašovičová, S.; Výrostková, J.; Bujok, J.; Pecka-Kielb, E. Maintaining Optimal Mammary Gland Health and Prevention of Mastitis. Front. Vet. Sci. 2021, 8, 607311. [Google Scholar] [CrossRef]
- Suriyasathaporn, W.; Heuer, C.; Noordhuizen-Stassen, E.N.; Schukken, Y. Hyperketonemia and the impairment of udder defense: A review. Vet. Res. 2000, 31, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Leelahapongsathon, K.; Schukken, Y.H.; Pinyopummintr, T.; Suriyasathaporn, W. Comparison of transmission dynamics between Streptococcus uberis and Streptococcus agalactiae intramammary infections. J. Dairy Sci. 2016, 99, 1418–1426. [Google Scholar] [CrossRef] [PubMed]
- Leelahapongsathon, K.; Schukken, Y.; Srithanasuwan, A.; Suriyasathaporn, W. Molecular epidemiology of Streptococcus uberis intramammary infections: Persistent and transient patterns of infection in a dairy herd. J. Dairy Sci. 2020, 103, 3565–3576. [Google Scholar] [CrossRef]
- Srithanasuwan, A.; Pangprasit, N.; Suriyasathaporn, W. Comparison of Virulence Patterns Between Streptococcus uberis Causing Transient and Persistent Intramammary Infection. Front. Vet. Sci. 2022, 9, 806674. [Google Scholar] [CrossRef]
- Schukken, Y.H.; González, R.N.; Tikofsky, L.L.; Schulte, H.F.; Santisteban, C.G.; Welcome, F.L.; Bennett, G.J.; Zurakowski, M.J.; Zadoks, R.N.J.V.M. CNS mastitis: Nothing to worry about? Vet. Microbiol. 2009, 134, 9–14. [Google Scholar] [CrossRef]
- Zadoks, R.N.; Watts, J.L. Species identification of coagulase-negative staphylococci: Genotyping is superior to phenotyping. Vet. Microbiol. 2009, 134, 20–28. [Google Scholar] [CrossRef]
- Supré, K.; Haesebrouck, F.; Zadoks, R.; Vaneechoutte, M.; Piepers, S.; De Vliegher, S. Some coagulase-negative Staphylococcus species affect udder health more than others. J. Dairy Sci. 2011, 94, 2329–2340. [Google Scholar] [CrossRef]
- Tomazi, T.; Gonçalves, J.L.; Barreiro, J.R.; Arcari, M.A.; dos Santos, M.V. Bovine subclinical intramammary infection caused by coagulase-negative staphylococci increases somatic cell count but has no effect on milk yield or composition. J. Dairy Sci. 2015, 98, 3071–3078. [Google Scholar] [CrossRef]
- Nair, N.; Biswas, R.; Götz, F.; Biswas, L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect. Immun. 2014, 82, 2162–2169. [Google Scholar] [CrossRef]
- Kasozi, K. High Prevalence of Subclinical Mastitis and Multidrug Resistant Staphylococcus aureus Are a Threat to Dairy Cattle Production in Kiboga District (Uganda). Open J. Vet. Med. 2014, 4, 35–43. [Google Scholar] [CrossRef]
- Vakkamäki, J.; Taponen, S.; Heikkilä, A.-M.; Pyörälä, S. Bacteriological etiology and treatment of mastitis in Finnish dairy herds. Acta Vet. Scand. 2017, 59, 33. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; Straight, P.D. Escape from Lethal Bacterial Competition through Coupled Activation of Antibiotic Resistance and a Mobilized Subpopulation. PLoS Genet. 2015, 11, e1005722. [Google Scholar] [CrossRef]
- Suriyasathaporn, W. Epidemiology of Subclinical Mastitis and Their Antibacterial Susceptibility in Smallholder Dairy Farms, Chiang Mai Province, Thailand. J. Anim. Vet. Adv. 2011, 10, 316–321. [Google Scholar] [CrossRef]
- Pangprasit, N.; Srithanasuwan, A.; Intanon, M.; Suriyasathaporn, W. Bacteriological cure rates of the primary intramammary infection due to the secondary intramammary infection of bovine mastitis. In Proceedings of the 19th AAAP (Asian-Australasian Association of Animal Production) Animal Science Congress, Jeju-do, Republic of Korea, 23–26 August 2022; p. 473. [Google Scholar]
- Keane, O.M.; Budd, K.E.; Flynn, J.; McCoy, F. Increased detection of mastitis pathogens by real-time PCR compared to bacterial culture. Vet. Rec. 2013, 173, 268. [Google Scholar] [CrossRef] [PubMed]
- Choe, H.; Kobayashi, I.Y.; Ike, N.; Iwamoto, H.; Ishida, N.; Yukizawa, T.; Fujimaki, H.; Tezuka, T.; Hirata, H.Y.; Saito, T. How long can dead bacterial DNA be detected by real-time PCR? In Proceedings of the 56th Annual Meeting of the Orthopaedic Research Society. Papers and Posters from the McKay Orthopaedic Laboratory, Poster No. 2144, New Orleans, LA, USA, 6–9 March 2010. [Google Scholar]
- Derakhshani, H.; Plaizier, J.C.; De Buck, J.; Barkema, H.W.; Khafipour, E. Composition and co-occurrence patterns of the microbiota of different niches of the bovine mammary gland: Potential associations with mastitis susceptibility, udder inflammation, and teat-end hyperkeratosis. Anim. Microbiome 2020, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, E.; Djeddi, A.N.; Mousavi, S.A. Prevalence of coagulase negative Staphylococcus including methicillin resistant strains in buffalo subclinical mastitis in northwest of Iran. Buffalo Bull. 2020, 39, 17–26. [Google Scholar]
- Klimiene, I.; Virgailis, M.; Pavilonis, A.; Siugzdiniene, R.; Mockeliunas, R.; Ruzauskas, M. Phenotypical and Genotypical Antimicrobial Resistance of Coagulase-negative staphylococci Isolated from Cow Mastitis. Pol. J. Vet. Sci. 2016, 19, 639–646. [Google Scholar] [CrossRef]
- Duse, A.; Persson-Waller, K.; Pedersen, K. Microbial Aetiology, Antibiotic Susceptibility and Pathogen-Specific Risk Factors for Udder Pathogens from Clinical Mastitis in Dairy Cows. Animals 2021, 11, 2113. [Google Scholar] [CrossRef] [PubMed]
- Ohn, H.M.; Mizuno, T.; Miyoshi, S.I. Inhibitory Effects of Escherichia coli on the Formation and Development of Staphylococcus epidermidis Biofilm. Biocontrol Sci. 2021, 26, 113–118. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Tagg, J.R. Purification and Characterization of Streptin, a Type A1 Lantibiotic Produced by Streptococcus pyogenes. Appl. Environ. Microbiol. 2003, 69, 2737–2747. [Google Scholar] [CrossRef] [PubMed]
- Karaya, K.; Shimizu, T.; Taketo, A. New Gene Cluster for Lantibiotic Streptin Possibly Involved in Streptolysin S Formation. J. Biochem. 2001, 129, 769–775. [Google Scholar] [CrossRef]
- Vidal Amaral, J.R.; Jucá Ramos, R.T.; Almeida Araújo, F.; Bentes Kato, R.; Figueira Aburjaile, F.; de Castro Soares, S.; Góes-Neto, A.; da Costa, M.M.; Azevedo, V.; Brenig, B.; et al. Bacteriocin Producing Streptococcus agalactiae Strains Isolated from Bovine Mastitis in Brazil. Microorganisms 2022, 10, 588. [Google Scholar] [CrossRef]
- Vogel, V.; Spellerberg, B. Bacteriocin Production by Beta-Hemolytic Streptococci. Pathogens 2021, 10, 867. [Google Scholar] [CrossRef]
- Mahmmod, Y.S.; Klaas, I.C.; Svennesen, L.; Pedersen, K.; Ingmer, H. Communications of Staphylococcus aureus and non-aureus Staphylococcus species from bovine intramammary infections and teat apex colonization. J. Dairy Sci. 2018, 101, 7322–7333. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Braem, G.; Stijlemans, B.; Van Haken, W.; De Vliegher, S.; De Vuyst, L.; Leroy, F. Antibacterial activities of coagulase-negative staphylococci from bovine teat apex skin and their inhibitory effect on mastitis-related pathogens. J. Appl. Microbiol. 2014, 116, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- Chin, D.; Goncheva, M.I.; Flannagan, R.S.; Deecker, S.R.; Guariglia-Oropeza, V.; Ensminger, A.W.; Heinrichs, D.E. Coagulase-negative staphylococci release a purine analog that inhibits Staphylococcus aureus virulence. Nat. Commun. 2021, 12, 1887. [Google Scholar] [CrossRef]
- Rahmdel, S.; Shekarforoush, S.S.; Hosseinzadeh, S.; Torriani, S.; Gatto, V. Antimicrobial spectrum activity of bacteriocinogenic Staphylococcus strains isolated from goat and sheep milk. J. Dairy Sci. 2019, 102, 2928–2940. [Google Scholar] [CrossRef] [PubMed]
- Carson, D.A.; Barkema, H.W.; Naushad, S.; Buck, J.D. Bacteriocins of Non-aureus Staphylococci Isolated from Bovine Milk. Appl. Environ. Microbiol 2017, 83, e01015–e01017. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Piccart, K.; Verbeke, J.; De Visscher, A.; Piepers, S.; Haesebrouck, F.; De Vliegher, S. Local host response following an intramammary challenge with Staphylococcus fleurettii and different strains of Staphylococcus chromogenes in dairy heifers. Vet. Res. 2016, 47, 56. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, J.; Piccart, K.; Piepers, S.; Van Poucke, M.; Peelman, L.; De Visscher, A.; De Vliegher, S. Somatic cell count and milk neutrophil viability of dairy heifers with specific CXCR1 genotypes following experimental intramammary infection with Staphylococcus chromogenes originating from milk. Vet. J. 2015, 204, 322–326. [Google Scholar] [CrossRef]
- Harmon, R.; Eberhart, R.; Jasper, D.; Langlois, B.; Wilson, R.J.I.; Arlington, V.A. Microbiological Procedures for the Diagnosis of Bovine Udder Infection; National Mastitis Council: Arlington, VA, USA, 1990; p. 34. [Google Scholar] [CrossRef]
- Kaspar, J.R.; Godwin, M.J.; Velsko, I.M.; Richards, V.P.; Burne, R.A. Spontaneously Arising Streptococcus mutans Variants with Reduced Susceptibility to Chlorhexidine Display Genetic Defects and Diminished Fitness. Antimicrob. Agents Chemother. 2019, 63, 528471. [Google Scholar] [CrossRef]


| No. | Species | Source |
|---|---|---|
| 1, 2 | Staphylococcus chromogenes | Mastitis quarter |
| 3, 4 | Staphylococcus epidermidis | Mastitis quarter |
| 5, 6 | Staphylococcus haemolyticus | Mastitis quarter |
| 7 | Staphylococcus simulans | Mastitis quarter |
| 8, 9 | Staphylococcus warneri | Mastitis quarter |
| 10 | Staphylococcus xylosus | Mastitis quarter |
| 11 | Staphylococcus epidermidis | ATCC |
| 12 | Staphylococcus hominis | Faculty of Science |
| 13 | Staphylococcus cohinii | Faculty of Science |
| 14 | Staphylococcus capitis | Faculty of Science |
| 15 | Staphylococcus sciuri | Faculty of Science |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srithanasuwan, A.; Intanon, M.; Chaisri, W.; Suriyasathaporn, W. In Vitro Bacterial Competition of Staphylococcus aureus, Streptococcus agalactiae, and Escherichia coli against Coagulase-Negative Staphylococci from Bovine Mastitis Milk. Antibiotics 2023, 12, 600. https://doi.org/10.3390/antibiotics12030600
Srithanasuwan A, Intanon M, Chaisri W, Suriyasathaporn W. In Vitro Bacterial Competition of Staphylococcus aureus, Streptococcus agalactiae, and Escherichia coli against Coagulase-Negative Staphylococci from Bovine Mastitis Milk. Antibiotics. 2023; 12(3):600. https://doi.org/10.3390/antibiotics12030600
Chicago/Turabian StyleSrithanasuwan, Anyaphat, Montira Intanon, Wasana Chaisri, and Witaya Suriyasathaporn. 2023. "In Vitro Bacterial Competition of Staphylococcus aureus, Streptococcus agalactiae, and Escherichia coli against Coagulase-Negative Staphylococci from Bovine Mastitis Milk" Antibiotics 12, no. 3: 600. https://doi.org/10.3390/antibiotics12030600
APA StyleSrithanasuwan, A., Intanon, M., Chaisri, W., & Suriyasathaporn, W. (2023). In Vitro Bacterial Competition of Staphylococcus aureus, Streptococcus agalactiae, and Escherichia coli against Coagulase-Negative Staphylococci from Bovine Mastitis Milk. Antibiotics, 12(3), 600. https://doi.org/10.3390/antibiotics12030600





