Antimicrobial Peptides Designed against the Ω-Loop of Class A β-Lactamases to Potentiate the Efficacy of β-Lactam Antibiotics
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids for Cellular and Molecular Studies
2.2. Media, Antibiotics, Peptides, and Culture Conditions for Biological Assays
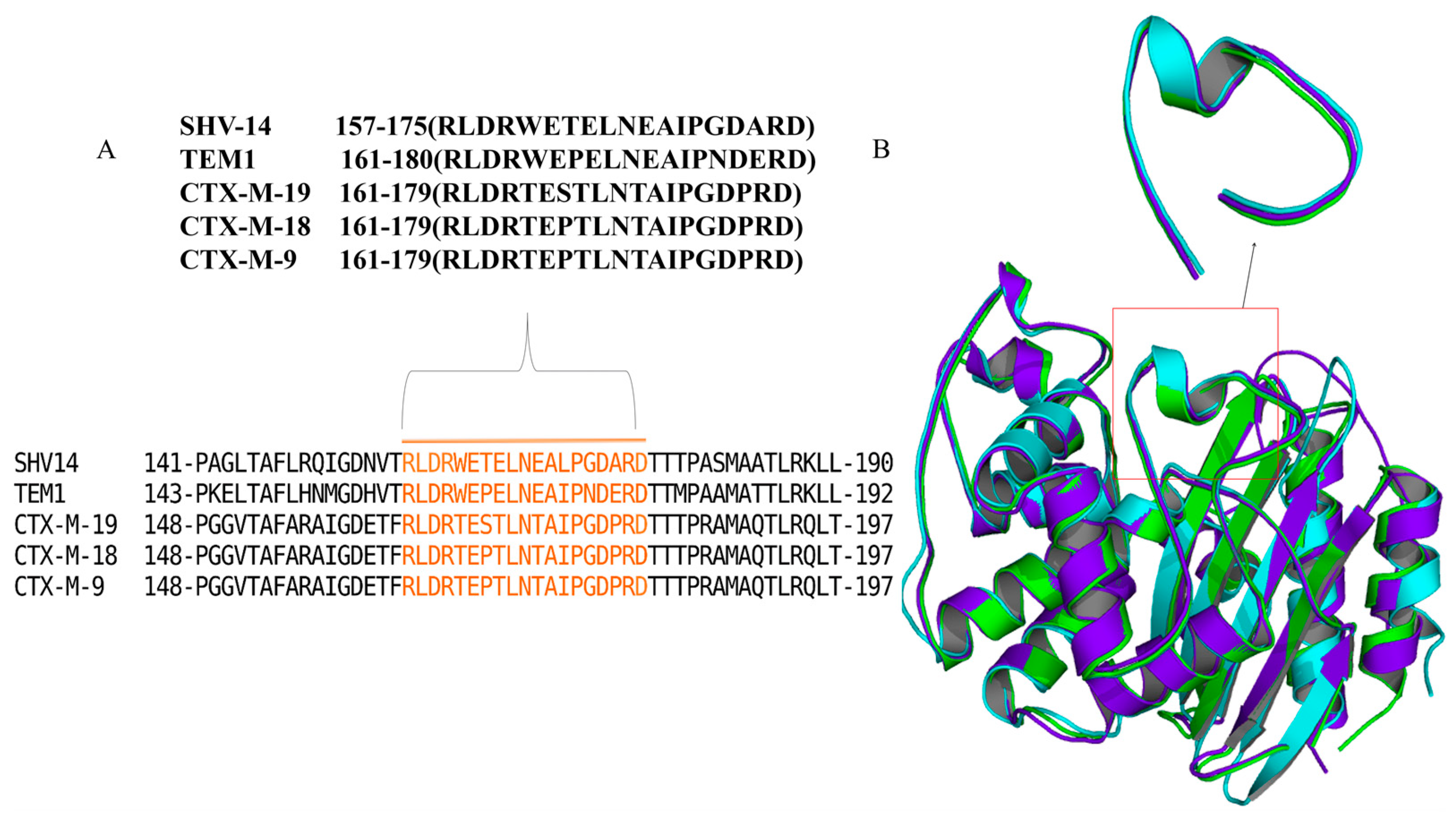
2.3. In Silico Modelling and Peptide Library Construction
2.4. Molecular Docking of Peptides into the Active Site Pocket of Class A β-Lactamases
2.5. Chemical Synthesis of Peptides by Solid-Phase Peptide Synthesis (SPPS)
2.6. High-Performance Liquid Chromatography (HPLC) and High-Resolution Mass Spectroscopy (HRMS)
2.7. Protein Purification
2.8. Antibiotic Susceptibility Testing
2.8.1. Whole-Cell Phenotypic Evaluation of Synthesized Peptides by HT-SPOTi
2.8.2. Determination of MIC Using Broth Microdilution Method
2.9. Kinetic Behaviour of β-Lactamases in the Presence of the Peptides
2.10. Molecular Dynamics Simulation Study
3. Results and Discussion
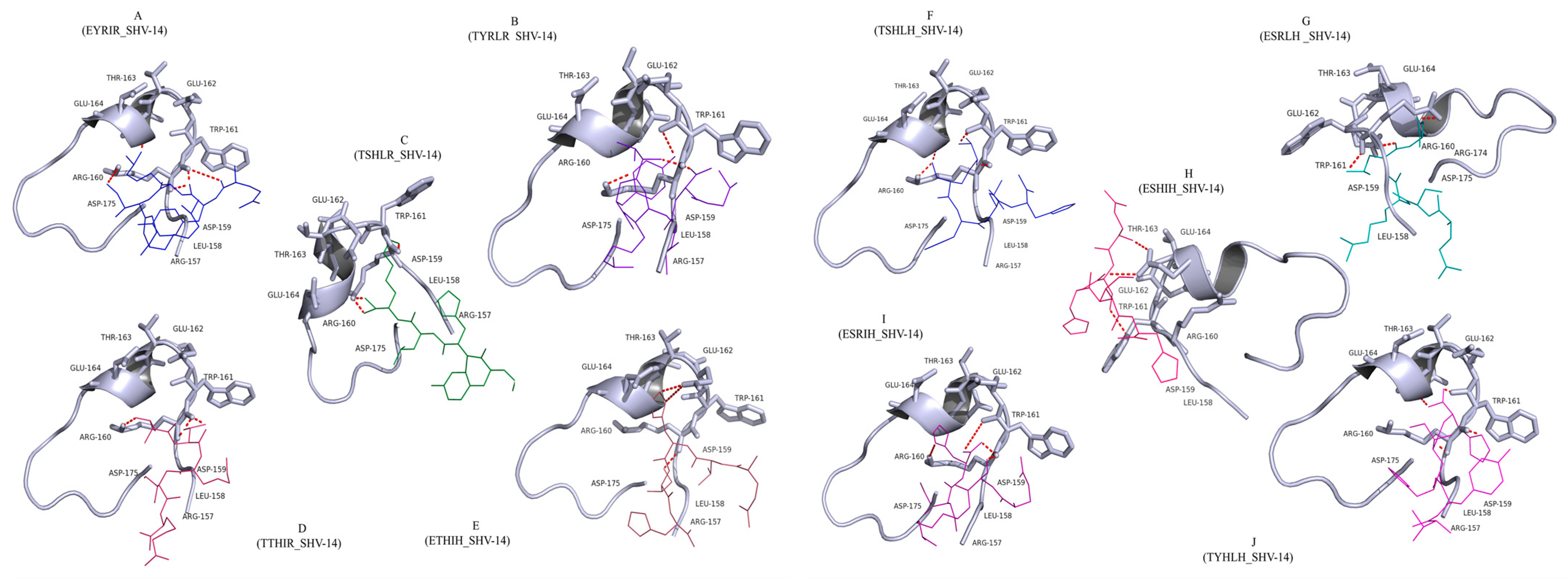
3.1. Docking Analysis Supported the Hypothesis That Peptides Could Inhibit β-Lactamase
3.2. The Antimicrobial Efficacy of the β-Lactams Was Enhanced in the Presence of the Peptides in Gram-Negative Organisms
3.3. Enhanced Antimicrobial Action of Peptides in Mycobacteria
3.4. Cells Expressing the β-Lactamases Confirmed Enhanced Sensitivity with Peptides
3.5. Peptides Have a Significant Inhibitory Impact on the Kinetic Behaviour of β-Lactamases
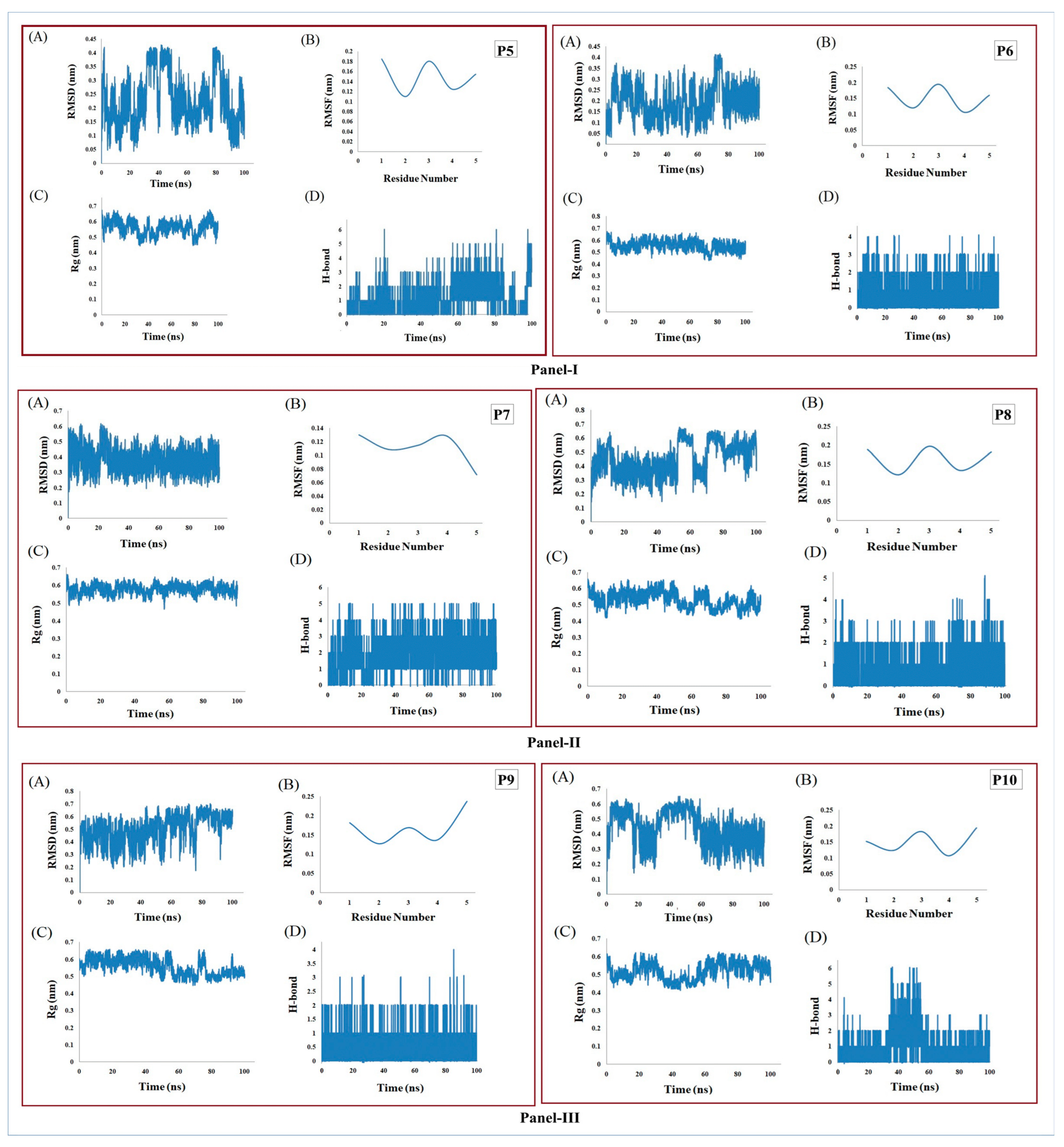
3.6. MD Simulation Supports the Stability of Peptides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banerjee, S.; Pieper, U.; Kapadia, G.; Pannell, L.K.; Herzbere, O. Role of the Ω-Loop in the Activity, Substrate Specificity, and Structure of Class A β-Lactamase. Biochemistry 1998, 37, 3286–3296. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.D.T.; Sothiselvam, S.; Lu, T.K.; de la Fuente-Nunez, C. Peptide Design Principles for Antimicrobial Applications. J. Mol. Biol. 2019, 431, 3547–3567. [Google Scholar] [CrossRef] [PubMed]
- Eiamphungporn, W.; Schaduangrat, N.; Malik, A.A.; Nantasenamat, C. Tackling the Antibiotic Resistance Caused by Class a β-Lactamases through the Use of β-Lactamase Inhibitory Protein. Int. J. Mol. Sci. 2018, 19, 2222. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Biswal, S.; Nathan, S.; Ghosh, A.S. Glutamate Residues at Positions 162 and 164 Influence the Beta-Lactamase Activity of SHV-14 Obtained from Klebsiella Pneumoniae. FEMS Microbiol. Lett. 2017, 365, fnx259. [Google Scholar] [CrossRef]
- Bansal, A.; Kar, D.; Pandey, S.D.; Matcha, A.; Kumar, N.G.; Nathan, S.; Ghosh, A.S. A Tyrosine Residue Along with a Glutamic Acid of the Omega-Like Loop Governs the Beta-Lactamase Activity of MSMEG_4455 in Mycobacterium Smegmatis. Protein J. 2017, 36, 220–227. [Google Scholar] [CrossRef]
- Majiduddin, F.K.; Materon, I.C.; Palzkill, T.G. Molecular Analysis of Beta-Lactamase Structure and Function. Int. J. Med. Microbiol. 2002, 292, 127–137. [Google Scholar] [CrossRef]
- Kazi, M.I.; Perry, B.W.; Card, D.C.; Schargel, R.D.; Ali, H.B.; Obuekwe, V.C.; Sapkota, M.; Kang, K.N.; Pellegrino, M.W.; Greenberg, D.E.; et al. Discovery and Characterization of New Delhi Metallo-beta-Lactamase-1 Inhibitor Peptides That Potentiate Meropenem-Dependent Killing of Carbapenemase-Producing Enterobacteriaceae. J. Antimicrob. Chemother. 2020, 75, 2843–2851. [Google Scholar] [CrossRef]
- Rotem, S.; Mor, A. Antimicrobial Peptide Mimics for Improved Therapeutic Properties. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1582–1592. [Google Scholar] [CrossRef]
- Kuczer, M.; Konopińska, D.; Rosiński, G. Insect Gonadotropic Peptide Hormones: Some Recent. J. Pept. Sci. 2007, 13, 16–26. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide Therapeutics: Current Status and Future Directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef]
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The Future of Peptide-Based Drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The Antimicrobial Peptide Thanatin Disrupts the Bacterial Outer Membrane and Inactivates the NDM-1 Metallo-β-Lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Lu, J.; Fang, C.; Zhou, Y.; Bai, H.; Zhang, X.; Xue, X.; Chen, Y.; Luo, X. Underlying Mechanism of in Vivo and in Vitro Activity of C-Terminal--Amidated Thanatin against Clinical Isolates of Extended-Spectrum β-Lactamase--Producing Escherichia Coli. J. Infect. Dis. 2011, 203, 273–282. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, A.; Vaudrey, J.; Vaiciunaite, B.; Moigboi, C.; McTavish, S.M.; Kearns, A.; Coates, A. Combinations of β-Lactam or Aminoglycoside Antibiotics with Plectasin Are Synergistic against Methicillin-Sensitive and Methicillin-Resistant Staphylococcus Aureus. PLoS ONE 2015, 10, e0117664. [Google Scholar] [CrossRef]
- Rishi, P.; Preet Singh, A.; Garg, N.; Rishi, M. Evaluation of Nisin--β-Lactam Antibiotics against Clinical Strains of Salmonella Enterica Serovar Typhi. J. Antibiot. 2014, 67, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.O.; Cardoso, M.H.; Franco, O.L. Development of Peptides That Inhibit Aminoglycoside-Modifying Enzymes and β-Lactamases for Control of Resistant Bacteria. Curr. Protein Pept. Sci. 2020, 21, 1011–1026. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.K.; Ghosh, A.S. Involvement of O8-Antigen in Altering β-Lactam Antibiotic Susceptibilities in Escherichia Coli. FEMS Microbiol. Lett. 2008, 282, 59–64. [Google Scholar] [CrossRef]
- Ghosh, A.S.; Young, K.D. Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 2005, 187, 1913–1922. [Google Scholar] [CrossRef]
- Grandjean, L.; Martin, L.; Gilman, R.H.; Valencia, T.; Herrera, B.; Quino, W.; Ramos, E.; Rivero, M.; Montoya, R.; Escombe, A.R.; et al. Tuberculosis diagnosis and multidrug resistance testing by direct sputum culture in selective broth without decontamination or centrifugation. J. Clin. Microbiol. 2008, 46, 2339–2344. [Google Scholar] [CrossRef]
- VLifeMDS®: Molecular Design Suite, Version 4.6; VLife Sciences Technologies (A division of NovaLead Pharma Pvt. Ltd.): Pune, India, 2018.
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Palomo, J.M. Solid-Phase Peptide Synthesis: An Overview Focused on the Preparation of Biologically Relevant Peptides. Rsc. Adv. 2014, 4, 32658–32672. [Google Scholar] [CrossRef]
- Danquah, C.A.; Maitra, A.; Gibbons, S.; Faull, J.; Bhakta, S. HT-SPOTi: A Rapid Drug Susceptibility Test (DST) to Evaluate Antibiotic Resistance Profiles and Novel Chemicals for Anti-Infective Drug Discovery. Curr. Protoc. Microbiol. 2016, 40, 17.8.1–17.8.12. [Google Scholar] [CrossRef] [PubMed]
- Guzman, J.D.; Evangelopoulos, D.; Gupta, A.; Birchall, K.; Mwaigwisya, S.; Saxty, B.; McHugh, T.D.; Gibbons, S.; Malkinson, J.; Bhakta, S. Antitubercular Specific Activity of Ibuprofen and the Other 2-Arylpropanoic Acids Using the HT-SPOTi Whole-Cell Phenotypic Assay. BMJ Open 2013, 3, e002672. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; 20th Informational Supplement; CLSI Document M100-SReplace M100; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2015. [Google Scholar]
- Sarkar, S.K.; Dutta, M.; Chowdhury, C.; Kumar, A.; Ghosh, A.S. PBP5, PBP6 and DacD Play Different Roles in Intrinsic β-Lactam Resistance of Escherichia Coli. Microbiology 2011, 157, 2702–2707. [Google Scholar] [CrossRef] [PubMed]
- Dowd, J.E.; Riggs, D.S. A Comparison of Estimates of Michaelis-Menten Kinetic Constants from Various Linear Transformations. J. Biol. Chem. 1965, 240, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B. GROMACS User Manual Version 5.0; The GROMACS Development Team in Sweden and Uppsala University: Uppsala, Sweden, 2014. [Google Scholar]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-Chain Torsion Potentials for the Amber Ff99SB Protein Force Field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Lmpey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Rudgers, G.W.; Huang, W.; Palzkill, T. Binding Properties of a Peptide Derived from β-Lactamase Inhibitory Protein. Antimicrob. Agents Chemother. 2001, 45, 3279–3286. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Kar, D.; Murugan, R.A.; Mallick, S.; Dutta, M.; Pandey, S.D.; Chowdhury, C.; Ghosh, A.S. A Putative Low-Molecular-Mass Penicillin-Binding Protein (PBP) of Mycobacterium Smegmatis Exhibits Prominent Physiological Characteristics of Dd-Carboxypeptidase and Beta-Lactamase. Microbiology 2015, 161, 1081–1091. [Google Scholar] [CrossRef]
- Sauvage, E.; Fonzé, E.; Quinting, B.; Galleni, M.; Frère, J.M.; Charlier, P. Crystal Structure of the Mycobacterium Fortuitum Class a β-Lactamase: Structural Basis for Broad Substrate Specificity. Antimicrob. Agents Chemother. 2006, 50, 2516–2521. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Yang, K.L. Identification of Peptide Inhibitors of Penicillinase Using a Phage Display Library. Anal. Biochem. 2016, 494, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Voladri, R.K.R.; Lakey, D.L.; Hennigan, S.H.; Menzies, B.E.; Edwards, K.M.; Kernodle, D.S. Recombinant expression and characterization of the major $β$-lactamase of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1998, 42, 1375–1381. [Google Scholar] [CrossRef]
- Soroka, D.; De La Sierra-Gallay, I.L.; Dubée, V.; Triboulet, S.; Van Tilbeurgh, H.; Compain, F.; Ballell, L.; Barros, D.; Mainardi, J.L.; Hugonnet, J.E.; et al. Hydrolysis of Clavulanate by Mycobacterium Tuberculosis β-Lactamase BlaC Harboring a Canonical SDN Motif. Antimicrob. Agents Chemother. 2015, 59, 5714–5720. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.R.; Parsons, L.M.; Pavelka, M.S. Genetic Analysis of the Beta-Lactamases of Mycobacterium Tuberculosis and Mycobacterium Smegmatis and Susceptibility to beta-Lactam Antibiotics. Microbiology 2005, 151, 521–532. [Google Scholar] [CrossRef]
- Phichith, D.; Bun, S.; Padiolleau-Lefevre, S.; Guellier, A.; Banh, S.; Galleni, M.; Frere, J.M.; Thomas, D.; Friboulet, A.; Avalle, B. Novel Peptide Inhibiting Both TEM-1 β-Lactamase and Penicillin-Binding Proteins. FEBS J. 2010, 277, 4965–4972. [Google Scholar] [CrossRef]
- Hale, J.D.F.; Hancock, R.E.W. Alternative Mechanisms of Action of Cationic Antimicrobial Peptides on Bacteria. Expert Rev. Anti-Infect. Ther. 2007, 5, 951–959. [Google Scholar] [CrossRef]
- Copolovici, D.M.; Langel, K.; Eriste, E.; Langel, Ü. Cell-Penetrating Peptides: Design, Synthesis, and Applications. ACS Nano 2014, 8, 1972–1994. [Google Scholar] [CrossRef] [PubMed]
| Strains/Plasmids | Source or Reference | |
|---|---|---|
| E. coli CS109 | C. Schnaitman; Ghosh and Young, 2005 [18] | |
| S. aureus | ATCC25923 | |
| Klebsiella pneumoniae | NCTC Number: 12463 | |
| Klebsiella pneumoniae | MTCC 3384 (Kumar et al., 2018) [4] | |
| M. tuberculosis MDR strain | Peru isolate (Grandjean et al., 2008) [19] | |
| M. tuberculosis H37Rv | ATCC25923 | |
| M. smegmatis mc2155 | ATCC700084 | |
| pBAD18Cam | Expression vector with arabinose-inducible promoter | Stratagene |
| pET28a(+) | E. coli expression vector generating His6 fusion proteins for overexpression | Stratagene |
| BL21 (DE3) pLysS | F–, ompT, hsdSB (rB –, mB –), dcm, gal, λ(DE3), pLysS (Cmr) | Promega |
| Peptide Sequence | Binding Energy kcal/mol | Hydrogen Bonds | Interacting Amino Acids |
|---|---|---|---|
| EYRIR (P1) | −3.28 | 3 | Arg160, Trp161 (2) |
| TYRLR(P2) | −6.56 | 3 | Arg160 (2), Trp161 |
| TSHLR (P3) | −5.88 | 3 | Arg160, Trp161 |
| TTHIR (P4) | −4.56 | 3 | Leu158, Asp159, Asp175 |
| ETHIH (P5) | −3.42 | 4 | Arg160 (3), Trp161 |
| TSHLH (P6) | −4.68 | 5 | Arg160 (4), Trp161, |
| ESRLH (P7) | −5.84 | 5 | Arg160 (2),Trp161, Glu164 |
| ESHIH (P8) | −4.25 | 4 | Arg160 (3), Trp161 |
| ESRIH (P9) | −3.68 | 4 | Arg160 (3), Trp161 |
| TYHLH (P10) | −6.78 | 3 | Trp161, Glu162, Thr163 |
| (a) | ||||
| Strain | MIC Value Antibiotics (mg/L) | |||
| Pen | Amp | Amx | Pip | |
| E. coli CS109 | 32 | 4 | 8 | 16 |
| E. coli CS109 + P1 | 16 | 4 | 4 | 16 |
| E. coli CS109 + P2 | 16 | 2 | 8 | 4 |
| E. coli CS109 + P3 | 32 | 8 | 8 | 8 |
| E. coli CS109 + P4 | 16 | 4 | 4 | 16 |
| E. coli CS109 + P5 | 32 | 2 | 2 | 2 |
| E. coli CS109 + P6 | 16 | 1 | 2 | 8 |
| E. coli CS109 + P7 | 16 | 4 | 2 | 4 |
| E. coli CS109 + P8 | 32 | 4 | 4 | 4 |
| E. coli CS109 + P9 | 16 | 2 | 8 | 4 |
| E. coli CS109 + P10 | 16 | 2 | 8 | 4 |
| DMSO | >500 | >500 | >500 | >500 |
| Kanamycin | 64 | 64 | 64 | >500 |
| (b) | ||||
| Strain | MIC Value Antibiotics (mg/L) | |||
| Pen | Amp | Amx | Pip | |
| K. pneumoniae | 32 | 32 | 64 | 64 |
| K. pneumoniae + P5 | 8 | 8 | 16 | 32 |
| K. pneumoniae + P6 | 8 | 4 | 32 | 16 |
| K. pneumoniae + P7 | 16 | 8 | 16 | 16 |
| K. pneumoniae + P8 | 8 | 8 | 16 | 16 |
| K. pneumoniae + P9 | 8 | 4 | 32 | 16 |
| K. pneumoniae + P10 | 8 | 4 | 32 | 32 |
| DMSO | >500 | >500 | >500 | >500 |
| Kanamycin | 64 | 64 | 64 | 64 |
| (c) | ||||
| Strain | MIC Value Antibiotics (mg/L) | |||
| Pen | Amp | Amx | Pip | |
| M. smegmatis | 125 | 4 | 2 | >250 |
| M. smegmatis + P1 | 125 | 4 | 2 | >250 |
| M. smegmatis + P2 | 64 | 4 | 2 | >250 |
| M. smegmatis + P3 | 125 | 8 | 8 | >250 |
| M. smegmatis + P4 | 64 | 8 | 4 | >250 |
| M. smegmatis + P5 | 32 | 1 | 1 | >250 |
| M. smegmatis + P6 | 32 | 0.5 | 0.5 | >250 |
| M. smegmatis + P7 | 64 | 1 | 1 | >250 |
| M. smegmatis + P8 | 32 | 4 | 1 | >250 |
| M. smegmatis + P9 | 32 | 2 | 2 | >250 |
| M. smegmatis + P10 | 64 | 2 | 2 | >250 |
| DMSO | >500 | >500 | >500 | >250 |
| Isoniazid | 64 | 64 | 64 | >250 |
| Strain | MIC Value of Antibiotics (mg/L) | |||
|---|---|---|---|---|
| Pen | Amp | Amx | Pip | |
| E. coil CS109 | 32 | 4 | 8 | 16 |
| E. coli CS109 + pBAD-TEM1 | 64 | 16 | 32 | 64 |
| E. coil CS109 + pBAD-TEM1 + P5 | 16 | 4 | 8 | 8 |
| E. coil CS109 + pBAD-TEM1 + P6 | 16 | 2 | 16 | 8 |
| E. coil CS109 + pBAD-TEM1 + P7 | 16 | 4 | 4 | 8 |
| E. coil CS109 + pBAD-TEM1 + P8 | 32 | 4 | 16 | 8 |
| E. coil CS109 + pBAD-TEM1 + P9 | 16 | 8 | 8 | 8 |
| E. coil CS109 + pBAD-TEM1 + P10 | 16 | 8 | 8 | 8 |
| E. coli CS109 + pBAD-SHV-14 | 250 | 32 | 125 | 64 |
| E. coli CS109 + pBAD-SHV-14 + P5 | 64 | 32 | 16 | 16 |
| E. coli CS109 + pBAD-SHV-14 + P6 | 125 | 4 | 16 | 8 |
| E. coli CS109 + pBAD-SHV-14 + P7 | 64 | 4 | 32 | 16 |
| E. coli CS109 + pBAD-SHV-14 + P8 | 32 | 4 | 32 | 32 |
| E. coli CS109 + pBAD-SHV-14 + P9 | 125 | 8 | 32 | 32 |
| E. coli CS109 + pBAD-SHV-14 + P10 | 32 | 4 | 32 | 32 |
| DMSO | >500 | >500 | >500 | >500 |
| Kanamycin | 64 | 64 | 64 | 64 |
| Strain | MIC Value of Antibiotics (mg/L) | |||
|---|---|---|---|---|
| Pen | Amp | Amx | Pip | |
| E. coil CS109 | 64 | 8 | 16 | 16 |
| E. coli CS109 + pBAD-TEM1 | 125 | 32 | 64 | 125 |
| E. coil CS109 + pBAD-TEM1 + P5 | 32 | 8 | 16 | 16 |
| E. coil CS109 + pBAD-TEM1 + P6 | 32 | 4 | 32 | 16 |
| E. coil CS109 + pBAD-TEM1 + P7 | 32 | 8 | 8 | 16 |
| E. coil CS109 + pBAD-TEM1 + P8 | 64 | 8 | 16 | 16 |
| E. coil CS109 + pBAD-TEM1 + P9 | 32 | 16 | 16 | 16 |
| E. coil CS109 + pBAD-TEM1 + P10 | 32 | 16 | 16 | 16 |
| E. coli CS109 + pBAD-SHV-14 | 500 | 64 | 125 | 125 |
| E. coli CS109 + pBAD-SHV-14 + P5 | 125 | 64 | 32 | 32 |
| E. coli CS109 + pBAD-SHV-14 + P6 | 250 | 8 | 32 | 16 |
| E. coli CS109 + pBAD-SHV-14 + P7 | 125 | 8 | 64 | 32 |
| E. coli CS109 + pBAD-SHV-14 + P8 | 250 | 8 | 64 | 64 |
| E. coli CS109 + pBAD-SHV-14 + P9 | 250 | 16 | 64 | 64 |
| E. coli CS109 + pBAD-SHV-14 + P10 | 250 | 8 | 64 | 64 |
| (a) | |||
| TEM-1 | Km (μM) | kcat (s−1) | kcat/Km (s−1μM−1) |
| Pen | 67.20 ± 4.43 | 386.95 ± 15.97 | 5.4 |
| Pen + P5 | 504.34 ± 72.56 | 14.89 ± 0.74 | 0.027 |
| Pen + P6 | 331.78 ± 29.30 | 12.28 ± 1.87 | 0.036 |
| Pen + P7 | 324.04 ± 20.20 | 11.42 ± 1.52 | 0.033 |
| Pen + P8 | 290.48 ± 20.81 | 31.1 ± 1.46 | 0.106 |
| Pen + P9 | 281.01 ± 47.47 | 19.03 ± 2.5 | 0.67 |
| Pen + P10 | 240.38 ± 50.54 | 30.05 ± 0.5 | 0.125 |
| TEM-1 | Km (μM) | kcat (s−1) | kcat/Km (μM) |
| Amp | 66.04 ± 12.16 | 290 ± 85.55 | 4.3 |
| Amp + P5 | 442.54 ± 37.75 | 23.43 ± 1.09 | 0.052 |
| Amp + P6 | 358.03± 47.30 | 17.33 ± 0.03 | 0.047 |
| Amp + P7 | 602.09 ± 26.36 | 5.99 ± 0.05 | 0.009 |
| Amp + P8 | 328.99 ± 67.04 | 14.56 ± 2.03 | 0.042 |
| Amp + P9 | 289.40 ± 31.05 | 24.12 ± 0.87 | 0.081 |
| Amp + P10 | 179.67 ± 30.07 | 51.08 ± 1.44 | 0.28 |
| TEM-1 | Km (μM) | kcat (s−1) | kcat/Km (μM) |
| Amx | 105.09 ± 12.16 | 427 ± 20.99 | 4 |
| Amx + P5 | 410.62 ± 102.46 | 31.91 ± 6.27 | 0.07 |
| Amx + P6 | 262.35 ± 93.87 | 20 ± 2.02 | 0.70 |
| Amx + P7 | 668.78 ± 59.91 | 14.29 ± 1.46 | 0.02 |
| Amx + P8 | 452.78 ± 45.01 | 23.01 ± 0.87 | 0.05 |
| Amx + P9 | 342.45 ± 23.11 | 2.1 ± 0.09 | 0.005 |
| Amx + P10 | 487.32 ± 14.21 | 14.34 ± 3.02 | 0.02 |
| TEM-1 | Km (μM) | kcat (s−1) | kcat /Km (μM) |
| Pip | 136.32 ± 12.16 | 331.26 ± 12.22 | 2.4 |
| Pip + P5 | 787.11 ± 122.49 | 14.25 ± 1.09 | 0.01 |
| Pip + P6 | 521.08 ± 34.79 | 23.9 ± 1.41 | 0.044 |
| Pip + P7 | 387.78 ± 129.8 | 32.01± 6.43 | 0.023 |
| Pip + P8 | 737.46 ± 67.98 | 12.12± 4.11 | 0.016 |
| Pip + P9 | 814.67 ± 33.19 | 27.32 ± 5.56 | 0.033 |
| Pip + P10 | 768.09 ± 78.66 | 19.09 ± 3.03 | 0.024 |
| (b) | |||
| SHV-14 | Km (μM) | kcat (s−1) | kcat/Km (s−1μM−1) |
| Pen | 144.20 ± 4.43 | 586.95 ± 15.97 | 4.07 |
| Pen + P5 | 343.98 ± 19.94 | 42.32 ± 3.01 | 0.12 |
| Pen + P6 | 453.05 ± 8.97 | 18.14 ± 2.32 | 0.04 |
| Pen + P7 | 564.01 ± 6.89 | 15.2 ± 1.21 | 0.02 |
| Pen + P8 | 612.34 ± 11.32 | 34.13 ± 3.12 | 0.05 |
| Pen + P9 | 432.28 ± 32.9 | 38.81 ± 0.1 | 0.08 |
| Pen + P10 | 461.32 ± 22.01 | 11.24 ± 2.1 | 0.02 |
| SHV-14 | Km (μM) | kcat (s−1) | kcat /Km (μM) |
| Amp | 186.74 ± 12.16 | 644.95 ± 10.97 | 3.4 |
| Amp + P5 | 603.42 ± 7.02 | 14.11 ± 1.03 | 0.02 |
| Amp + P6 | 560.99 ± 43.12 | 19.5 ± 2.05 | 0.03 |
| Amp + P7 | 667.51 ± 16.43 | 12.09 ± 1.01 | 0.01 |
| Amp + P8 | 340.21 ± 6.86 | 54.7 ± 0.09 | 0.1 |
| Amp + P9 | 365.34 ± 35.1 | 34.43 ± 0.98 | 0.07 |
| Amp + P10 | 276.56 ± 34.09 | 34.03 ± 4.33 | 0.01 |
| SHV-14 | Km (μM) | kcat (s−1) | kcat/Km (μM) |
| Amx | 111.74 ± 12.16 | 416.95 ± 5.02 | 3.7 |
| Amx + P5 | 626.02 ± 24.01 | 23.02 ± 0.04 | 0.03 |
| Amx + P6 | 564.55 ± 50.3 | 14.67 ± 0.78 | 0.02 |
| Amx + P7 | 343.23 ± 13.9 | 12.3 ± 1.2 | 0.03 |
| Amx + P8 | 266.04 ± 23.06 | 24.32 ± 4.45 | 0.09 |
| Amx + P9 | 353.34 ± 5.45 | 29.87 ± 5.7 | 0.06 |
| Amx + P10 | 344.18 ± 29.09 | 16.03 ± 1.03 | 0.04 |
| SHV-14 | Km (μM) | kcat (s−1) | kcat/Km (μM) |
| Pip | 98.74 ± 12.16 | 432.09 ± 5.97 | 4.4 |
| Pip + P5 | 373.98 ± 19.94 | 35.02 ± 7.01 | 0.09 |
| Pip + P6 | 537.05 ± 8.97 | 31.21 ± 6.32 | 0.06 |
| Pip + P7 | 424.01 ± 6.89 | 32.01± 6.43 | 0.03 |
| Pip + P8 | 542.34 ± 11.32 | 34.13 ± 3.12 | 0.05 |
| Pip + P9 | 332.28 ± 32.9 | 28.81 ± 0.1 | 0.06 |
| Pip + P10 | 411.32 ± 15.01 | 18.24 ± 2.1 | 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswal, S.; Caetano, K.; Jain, D.; Sarrila, A.; Munshi, T.; Dickman, R.; Tabor, A.B.; Rath, S.N.; Bhakta, S.; Ghosh, A.S. Antimicrobial Peptides Designed against the Ω-Loop of Class A β-Lactamases to Potentiate the Efficacy of β-Lactam Antibiotics. Antibiotics 2023, 12, 553. https://doi.org/10.3390/antibiotics12030553
Biswal S, Caetano K, Jain D, Sarrila A, Munshi T, Dickman R, Tabor AB, Rath SN, Bhakta S, Ghosh AS. Antimicrobial Peptides Designed against the Ω-Loop of Class A β-Lactamases to Potentiate the Efficacy of β-Lactam Antibiotics. Antibiotics. 2023; 12(3):553. https://doi.org/10.3390/antibiotics12030553
Chicago/Turabian StyleBiswal, Sarmistha, Karina Caetano, Diamond Jain, Anusha Sarrila, Tulika Munshi, Rachael Dickman, Alethea B. Tabor, Surya Narayan Rath, Sanjib Bhakta, and Anindya S. Ghosh. 2023. "Antimicrobial Peptides Designed against the Ω-Loop of Class A β-Lactamases to Potentiate the Efficacy of β-Lactam Antibiotics" Antibiotics 12, no. 3: 553. https://doi.org/10.3390/antibiotics12030553
APA StyleBiswal, S., Caetano, K., Jain, D., Sarrila, A., Munshi, T., Dickman, R., Tabor, A. B., Rath, S. N., Bhakta, S., & Ghosh, A. S. (2023). Antimicrobial Peptides Designed against the Ω-Loop of Class A β-Lactamases to Potentiate the Efficacy of β-Lactam Antibiotics. Antibiotics, 12(3), 553. https://doi.org/10.3390/antibiotics12030553







