The Role of Coagulase-Negative Staphylococci Biofilms on Late-Onset Sepsis: Current Challenges and Emerging Diagnostics and Therapies
Abstract
1. Introduction
2. CoNS Role as Commensal in Newborns
3. CoNS Role as a Pathogen in Newborns
3.1. Risk Factors for LOS Caused by CoNS
3.2. CoNS Virulence Factors
3.2.1. Toxins and Exoenzymes
3.2.2. CoNS Biofilm Formation
Biofilm Formation as an Antibiotic Tolerance/Resistance Mechanism
Biofilm Formation as an Immune System Resistance Mechanism
4. LOS Diagnosis and Treatment
4.1. Diagnosis of LOS
4.1.1. Clinical Symptoms
4.1.2. Biological Samples Culture
4.1.3. Biomarkers—Inflammatory Molecules and Hematologic Indices
4.1.4. Molecular Biology Methods—Nucleic Acids Analysis
4.1.5. Molecular Methods—Proteins Analysis
4.1.6. Diagnosis Approaches under Development
4.2. LOS Treatment
4.2.1. Currently Available Therapies—Antibiotics
Improved Antibiotic-Based Treatment Approaches
4.2.2. Antibiotic-Independent Treatment Approaches under Development
Biofilm-Degrading Enzymes
Bacteriophages and Lysins
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Definition |
| NS | Neonatal sepsis |
| EOS | Early-onset sepsis |
| LOS | Late-onset sepsis |
| CoNS | Coagulase-negative staphylococci |
| GBS | Group B Streptococcus |
| NICU | Neonatal intensive care unit |
| MSCRAMMs | Microbial surface components recognizing matrix molecules |
| Atl | Autolysins |
| TA | Teichoic acids |
| eDNA | Extracellular DNA |
| PNAG | Poly-N-acetyl-glucosamine |
| Embp | Extracellular matrix binding protein |
| HGT | Horizontal gene transfer |
| Srd | Serine-aspartate repeat proteins |
| Aap | Accumulation-associated protein |
| Bhp | Biofilm-homologue protein |
| SepA | S. epidermidis metalloprotease A |
| Esp | S. epidermidis serine protease |
| EcpA | S. epidermidis protease A |
| PSM | Phenol-soluble modulins |
| AMPs | Antimicrobial peptides |
| CRP | C-reactive protein |
| PCT | Procalcitonin |
| PCR | Polymerase chain reaction |
| qPCR | Quantitative-polymerase chain reaction |
| FISH | Fluorescence in situ hybridization |
| PNA FISH | Peptide nucleic acid fluorescence in situ hybridization |
| MALDI-TOF | Matrix-assisted laser desorption-ionization/time-of-flight |
| CRISPR-Cas | Clustered regularly interspaced short palindromic repeats |
References
- Preterm Birth. Available online: https://www.who.int/news-room/fact-sheets/detail/preterm-birth (accessed on 1 November 2022).
- Liu, L.; Oza, S.; Hogan, D.; Chu, Y.; Perin, J.; Zhu, J.; Lawn, J.E.; Cousens, S.; Mathers, C.; Black, R.E. Global, Regional, and National Causes of under-5 Mortality in 2000–15: An Updated Systematic Analysis with Implications for the Sustainable Development Goals. Lancet 2016, 388, 3027–3035. [Google Scholar] [CrossRef] [PubMed]
- Schlapbach, L.J.; Aebischer, M.; Adams, M.; Natalucci, G.; Bonhoeffer, J.; Latzin, P.; Nelle, M.; Bucher, H.U.; Latal, B.; Zeilinger, G.; et al. Impact of Sepsis on Neurodevelopmental Outcome in a Swiss National Cohort of Extremely Premature Infants. Pediatrics 2011, 128, e348–e357. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann-Struzek, C.; Goldfarb, D.M.; Schlattmann, P.; Schlapbach, L.J.; Reinhart, K.; Kissoon, N. The Global Burden of Paediatric and Neonatal Sepsis: A Systematic Review. Lancet. Respir. Med. 2018, 6, 223–230. [Google Scholar] [CrossRef]
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 354 Diseases and Injuries for 195 Countries and Territories, 1990-2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980-2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Deutschman, C.S.; Seymour, C.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801. [Google Scholar] [CrossRef]
- Molloy, E.J.; Wynn, J.L.; Bliss, J.; Koenig, J.M.; Keij, F.M.; McGovern, M.; Kuester, H.; Turner, M.A.; Giannoni, E.; Mazela, J.; et al. Neonatal Sepsis: Need for Consensus Definition, Collaboration and Core Outcomes. Pediatr. Res. 2020, 88, 2–4. [Google Scholar] [CrossRef]
- Ershad, M.; Mostafa, A.; Cruz, M.D.; Vearrier, D. Neonatal Sepsis Risk Factor American Academy of Pediatrics. Curr. Emerg. Hosp. Med. Rep. 2019, 7, 83–90. [Google Scholar] [CrossRef]
- Camacho-Gonzalez, A.; Spearman, P.W.; Stoll, B.J. Neonatal Infectious Diseases: Evaluation of Neonatal Sepsis. Pediatr. Clin. North Am. 2013, 60, 367–389. [Google Scholar] [CrossRef]
- Jyoti, A.; Kumar, S.; Kumar Srivastava, V.; Kaushik, S.; Govind Singh, S. Neonatal Sepsis at Point of Care. Clin. Chim. Acta 2021, 521, 45–58. [Google Scholar] [CrossRef]
- Singh, M.; Alsaleem, M.; Gray, C.P. Neonatal Sepsis. StatPearls 2022, 390, 1770–1780. [Google Scholar]
- Bizzarro, M.J.; Raskind, C.; Baltimore, R.S.; Gallagher, P.G. Seventy-Five Years of Neonatal Sepsis at Yale: 1928-2003. Pediatrics 2005, 116, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.J.; Hansen, N.; Fanaroff, A.A.; Wright, L.L.; Carlo, W.A.; Ehrenkranz, R.A.; Lemons, J.A.; Donovan, E.F.; Stark, A.R.; Tyson, J.E.; et al. Late-Onset Sepsis in Very Low Birth Weight Neonates: The Experience of the NICHD Neonatal Research Network. Pediatrics 2002, 110, 285–291. [Google Scholar] [CrossRef]
- Shane, A.L.; Sánchez, P.J.; Stoll, B.J. Neonatal Sepsis. Lancet 2017, 390, 1770–1780. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.A.; Padbury, J.F. Neonatal Sepsis: An Old Problem with New Insights. Virulence 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, K.A.; Anderson-Berry, A.L.; Delair, S.F.; Dele Davies, H. Early-Onset Neonatal Sepsis. Clin. Microbiol. Rev. 2014, 27, 21–47. [Google Scholar] [CrossRef]
- Lawn, J.E.; Bianchi-Jassir, F.; Russell, N.J.; Kohli-Lynch, M.; Tann, C.J.; Hall, J.; Madrid, L.; Baker, C.J.; Bartlett, L.; Cutland, C.; et al. Group B Streptococcal Disease Worldwide for Pregnant Women, Stillbirths, and Children: Why, What, and How to Undertake Estimates? Clin. Infect. Dis. 2017, 65, S89–S99. [Google Scholar] [CrossRef]
- Stoll, B.J.; Puopolo, K.M.; Hansen, N.I.; Sánchez, P.J.; Bell, E.F.; Carlo, W.A.; Cotten, C.M.; D’Angio, C.T.; Kazzi, S.N.J.; Poindexter, B.B.; et al. Early-Onset Neonatal Sepsis 2015 to 2017, the Rise of Escherichia Coli, and the Need for Novel Prevention Strategies. JAMA Pediatr. 2020, 174, e200593. [Google Scholar] [CrossRef]
- Cortese, F.; Scicchitano, P.; Gesualdo, M.; Filaninno, A.; De Giorgi, E.; Schettini, F.; Laforgia, N.; Ciccone, M.M. Early and Late Infections in Newborns: Where Do We Stand? A Review. Pediatr. Neonatol. 2016, 57, 265–273. [Google Scholar] [CrossRef]
- Dong, Y.; Speer, C.P. Late-Onset Neonatal Sepsis:Recent Developments. Arch. Dis. Child. Fetal Neonatal Ed. 2015, 100, F257–F263. [Google Scholar] [CrossRef]
- Shane, A.L.; Stoll, B.J. Neonatal Sepsis: Progress towards Improved Outcomes. J. Infect. 2014, 68 (Suppl 1), S24–S32. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.L.; Lim, C.S.E.; Sultana, R.; De La Puerta, R.; Rajadurai, V.S.; Yeo, K.T. Risk Factors for Mortality From Late-Onset Sepsis Among Preterm Very-Low-Birthweight Infants: A Single-Center Cohort Study From Singapore. Front. Pediatr. 2022, 9, 801955. [Google Scholar] [CrossRef]
- Dong, Y.; Speer, C.P.; Glaser, K. Beyond Sepsis: Staphylococcus Epidermidis Is an Underestimated but Significant Contributor to Neonatal Morbidity. Virulence 2018, 9, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Boghossian, N.S.; Page, G.P.; Bell, E.F.; Stoll, B.J.; Murray, J.C.; Cotten, C.M.; Shankaran, S.; Walsh, M.C.; Laptook, A.R.; Newman, N.S.; et al. Late-Onset Sepsis in Very Low Birth Weight Infants from Singleton and Multiple-Gestation Births. J. Pediatr. 2013, 162, 1120–1124. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Speer, C.P. The Role of Staphylococcus Epidermidis in Neonatal Sepsis: Guarding Angel or Pathogenic Devil? Int. J. Med. Microbiol. 2014, 304, 513–520. [Google Scholar] [CrossRef]
- Berardi, A.; Sforza, F.; Baroni, L.; Spada, C.; Ambretti, S.; Biasucci, G.; Bolognesi, S.; Capretti, M.; Carretto, E.; Ciccia, M.; et al. Epidemiology and Complications of Late-Onset Sepsis: An Italian Area-Based Study. PLoS ONE 2019, 14, e0225407. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Sousa, E.; Freitas, J.; Viana, M.; Miranda, F.; Silva, F.P. da Positive Blood Culture and Neonatal Sepsis—A Five-Year Study. NASCER E CRESCER BIRTH GROWTH Med. J. 2022, 31, 106–114. [Google Scholar] [CrossRef]
- Størdal, E.H.; Solevåg, A.L.; Bjørnholt, J.V.; Rønnestad, A.; Stensvold, H.J. Sepsis Treatment Options Identified by 10-Year Study of Microbial Isolates and Antibiotic Susceptibility in a Level-Four Neonatal Intensive Care Unit. Acta Paediatr. Int. J. Paediatr. 2022, 111, 519–526. [Google Scholar] [CrossRef]
- Flannery, D.D.; Edwards, E.M.; Coggins, S.A.; Horbar, J.D.; Puopolo, K.M. Late-Onset Sepsis Among Very Preterm Infants. Pediatrics 2022, 150, e2022058813. [Google Scholar] [CrossRef]
- Sands, K.; Carvalho, M.J.; Spiller, O.B.; Portal, E.A.R.; Thomson, K.; Watkins, W.J.; Mathias, J.; Dyer, C.; Akpulu, C.; Andrews, R.; et al. Characterisation of Staphylococci Species from Neonatal Blood Cultures in Low- and Middle-Income Countries. BMC Infect. Dis. 2022, 22, 593. [Google Scholar] [CrossRef]
- Okomo, U.; Akpalu, E.N.K.; Le Doare, K.; Roca, A.; Cousens, S.; Jarde, A.; Sharland, M.; Kampmann, B.; Lawn, J.E. Aetiology of Invasive Bacterial Infection and Antimicrobial Resistance in Neonates in Sub-Saharan Africa: A Systematic Review and Meta-Analysis in Line with the STROBE-NI Reporting Guidelines. Lancet. Infect. Dis. 2019, 19, 1219–1234. [Google Scholar] [CrossRef] [PubMed]
- Medugu, N.; Iregbu, K.; Tam, P.Y.I.; Obaro, S. Aetiology of Neonatal Sepsis in Nigeria, and Relevance of Group b Streptococcus: A Systematic Review. PLoS ONE 2018, 13, e0200350. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Boxberger, M.; Cenizo, V.; Cassir, N.; La Scola, B. Challenges in Exploring and Manipulating the Human Skin Microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef]
- Flowers, L.; Grice, E.A. The Skin Microbiota: Balancing Risk and Reward. Cell Host Microbe 2020, 28, 190–200. [Google Scholar] [CrossRef]
- França, A.; Gaio, V.; Lopes, N.; Melo, L.D.R.L.D.R. Virulence Factors in Coagulase-Negative Staphylococci. Pathogens 2021, 10, 170. [Google Scholar] [CrossRef]
- Rasigade, J.P.; Raulin, O.; Picaud, J.C.; Tellini, C.; Bes, M.; Grando, J.; Saïd, M.B.; Claris, O.; Etienne, J.; Tigaud, S.; et al. Methicillin-Resistant Staphylococcus Capitis with Reduced Vancomycin Susceptibility Causes Late-Onset Sepsis in Intensive Care Neonates. PLoS ONE 2012, 7, e31548. [Google Scholar] [CrossRef]
- Ben Said, M.; Hays, S.; Bonfils, M.; Jourdes, E.; Rasigade, J.P.; Laurent, F.; Picaud, J.C. Late-Onset Sepsis Due to Staphylococcus Capitis “neonatalis” in Low-Birthweight Infants: A New Entity? J. Hosp. Infect. 2016, 94, 95–98. [Google Scholar] [CrossRef]
- Björkqvist, M.; Söderquist, B.; Törnqvist, E.; Sjöberg, L.; Fredlund, H.; Kühn, I.; Colque-Navarro, P.; Schollin, J. Phenotypic and Genotypic Characterisation of Blood Isolates of Coagulase-Negative Staphylococci in the Newborn. APMIS 2002, 110, 332–339. [Google Scholar] [CrossRef]
- Foka, A.; Chini, V.; Petinaki, E.; Kolonitsiou, F.; Anastassiou, E.D.; Dimitracopoulos, G.; Spiliopoulou, I. Clonality of Slime-Producing Methicillin-Resistant Coagulase-Negative Staphylococci Disseminated in the Neonatal Intensive Care Unit of a University Hospital. Clin. Microbiol. Infect. 2006, 12, 1230–1233. [Google Scholar] [CrossRef]
- Neumeister, B.; Kastner, S.; Conrad, S.; Klotz, G.; Bartmann, P. Characterization of Coagulase-Negative Staphylococci Causing Nosocomial Infections in Preterm Infants. Eur. J. Clin. Microbiol. Infect. Dis. 1995, 14, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Al-Haqan, A.; Boswihi, S.S.; Pathan, S.; Udo, E.E. Antimicrobial Resistance and Virulence Determinants in Coagulase-Negative Staphylococci Isolated Mainly from Preterm Neonates. PLoS ONE 2020, 15, e0236713. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.M.A.; Binatti, V.B.; Sued, B.P.R.; Ramos, J.N.; Peixoto, R.S.; Simões, C.; de Castro, E.A.; Duarte, J.L.M.B.; Vieira, V.Ô.V.; Hirata, R.; et al. Staphylococcus Haemolyticus Disseminated among Neonates with Bacteremia in a Neonatal Intensive Care Unit in Rio de Janeiro, Brazil. Diagn. Microbiol. Infect. Dis. 2014, 78, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Butin, M.; Rasigade, J.P.; Martins-Simões, P.; Meugnier, H.; Lemriss, H.; Goering, R.V.; Kearns, A.; Deighton, M.A.; Denis, O.; Ibrahimi, A.; et al. Wide Geographical Dissemination of the Multiresistant Staphylococcus Capitis NRCS-A Clone in Neonatal Intensive-Care Units. Clin. Microbiol. Infect. 2016, 22, 46–52. [Google Scholar] [CrossRef]
- Penesyan, A.; Paulsen, I.T.; Kjelleberg, S.; Gillings, M.R. Three Faces of Biofilms: A Microbial Lifestyle, a Nascent Multicellular Organism, and an Incubator for Diversity. npj Biofilms Microbiomes 2021, 7, 80. [Google Scholar] [CrossRef]
- Heilmann, C.; Ziebuhr, W.; Becker, K. Are Coagulase-Negative Staphylococci Virulent? Clin. Microbiol. Infect. 2019, 25, 1071–1080. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-Negative Staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef]
- Becker, K.; Both, A.; Weißelberg, S.; Heilmann, C.; Rohde, H. Emergence of Coagulase-Negative Staphylococci. Expert Rev. Anti. Infect. Ther. 2020, 18, 349–366. [Google Scholar] [CrossRef]
- Klingenberg, C.; Aarag, E.; Rønnestad, A.; Sollid, J.E.; Abrahamsen, T.G.; Kjeldsen, G.; Flœgstad, T. Coagulase-Negative Staphylococcal Sepsis in Neonates: Association between Antibiotic Resistance, Biofilm Formation and the Host Inflammatory Response. Pediatr. Infect. Dis. J. 2005, 24, 817–822. [Google Scholar] [CrossRef]
- Oliveira, F.; Cerca, N. Antibiotic Resistance and Biofilm Formation Ability among Coagulase-Negative Staphylococci in Healthy Individuals from Portugal. J. Antibiot. 2013, 66, 739–741. [Google Scholar] [CrossRef]
- Cerca, N.; Martins, S.; Cerca, F.; Jefferson, K.K.; Pier, G.B.; Oliveira, R.; Azeredo, J. Comparative Assessment of Antibiotic Susceptibility of Coagulase-Negative Staphylococci in Biofilm versus Planktonic Culture as Assessed by Bacterial Enumeration or Rapid XTT Colorimetry. J. Antimicrob. Chemother. 2005, 56, 331–336. [Google Scholar] [CrossRef] [PubMed]
- França, A.; Carvalhais, V.; Vilanova, M.; Pier, G.B.; Cerca, N. Characterization of an in Vitro Fed-Batch Model to Obtain Cells Released from S. Epidermidis Biofilms. AMB Express 2016, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- O’ Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; HM Government: London, UK, 2014.
- Production, A.P.; Meng, H.; Dai, Y.; Otto, M.; Li, M.; Liu, Q.; Liu, Q.; Meng, H.; Lv, H.; Liu, Y.; et al. Staphylococcus Epidermidis Contributes to Healthy Maturation of the Nasal Microbiome by Stimulating Article Staphylococcus Epidermidis Contributes to Healthy Maturation of the Nasal Microbiome by Stimulating Antimicrobial Peptide Production. Cell Host Microbe 2020, 27, 68–78.e5. [Google Scholar] [CrossRef]
- Lai, Y.; Di Nardo, A.; Nakatsuji, T.; Leichtle, A.; Yang, Y.; Cogen, A.L.; Wu, Z.; Hooper, L.V.; Von Aulock, S.; Radek, K.A.; et al. Commensal Bacteria Regulate TLR3-Dependent Inflammation Following Skin Injury. Nat. Med. 2009, 15, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from Human Skin Commensal Bacteria Protect against Staphylococcus Aureus and Are Deficient in Atopic Dermatitis. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Bitschar, K.; Staudenmaier, L.; Klink, L.; Focken, J.; Sauer, B.; Fehrenbacher, B.; Herster, F.; Bittner, Z.; Bleul, L.; Schaller, M.; et al. Staphylococcus Aureus Skin Colonization Is Enhanced by the Interaction of Neutrophil Extracellular Traps with Keratinocytes. J. Invest. Dermatol. 2020, 140, 1054–1065.e4. [Google Scholar] [CrossRef]
- Yoon, H.S. Neonatal Innate Immunity and Toll-like Receptor. Korean J. Pediatr. 2010, 53, 985–988. [Google Scholar] [CrossRef]
- Kai-Larsen, Y.; Gudmundsson, G.H.; Agerberth, B. A Review of the Innate Immune Defence of the Human Foetus and Newborn, with the Emphasis on Antimicrobial Peptides. Acta Paediatr. 2014, 103, 1000–1008. [Google Scholar] [CrossRef]
- Tsafaras, G.P.; Ntontsi, P.; Xanthou, G. Advantages and Limitations of the Neonatal Immune System. Front. Pediatr. 2020, 8, 1–10. [Google Scholar] [CrossRef]
- Kumar, S.K.M.; Bhat, B.V. Distinct Mechanisms of the Newborn Innate Immunity. Immunol. Lett. 2016, 173, 42–54. [Google Scholar] [CrossRef]
- Kan, B.; Razzaghian, H.R.; Lavoie, P.M. An Immunological Perspective on Neonatal Sepsis. Trends Mol. Med. 2016, 22, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Harbeson, D.; Ben-Othman, R.; Amenyogbe, N.; Kollmann, T.R. Outgrowing the Immaturity Myth: The Cost of Defending From Neonatal Infectious Disease. Front. Immunol. 2018, 9, 1077. [Google Scholar] [CrossRef] [PubMed]
- Kollmann, T.R.; Kampmann, B.; Mazmanian, S.K.; Marchant, A.; Levy, O. Protecting the Newborn and Young Infant from Infectious Diseases: Lessons from Immune Ontogeny. Immunity 2017, 46, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Brook, B.; Harbeson, D.; Ben-Othman, R.; Viemann, D.; Kollmann, T.R. Newborn Susceptibility to Infection vs. Disease Depends on Complex in Vivo Interactions of Host and Pathogen. Semin. Immunopathol. 2017, 39, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Schneider, D.S.; Soares, M.P. Disease Tolerance as a Defense Strategy. Science 2012, 335, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Rocamora-Reverte, L.; Melzer, F.L.; Würzner, R.; Weinberger, B. The Complex Role of Regulatory T Cells in Immunity and Aging. Front. Immunol. 2021, 11, 616949. [Google Scholar] [CrossRef]
- Leech, J.M.; Dhariwala, M.O.; Lowe, M.M.; Chu, K.; Merana, G.R.; Cornuot, C.; Weckel, A.; Ma, J.M.; Leitner, E.G.; Gonzalez, J.R.; et al. Toxin-Triggered Interleukin-1 Receptor Signaling Enables Early-Life Discrimination of Pathogenic versus Commensal Skin Bacteria. Cell Host Microbe 2019, 26, 795–809.e5. [Google Scholar] [CrossRef]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the Human Skin Microbiome Early in Life. J. Invest. Dermatol. 2011, 131, 2026–2032. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota across Multiple Body Habitats in Newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Joubert, I.A.; Otto, M.; Strunk, T.; Currie, A.J. Look Who’s Talking: Host and Pathogen Drivers of Staphylococcus Epidermidis Virulence in Neonatal Sepsis. Int. J. Mol. Sci. 2022, 23, 860. [Google Scholar] [CrossRef]
- Yasmin, F.; Tun, H.M.; Konya, T.B.; Guttman, D.S.; Chari, R.S.; Field, C.J.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; et al. Cesarean Section, Formula Feeding, and Infant Antibiotic Exposure: Separate and Combined Impacts on Gut Microbial Changes in Later Infancy. Front. Pediatr. 2017, 5, 200. [Google Scholar] [CrossRef] [PubMed]
- Gasparrini, A.J.; Wang, B.; Sun, X.; Kennedy, E.A.; Hernandez-Leyva, A.; Ndao, I.M.; Tarr, P.I.; Warner, B.B.; Dantas, G. Persistent Metagenomic Signatures of Early-Life Hospitalization and Antibiotic Treatment in the Infant Gut Microbiota and Resistome. Nat. Microbiol. 2019, 4, 2285–2297. [Google Scholar] [CrossRef] [PubMed]
- Gibson, M.K.; Wang, B.; Ahmadi, S.; Burnham, C.A.D.; Tarr, P.I.; Warner, B.B.; Dantas, G. Developmental Dynamics of the Preterm Infant Gut Microbiota and Antibiotic Resistome. Nat. Microbiol. 2016, 1, 16024. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.; Olm, M.R.; Firek, B.A.; Baker, R.; Thomas, B.C.; Morowitz, M.J.; Banfield, J.F. Strain-Resolved Analysis of Hospital Rooms and Infants Reveals Overlap between the Human and Room Microbiome. Nat. Commun. 2017, 8, 1814. [Google Scholar] [CrossRef]
- Brooks, B.; Olm, M.R.; Firek, B.A.; Baker, R.; Geller-McGrath, D.; Reimer, S.R.; Soenjoyo, K.R.; Yip, J.S.; Dahan, D.; Thomas, B.C.; et al. The Developing Premature Infant Gut Microbiome Is a Major Factor Shaping the Microbiome of Neonatal Intensive Care Unit Rooms. Microbiome 2018, 6, 112. [Google Scholar] [CrossRef]
- Altuntas, E.G. Isolation, Identification and Characterization of Staphylococcus Epidermidis in Human Milk. LWT Food Sci. Technol. 2015, 60, 36–41. [Google Scholar] [CrossRef]
- Damaceno, Q.S.; Souza, J.P.; Nicoli, J.R.; Paula, R.L.; Assis, G.B.; Figueiredo, H.C.; Azevedo, V.; Martins, F.S. Evaluation of Potential Probiotics Isolated from Human Milk and Colostrum. Probiotics Antimicrob. Proteins 2017, 9, 371–379. [Google Scholar] [CrossRef]
- Jiménez, E.; Delgado, S.; Fernández, L.; García, N.; Albújar, M.; Gómez, A.; Rodríguez, J.M. Assessment of the Bacterial Diversity of Human Colostrum and Screening of Staphylococcal and Enterococcal Populations for Potential Virulence Factors. Res. Microbiol. 2008, 159, 595–601. [Google Scholar] [CrossRef]
- Jiménez, E.; Delgado, S.; Maldonado, A.; Arroyo, R.; Albújar, M.; García, N.; Jariod, M.; Fernández, L.; Gómez, A.; Rodríguez, J.M. Staphylococcus Epidermidis: A Differential Trait of the Fecal Microbiota of Breast-Fed Infants. BMC Microbiol. 2008, 8, 143. [Google Scholar] [CrossRef]
- Martín, V.; Maldonado-Barragán, A.; Moles, L.; Rodriguez-Baños, M.; Del Campo, R.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Sharing of Bacterial Strains between Breast Milk and Infant Feces. J. Hum. Lact. 2012, 28, 36–44. [Google Scholar] [CrossRef]
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is Meconium from Healthy Newborns Actually Sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Moles, L.; Gómez, M.; Heilig, H.; Bustos, G.; Fuentes, S.; de Vos, W.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Bacterial Diversity in Meconium of Preterm Neonates and Evolution of Their Fecal Microbiota during the First Month of Life. PLoS ONE 2013, 8, e66986. [Google Scholar] [CrossRef] [PubMed]
- Soeorg, H.; Metsvaht, T.; Eelmäe, I.; Metsvaht, H.K.; Treumuth, S.; Merila, M.; Ilmoja, M.L.; Lutsar, I. Coagulase-Negative Staphylococci in Human Milk From Mothers of Preterm Compared With Term Neonates. J. Hum. Lact. 2017, 33, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Soeorg, H.; Treumuth, S.; Metsvaht, H.K.; Eelmäe, I.; Merila, M.; Ilmoja, M.L.; Lutsar, I.; Metsvaht, T. Higher Intake of Coagulase-Negative Staphylococci from Maternal Milk Promotes Gut Colonization with MecA-Negative Staphylococcus Epidermidis in Preterm Neonates. J. Perinatol. 2018, 38, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.J.; Hansen, N.I.; Adams-Chapman, I.; Fanaroff, A.A.; Hintz, S.R.; Vohr, B.; Higgins, R.D. Neurodevelopmental and Growth Impairment among Extremely Low-Birth-Weight Infants with Neonatal Infection. JAMA 2004, 292, 2357–2365. [Google Scholar] [CrossRef]
- Alshaikh, B.; Yee, W.; Lodha, A.; Henderson, E.; Yusuf, K.; Sauve, R. Coagulase-Negative Staphylococcus Sepsis in Preterm Infants and Long-Term Neurodevelopmental Outcome. J. Perinatol. 2014, 34, 125–129. [Google Scholar] [CrossRef]
- Alshaikh, B.; Yusuf, K.; Sauve, R. Neurodevelopmental Outcomes of Very Low Birth Weight Infants with Neonatal Sepsis: Systematic Review and Meta-Analysis. J. Perinatol. 2013, 33, 558–564. [Google Scholar] [CrossRef]
- Liljedahl, M.; Bodin, L.; Schollin, J. Coagulase-Negative Staphylococcal Sepsis as a Predictor of Bronchopulmonary Dysplasia. Acta Paediatr. 2004, 93, 211–215. [Google Scholar] [CrossRef]
- Shah, D.K.; Doyle, L.W.; Anderson, P.J.; Bear, M.; Daley, A.J.; Hunt, R.W.; Inder, T.E. Adverse Neurodevelopment in Preterm Infants with Postnatal Sepsis or Necrotizing Enterocolitis Is Mediated by White Matter Abnormalities on Magnetic Resonance Imaging at Term. J. Pediatr. 2008, 153, 170–175. [Google Scholar] [CrossRef]
- Anderson-Berry, A.; Brinton, B.; Lyden, E.; Faix, R.G. Risk Factors Associated with Development of Persistent Coagulase-Negative Staphylococci Bacteremia in the Neonate and Associated Short-Term and Discharge Morbidities. Neonatology 2011, 99, 23–31. [Google Scholar] [CrossRef]
- Jean-Baptiste, N.; Benjamin, D.K.; Cohen-Wolkowiez, M.; Fowler, V.G.; Laughon, M.; Clark, R.H.; Smith, P.B. Coagulase-Negative Staphylococcal Infections in the Neonatal Intensive Care Unit. Infect. Control Hosp. Epidemiol. 2011, 32, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Zea-Vera, A.; Ochoa, T.J. Challenges in the Diagnosis and Management of Neonatal Sepsis. J. Trop. Pediatr. 2015, 61, 1. [Google Scholar] [CrossRef] [PubMed]
- el Manouni el Hassani, S.; Berkhout, D.J.C.; Niemarkt, H.J.; Mann, S.; de Boode, W.P.; Cossey, V.; Hulzebos, C.V.; van Kaam, A.H.; Kramer, B.W.; van Lingen, R.A.; et al. Risk Factors for Late-Onset Sepsis in Preterm Infants: A Multicenter Case-Control Study. Neonatology 2019, 116, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Sobczak, A.; Klepacka, J.; Amrom, D.; Żak, I.; Kruczek, P.; Kwinta, P. Umbilical Catheters as Vectors for Generalized Bacterial Infection in Premature Infants Regardless of Antibiotic Use. J. Med. Microbiol. 2019, 68, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Hira, V.; Sluijter, M.; Estevão, S.; Horst-Kreft, D.; Ott, A.; De Groot, R.; Hermans, P.W.M.; Kornelisse, R.F. Clinical and Molecular Epidemiologic Characteristics of Coagulase-Negative Staphylococcal Bloodstream Infections in Intensive Care Neonates. Pediatr. Infect. Dis. J. 2007, 26, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Cortez, J.; Makker, K.; Kraemer, D.F.; Neu, J.; Sharma, R.; Hudak, M.L. Maternal Milk Feedings Reduce Sepsis, Necrotizing Enterocolitis and Improve Outcomes of Premature Infants. J. Perinatol. 2018, 38, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.L.; Johnson, T.J.; Engstrom, J.L.; Fogg, L.F.; Jegier, B.J.; Bigger, H.R.; Meier, P.P. Impact of Early Human Milk on Sepsis and Health-Care Costs in Very Low Birth Weight Infants. J. Perinatol. 2013, 33, 514–519. [Google Scholar] [CrossRef]
- Muk, T.; Brunse, A.; Henriksen, N.L.; Aasmul-Olsen, K.; Nguyen, D.N. Glucose Supply and Glycolysis Inhibition Shape the Clinical Fate of Staphylococcus Epidermidis-Infected Preterm Newborns. JCI Insight 2022, 7, e157234. [Google Scholar] [CrossRef]
- Golińska, E.; Strus, M.; Tomusiak-Plebanek, A.; Więcek, G.; Kozień, Ł.; Lauterbach, R.; Pawlik, D.; Rzepecka-Węglarz, B.; Kędzierska, J.; Dorycka, M.; et al. Coagulase-Negative Staphylococci Contained in Gut Microbiota as a Primary Source of Sepsis in Low- and Very Low Birth Weight Neonates. J. Clin. Med. 2020, 9, 2517. [Google Scholar] [CrossRef]
- Soeorg, H.; Huik, K.; Parm, Ü.; Ilmoja, M.L.; Metsvaht, T.; Lutsar, I. Molecular Epidemiology of Staphylococcus Epidermidis in Neonatal Intensive Care Units. APMIS 2017, 125, 63–73. [Google Scholar] [CrossRef]
- Tolo, I.; Thomas, J.C.; Fischer, R.S.B.; Brown, E.L.; Gray, B.M.; Robinson, D.A. Do Staphylococcus Epidermidis Genetic Clusters Predict Isolation Sources? J. Clin. Microbiol. 2016, 54, 1711–1719. [Google Scholar] [CrossRef] [PubMed]
- Soeorg, H.; Metsvaht, H.K.; Keränen, E.E.; Eelmäe, I.; Merila, M.; Ilmoja, M.L.; Metsvaht, T.; Lutsar, I. Genetic Relatedness of Staphylococcus Haemolyticus in Gut and Skin of Preterm Neonates and Breast Milk of Their Mothers. Pediatr. Infect. Dis. J. 2019, 38, 308–313. [Google Scholar] [CrossRef]
- Soeorg, H.; Metsvaht, T.; Eelmäe, I.; Merila, M.; Treumuth, S.; Huik, K.; Jürna-Ellam, M.; Ilmoja, M.L.; Lutsar, I. The Role of Breast Milk in the Colonization of Neonatal Gut and Skin with Coagulase-Negative Staphylococci. Pediatr. Res. 2017, 82, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Butin, M.; Rasigade, J.P.; Subtil, F.; Martins-Simões, P.; Pralong, C.; Freydière, A.M.; Vandenesch, F.; Tigaud, S.; Picaud, J.C.; Laurent, F. Vancomycin Treatment Is a Risk Factor for Vancomycin-Nonsusceptible Staphylococcus Capitis Sepsis in Preterm Neonates. Clin. Microbiol. Infect. 2017, 23, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Soeorg, H.; Huik, K.; Parm, Ü.; Ilmoja, M.L.; Metelskaja, N.; Metsvaht, T.; Lutsar, I. Genetic Relatedness of Coagulase-Negative Staphylococci from Gastrointestinal Tract and Blood of Preterm Neonates with Late-Onset Sepsis. Pediatr. Infect. Dis. J. 2013, 32, 389–393. [Google Scholar] [CrossRef]
- Yu, J.; Liu, Y.; Xu, Z. Hand Carriage, Antimicrobial Resistance and Molecular Characterisation of Methicillin-Resistant Coagulase-Negative Staphylococci Isolated from Gynaecological Surgical Staff. J. Obstet. Gynaecol. 2022, 1–6. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus Epidermidis – the “Accidental” Pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef]
- Le, K.Y.; Villaruz, A.E.; Zheng, Y.; He, L.; Fisher, E.L.; Nguyen, T.H.; Ho, T.V.; Yeh, A.J.; Joo, H.S.; Cheung, G.Y.C.; et al. Role of Phenol-Soluble Modulins in Staphylococcus Epidermidis Biofilm Formation and Infection of Indwelling Medical Devices. J. Mol. Biol. 2019, 431, 3015–3027. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Rigby, K.; Wang, R.; Queck, S.Y.; Braughton, K.R.; Whitney, A.R.; Teintze, M.; DeLeo, F.R.; Otto, M. Staphylococcus Epidermidis Strategies to Avoid Killing by Human Neutrophils. PLoS Pathog. 2010, 6, e1001133. [Google Scholar] [CrossRef]
- Wang, R.; Braughton, K.R.; Kretschmer, D.; Bach, T.H.L.; Queck, S.Y.; Li, M.; Kennedy, A.D.; Dorward, D.W.; Klebanoff, S.J.; Peschel, A.; et al. Identification of Novel Cytolytic Peptides as Key Virulence Determinants for Community-Associated MRSA. Nat. Med. 2007, 13, 1510–1514. [Google Scholar] [CrossRef]
- Kretschmer, D.; Gleske, A.-K.; Rautenberg, M.; Wang, R.; Köberle, M.; Bohn, E.; Schöneberg, T.; Rabiet, M.-J.; Boulay, F.; Klebanoff, S.J.; et al. Human Formyl Peptide Receptor 2 Senses Highly Pathogenic Staphylococcus Aureus. Cell Host Microbe 2010, 7, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Da, F.; Fisher, E.L.; Tan, D.C.S.; Nguyen, T.H.; Fu, C.L.; Tan, V.Y.; McCausland, J.W.; Sturdevant, D.E.; Joo, H.S.; et al. Toxin Mediates Sepsis Caused by Methicillin-Resistant Staphylococcus Epidermidis. PLoS Pathog. 2017, 13, e1006153. [Google Scholar] [CrossRef] [PubMed]
- Queck, S.Y.; Khan, B.A.; Wang, R.; Bach, T.H.L.; Kretschmer, D.; Chen, L.; Kreiswirth, B.N.; Peschel, A.; DeLeo, F.R.; Otto, M. Mobile Genetic Element-Encoded Cytolysin Connects Virulence to Methicillin Resistance in MRSA. PLoS Pathog. 2009, 5, e1000533. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Villaruz, A.E.; Li, M.; Cha, D.J.; Sturdevant, D.E.; Otto, M. The Human Anionic Antimicrobial Peptide Dermcidin Induces Proteolytic Defence Mechanisms in Staphylococci. Mol. Microbiol. 2007, 63, 497–506. [Google Scholar] [CrossRef]
- Dubin, G.; Chmiel, D.; Mak, P.; Rakwalska, M.; Rzychon, M.; Dubin, A. Molecular Cloning and Biochemical Characterization of Proteases from Staphylococcus Epidermidis. Biol. Chem. 2001, 382, 1575–1582. [Google Scholar] [CrossRef]
- Longshaw, C.M.; Farrell, A.M.; Wright, J.D.; Holland, K.T. Identification of a Second Lipase Gene, GehD, in Staphylococcus Epidermidis: Comparison of Sequence with Those of Other Staphylococcal Lipases. Microbiology 2000, 146 (Pt 6), 1419–1427. [Google Scholar] [CrossRef]
- Simons, J.W.F.A.; Van Kampen, M.D.; Riel, S.; Götz, F.; Egmond, M.R.; Verheij, H.M. Cloning, Purification and Characterisation of the Lipase from Staphylococcus Epidermidis--Comparison of the Substrate Selectivity with Those of Other Microbial Lipases. Eur. J. Biochem. 1998, 253, 675–683. [Google Scholar] [CrossRef]
- Gabriela Bowden, M.; Visai, L.; Longshaw, C.M.; Holland, K.T.; Speziale, P.; Höök, M. Is the GehD Lipase from Staphylococcus Epidermidis a Collagen Binding Adhesin? J. Biol. Chem. 2002, 277, 43017–43023. [Google Scholar] [CrossRef]
- Chamberlain, N.R.; Brueggemann, S.A. Characterisation and Expression of Fatty Acid Modifying Enzyme Produced by Staphylococcus Epidermidis. J. Med. Microbiol. 1997, 46, 693–697. [Google Scholar] [CrossRef][Green Version]
- Flemming, H.C.; Wuertz, S. Bacteria and Archaea on Earth and Their Abundance in Biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Bowler, P.; Murphy, C.; Wolcott, R. Biofilm Exacerbates Antibiotic Resistance: Is This a Current Oversight in Antimicrobial Stewardship? Antimicrob. Resist. Infect. Control 2020, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Scherr, T.D.; Heim, C.E.; Morrison, J.M.; Kielian, T. Hiding in Plain Sight: Interplay between Staphylococcal Biofilms and Host Immunity. Front. Immunol. 2014, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Hogan, D.; Kolter, R. Why Are Bacteria Refractory to Antimicrobials? Curr. Opin. Microbiol. 2002, 5, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.; Otto, M. Understanding the Significance of Staphylococcus Epidermidis Bacteremia in Babies and Children. Curr. Opin. Infect. Dis. 2010, 23, 208–216. [Google Scholar] [CrossRef]
- Wang, R.; Khan, B.A.; Cheung, G.Y.C.; Bach, T.-H.L.; Jameson-Lee, M.; Kong, K.-F.; Queck, S.Y.; Otto, M. Staphylococcus Epidermidis Surfactant Peptides Promote Biofilm Maturation and Dissemination of Biofilm-Associated Infection in Mice. J. Clin. Invest. 2011, 121, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 27. [Google Scholar] [CrossRef]
- Davey, M.E.; O’toole, G.A. Microbial Biofilms: From Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- Heilmann, C. Adhesion Mechanisms of Staphylococci. Adv. Exp. Med. Biol. 2011, 7, 105–123. [Google Scholar]
- Patti, J.M.; Allen, B.L.; McGavin, M.J.; Hook, M. MSCRAMM-Medicated Adherence of Microorganisms to Host Tissues. Annu. Rev. Microbiol. 1994, 48, 585–617. [Google Scholar] [CrossRef]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; DeBoy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on Evolution of Virulence and Resistance from the Complete Genome Analysis of an Early Methicillin-Resistant Staphylococcus Aureus Strain and a Biofilm-Producing Methicillin-Resistant Staphylococcus Epidermidis Strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef]
- McCrea, K.W.; Hartford, O.; Davis, S.; Eidhin, D.N.; Lina, G.; Speziale, P.; Foster, T.J.; Höök, M. The Serine-Aspartate Repeat (Sdr) Protein Family in Staphylococcus Epidermidis. Microbiology 2000, 146, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, C.; Hussain, M.; Peters, G.; Götz, F. Evidence for Autolysin-Mediated Primary Attachment of Staphylococcus Epidermidis to a Polystyrene Surface. Mol. Microbiol. 1997, 24, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Christner, M.; Franke, G.C.; Schommer, N.N.; Wendt, U.; Wegert, K.; Pehle, P.; Kroll, G.; Schulze, C.; Buck, F.; Mack, D.; et al. The Giant Extracellular Matrix-Binding Protein of Staphylococcus Epidermidis Mediates Biofilm Accumulation and Attachment to Fibronectin. Mol. Microbiol. 2010, 75, 187–207. [Google Scholar] [CrossRef] [PubMed]
- Weidenmaier, C.; Kokai-Kun, J.F.; Kristian, S.A.; Chanturiya, T.; Kalbacher, H.; Gross, M.; Nicholson, G.; Neumeister, B.; Mond, J.J.; Peschel, A. Role of Teichoic Acids in Staphylococcus Aureus Nasal Colonization, a Major Risk Factor in Nosocomial Infections. Nat. Med. 2004, 10, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Schilcher, K.; Horswill, A.R. Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- Schaeffer, C.R.; Hoang, T.-M.N.; Sudbeck, C.M.; Alawi, M.; Tolo, I.E.; Robinson, D.A.; Horswill, A.R.; Rohde, H.; Fey, P.D. Versatility of Biofilm Matrix Molecules in Staphylococcus Epidermidis Clinical Isolates and Importance of Polysaccharide Intercellular Adhesin Expression during High Shear Stress. mSphere 2016, 1, e00165-16. [Google Scholar] [CrossRef]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The Intercellular Adhesin Involved in Biofilm Accumulation of Staphylococcus Epidermidis Is a Linear β-1,6-Linked Glucosaminoglycan: Purification and Structural Analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar] [CrossRef]
- Cherifi, S.; Byl, B.; Deplano, A.; Nonhoff, C.; Denis, O.; Hallin, M. Comparative Epidemiology of Staphylococcus Epidermidis Isolates from Patients with Catheter-Related Bacteremia and from Healthy Volunteers. J. Clin. Microbiol. 2013, 51, 1541–1547. [Google Scholar] [CrossRef]
- Harris, L.G.; Murray, S.; Pascoe, B.; Bray, J.; Meric, G.; Magerios, L.; Wilkinson, T.S.; Jeeves, R.; Rohde, H.; Schwarz, S.; et al. Biofilm Morphotypes and Population Structure among Staphylococcus Epidermidis from Commensal and Clinical Samples. PLoS ONE 2016, 11, e0154510. [Google Scholar] [CrossRef]
- Ahmed, D.M.; Abel Messih, M.A.W.; Ibrahim, N.H.; Meabed, M.H.; Abdel-Salam, S.M. Frequency of IcaA and IcaD Determinants and Biofilm Formation among Coagulase-Negative Staphylococci Associated with Nasal Carriage in Neonatal Intensive Care Units. Germs 2019, 9, 61. [Google Scholar] [CrossRef]
- Sheikh, A.F.; Dezfuli, A.A.Z.; Navidifar, T.; Fard, S.S.; Dehdashtian, M. Association between Biofilm Formation, Structure and Antibiotic Resistance in Staphylococcus Epidermidis Isolated from Neonatal Septicemia in Southwest Iran. Infect. Drug Resist. 2019, 12, 1771–1782. [Google Scholar] [CrossRef]
- Martínez-Meléndez, A.; Morfín-Otero, R.; Villarreal-Treviño, L.; Camacho-Ortíz, A.; González-González, G.; Llaca-Díaz, J.; Rodríguez-Noriega, E.; Garza-González, E. Molecular Epidemiology of Coagulase-Negative Bloodstream Isolates: Detection of Staphylococcus Epidermidis ST2, ST7 and Linezolid-Resistant ST23. Brazilian J. Infect. Dis. 2016, 20, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, C.; Schweitzer, O.; Gerke, C.; Vanittanakom, N.; Mack, D.; Götz, F. Molecular Basis of Intercellular Adhesion in the Biofilm-Forming Staphylococcus Epidermidis. Mol. Microbiol. 1996, 20, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Herrmann, M.; Von Eiff, C.; Perdreau-Remington, F.; Peters, G. A 140-Kilodalton Extracellular Protein Is Essential for the Accumulation of Staphylococcus Epidermidis Strains on Surfaces. Infect. Immun. 1997, 65, 519–524. [Google Scholar] [CrossRef]
- Bowden, M.G.; Chen, W.; Singvall, J.; Xu, Y.; Peacock, S.J.; Valtulina, V.; Speziale, P.; Höök, M. Identification and Preliminary Characterization of Cell-Wall-Anchored Proteins of Staphylococcus Epidermidis. Microbiology 2005, 151, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Paharik, A.E.; Kotasinska, M.; Both, A.; Hoang, T.M.N.; Büttner, H.; Roy, P.; Fey, P.D.; Horswill, A.R.; Rohde, H. The Metalloprotease SepA Governs Processing of Accumulation-Associated Protein and Shapes Intercellular Adhesive Surface Properties in Staphylococcus Epidermidis. Mol. Microbiol. 2017, 103, 860–874. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus Epidermidis Esp Inhibits Staphylococcus Aureus Biofilm Formation and Nasal Colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef]
- Sugimoto, S.; Iwamoto, T.; Takada, K.; Okuda, K.I.; Tajima, A.; Iwase, T.; Mizunoe, Y. Staphylococcus Epidermidis Esp Degrades Specific Proteins Associatedwith Staphylococcus Aureus Biofilm Formation and Host-Pathogen Interaction. J. Bacteriol. 2013, 195, 1645–1655. [Google Scholar] [CrossRef]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How Staphylococcus Aureus Biofilms Develop Their Characteristic Structure. Proc. Natl. Acad. Sci. 2012, 109, 1281–1286. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and Guidelines for Research on Antibiotic Persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J. Clin. Microbiol. 1999, 37, 1771. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Mechanisms of Antibiotic Resistance in Bacterial Biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J. Physiological Heterogeneity in Biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.A.; Pitts, B.; Beyenal, H.; Veluchamy, R.A.; Lewandowski, Z.; Davison, W.M.; Buckingham-Meyer, K.; Stewart, P.S. Spatial Patterns of DNA Replication, Protein Synthesis, and Oxygen Concentration within Bacterial Biofilms Reveal Diverse Physiological States. J. Bacteriol. 2007, 189, 4223–4233. [Google Scholar] [CrossRef]
- García-Betancur, J.C.; Lopez, D. Cell Heterogeneity in Staphylococcal Communities. J. Mol. Biol. 2019, 431, 4699–4711. [Google Scholar] [CrossRef]
- Kranjec, C.; Angeles, D.M.; Mårli, M.T.; Fernández, L.; García, P.; Kjos, M.; Diep, D.B. Staphylococcal Biofilms: Challenges and Novel Therapeutic Perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef]
- Dunne, W.M.; Mason, E.O.; Kaplan, S.L. Diffusion of Rifampin and Vancomycin through a Staphylococcus Epidermidis Biofilm. Antimicrob. Agents Chemother. 1993, 37, 2522–2526. [Google Scholar] [CrossRef]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Penetration of Antibiotics through Staphylococcus Aureus and Staphylococcus Epidermidis Biofilms. J. Antimicrob. Chemother. 2010, 65, 1955–1958. [Google Scholar] [CrossRef]
- Gebreyohannes, G.; Nyerere, A.; Bii, C.; Sbhatu, D.B. Challenges of Intervention, Treatment, and Antibiotic Resistance of Biofilm-Forming Microorganisms. Heliyon 2019, 5, e02192. [Google Scholar] [CrossRef]
- Fluit, A.C.; Visser, M.R.; Schmitz, F. Molecular Detection of Antimicrobial Resistance. Clin. Microbiol. Rev. 2001, 14, 836–871. [Google Scholar] [CrossRef]
- Van Acker, H.; Van Dijck, P.; Coenye, T. Molecular Mechanisms of Antimicrobial Tolerance and Resistance in Bacterial and Fungal Biofilms. Trends Microbiol. 2014, 22, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.E.; Christensen, H.; Aarestrup, F.M. Diversity and Evolution of BlaZ from Staphylococcus Aureus and Coagulase-Negative Staphylococci. J. Antimicrob. Chemother. 2006, 57, 450–460. [Google Scholar] [CrossRef]
- Vega, N.M.; Gore, J. Collective Antibiotic Resistance: Mechanisms and Implications. Curr. Opin. Microbiol. 2014, 21, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, H.; Pourhajibagher, M.; Chiniforush, N.; Soltanian, A.R.; Alikhani, M.Y.; Bahador, A. Biofilm Formation and Antibiotic Resistance in Meticillin-Resistant and Meticillin-Sensitive Staphylococcus Aureus Isolated from Burns. J. Wound Care 2019, 28, 66–73. [Google Scholar] [CrossRef]
- Qin, Z.; Yang, X.; Yang, L.; Jiang, J.; Ou, Y.; Molin, S.; Qu, D. Formation and Properties of in Vitro Biofilms of Ica-Negative Staphylococcus Epidermidis Clinical Isolates. J. Med. Microbiol. 2007, 56, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Cafiso, V.; Bertuccio, T.; Santagati, M.; Campanile, F.; Amicosante, G.; Perilli, M.G.; Selan, L.; Artini, M.; Nicoletti, G.; Stefani, S. Presence of the Ica Operon in Clinical Isolates of Staphylococcus Epidermidis and Its Role in Biofilm Production. Clin. Microbiol. Infect. 2004, 10, 1081–1088. [Google Scholar] [CrossRef]
- Rachid, S.; Ohlsen, K.; Witte, W.; Hacker, J.; Ziebuhr, W. Effect of Subinhibitory Antibiotic Concentrations on Polysaccharide Intercellular Adhesin Expression in Biofilm-Forming Staphylococcus Epidermidis. Antimicrob. Agents Chemother. 2000, 44, 3357–3363. [Google Scholar] [CrossRef]
- Águila-Arcos, S.; Álvarez-Rodríguez, I.; Garaiyurrebaso, O.; Garbisu, C.; Grohmann, E.; Alkorta, I. Biofilm-Forming Clinical Staphylococcus Isolates Harbor Horizontal Transfer and Antibiotic Resistance Genes. Front. Microbiol. 2017, 8, 2018. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal Transfer of Antibiotic Resistance Genes in Clinical Environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Woodford, N.; Ellington, M.J. The Emergence of Antibiotic Resistance by Mutation. Clin. Microbiol. Infect. 2007, 13, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Ryder, V.J.; Chopra, I.; O’Neill, A.J. Increased Mutability of Staphylococci in Biofilms as a Consequence of Oxidative Stress. PLoS ONE 2012, 7, e47695. [Google Scholar] [CrossRef] [PubMed]
- Le, K.Y.; Park, M.D.; Otto, M. Immune Evasion Mechanisms of Staphylococcus Epidermidis Biofilm Infection. Front. Microbiol. 2018, 9, 359. [Google Scholar] [CrossRef] [PubMed]
- Cerca, F.; França, A.; Perez-Cabezas, B.; Carvalhais, V.; Ribeiro, A.; Azeredo, J.; Pier, G.; Cerca, N.; Vilanova, M. Dormant Bacteria within Staphylococcus Epidermidis Biofilms Have Low Inflammatory Properties and Maintain Tolerance to Vancomycin and Penicillin after Entering Planktonic Growth. J. Med. Microbiol. 2014, 63, 1274–1283. [Google Scholar] [CrossRef]
- Cerca, F.; Andrade, F.; Franca, A.; Andrade, E.B.; Ribeiro, A.; Almeida, A.A.; Cerca, N.; Pier, G.; Azeredo, J.; Vilanova, M. Staphylococcus Epidermidis Biofilms with Higher Proportions of Dormant Bacteria Induce a Lower Activation of Murine Macrophages. J. Med. Microbiol. 2011, 60, 1717–1724. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Yao, Y.; Carmody, A.B.; Otto, M. Increased Colonization of Indwelling Medical Devices by Quorum-Sensing Mutants of Staphylococcus Epidermidis in Vivo. J. Infect. Dis. 2004, 190, 1498–1505. [Google Scholar] [CrossRef]
- Yao, Y.; Vuong, C.; Kocianova, S.; Villaruz, A.E.; Lai, Y.; Sturdavant, D.E.; Otto, M. Characterization of the Staphylococcus Epidermidis Accessory-Gene Regulator Response: Quorum-Sensing Regulation of Resistance to Human Innate Host Defense. J. Infect. Dis. 2006, 193, 841–848. [Google Scholar] [CrossRef]
- Dengler Haunreiter, V.; Boumasmoud, M.; Häffner, N.; Wipfli, D.; Leimer, N.; Rachmühl, C.; Kühnert, D.; Achermann, Y.; Zbinden, R.; Benussi, S.; et al. In-Host Evolution of Staphylococcus Epidermidis in a Pacemaker-Associated Endocarditis Resulting in Increased Antibiotic Tolerance. Nat. Commun. 2019, 10, 1149. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; DeLeo, F.R.; Otto, M. A Crucial Role for Exopolysaccharide Modification in Bacterial Biofilm Formation, Immune Evasion, and Virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [CrossRef]
- Cerca, N.; Jefferson, K.K.; Oliveira, R.; Pier, G.B.; Azeredo, J. Comparative Antibody-Mediated Phagocytosis of Staphylococcus Epidermidis Cells Grown in a Biofilm or in the Planktonic State. Infect. Immun. 2006, 74, 4849–4855. [Google Scholar] [CrossRef]
- Kristian, S.A.; Birkenstock, T.A.; Sauder, U.; Mack, D.; Götz, F.; Landmann, R. Biofilm Formation Induces C3a Release and Protects Staphylococcus Epidermidis from IgG and Complement Deposition and from Neutrophil-Dependent Killing. J. Infect. Dis. 2008, 197, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- Rupp, M.E.; Ulphani, J.S.; Fey, P.D.; Bartscht, K.; Mack, D. Characterization of the Importance of Polysaccharide Intercellular Adhesin/Hemagglutinin of Staphylococcus Epidermidis in the Pathogenesis of Biomaterial-Based Infection in a Mouse Foreign Body Infection Model. Infect. Immun. 1999, 67, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Whitney, A.R.; DeLeo, F.R.; Otto, M. Polysaccharide Intercellular Adhesin (PIA) Protects Staphylococcus Epidermidis against Major Components of the Human Innate Immune System. Cell. Microbiol. 2004, 6, 269–275. [Google Scholar] [CrossRef]
- Rupp, M.E.; Ulphani, J.S.; Fey, P.D.; Mack, D. Characterization of Staphylococcus Epidermidis Polysaccharide Intercellular Adhesin/Hemagglutinin in the Pathogenesis of Intravascular Catheter-Associated Infection in a Rat Model. Infect. Immun. 1999, 67, 2656–2659. [Google Scholar] [CrossRef]
- Härtel, C.; Osthues, I.; Rupp, J.; Haase, B.; Röder, K.; Göpel, W.; Herting, E.; Schultz, C. Characterisation of the Host Inflammatory Response to Staphylococcus Epidermidis in Neonatal Whole Blood. Arch. Dis. Child. Fetal Neonatal Ed. 2008, 93, F140–F145. [Google Scholar] [CrossRef] [PubMed]
- Fredheim, E.G.A.; Granslo, H.N.; Flægstad, T.; Figenschau, Y.; Rohde, H.; Sadovskaya, I.; Mollnes, T.E.; Klingenberg, C.; Aarag Fredheim, E.G.; Granslo, H.N.; et al. Staphylococcus Epidermidis Polysaccharide Intercellular Adhesin Activates Complement. FEMS Immunol. Med. Microbiol. 2011, 63, 269–280. [Google Scholar] [CrossRef]
- Hogan, S.; Stevens, N.T.; Humphreys, H.; O’Gara, J.P.; O’Neill, E. Current and Future Approaches to the Prevention and Treatment of Staphylococcal Medical Device-Related Infections. Curr. Pharm. Des. 2014, 21, 100–113. [Google Scholar] [CrossRef]
- Zandri, G.; Pasquaroli, S.; Vignaroli, C.; Talevi, S.; Manso, E.; Donelli, G.; Biavasco, F. Detection of Viable but Non-Culturable Staphylococci in Biofilms from Central Venous Catheters Negative on Standard Microbiological Assays. Clin. Microbiol. Infect. 2012, 18, E259–E261. [Google Scholar] [CrossRef]
- Ferrer, R.; Martin-Loeches, I.; Phillips, G.; Osborn, T.M.; Townsend, S.; Dellinger, R.P.; Artigas, A.; Schorr, C.; Levy, M.M. Empiric Antibiotic Treatment Reduces Mortality in Severe Sepsis and Septic Shock from the First Hour: Results from a Guideline-Based Performance Improvement Program. Crit. Care Med. 2014, 42, 1749–1755. [Google Scholar] [CrossRef]
- Iroh Tam, P.Y.; Bendel, C.M. Diagnostics for Neonatal Sepsis: Current Approaches and Future Directions. Pediatr. Res. 2017, 82, 574–583. [Google Scholar] [CrossRef]
- Schelonka, R.L.; Chai, M.K.; Yoder, B.A.; Hensley, D.; Brockett, R.M.; Ascher, D.P. Volume of Blood Required to Detect Common Neonatal Pathogens. J. Pediatr. 1996, 129, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Bury, G.; Leroux, S.; Borrego, C.L.; Leguen, C.G.; Mitanchez, D.; Gascoin, G.; Thollot, A.; Roué, J.M.; Carrault, G.; Pladys, P.; et al. Diagnosis of Neonatal Late-Onset Infection in Very Preterm Infant: Inter-Observer Agreement and International Classifications. Int. J. Environ. Res. Public Health 2021, 18, 882. [Google Scholar] [CrossRef] [PubMed]
- Wynn, J.L.; Wong, H.R.; Shanley, T.P.; Bizzarro, M.J.; Saiman, L.; Polin, R.A. Time for a Neonatal-Specific Consensus Definition for Sepsis. Pediatr. Crit. Care Med. 2014, 15, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, S.L.; Schrag, S.J. Clinical Sepsis in Neonates and Young Infants, United States, 1988-2006. J. Pediatr. 2012, 160. [Google Scholar] [CrossRef]
- Sofouli, G.A.; Kanellopoulou, A.; Vervenioti, A.; Dimitriou, G.; Gkentzi, D. Predictive Scores for Late-Onset Neonatal Sepsis as an Early Diagnostic and Antimicrobial Stewardship Tool: What Have We Done So Far? Antibiotics 2022, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- Ohlin, A.; Björkqvist, M.; Montgomery, S.M.; Schollin, J. Clinical Signs and CRP Values Associated with Blood Culture Results in Neonates Evaluated for Suspected Sepsis. Acta Paediatr. Int. J. Paediatr. 2010, 99, 1635–1640. [Google Scholar] [CrossRef]
- Nelson, J.C.; Rizwan-uddin; Griffin, M.P.; Moorman, J.R. Probing the Order within Neonatal Heart Rate Variability. Pediatr. Res. 1998, 43, 823–831. [Google Scholar] [CrossRef]
- Griffin, M.P.; O’Shea, T.M.; Bissonette, E.A.; Harrell, F.E.; Lake, D.E.; Moorman, J.R. Abnormal Heart Rate Characteristics Preceding Neonatal Sepsis and Sepsis-like Illness. Pediatr. Res. 2003, 53, 920–926. [Google Scholar] [CrossRef]
- Healy, C.M.; Baker, C.J.; Palazzi, D.L.; Campbell, J.R.; Edwards, M.S. Distinguishing True Coagulase-Negative Staphylococcus Infections from Contaminants in the Neonatal Intensive Care Unit. J. Perinatol. 2013, 33, 52–58. [Google Scholar] [CrossRef]
- Sharma, D.; Farahbakhsh, N.; Shastri, S.; Sharma, P. Biomarkers for Diagnosis of Neonatal Sepsis: A Literature Review. J. Matern. Neonatal Med. 2018, 31, 1646–1659. [Google Scholar] [CrossRef]
- Ng, P.C.; Lam, H.S. Biomarkers for Late-Onset Neonatal Sepsis: Cytokines and Beyond. Clin. Perinatol. 2010, 37, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Celik, I.H.; Hanna, M.; Canpolat, F.E. Mohan Pammi Diagnosis of Neonatal Sepsis: The Past, Present and Future. Pediatr. Res. 2021, 91, 337–350. [Google Scholar] [CrossRef]
- Gilfillan, M.; Bhandari, V. Biomarkers for the Diagnosis of Neonatal Sepsis and Necrotizing Enterocolitis: Clinical Practice Guidelines. Early Hum. Dev. 2017, 105, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, N.; Tiwari, S.; Jain, U. Potential Biomarkers for Effective Screening of Neonatal Sepsis Infections: An Overview. Microb. Pathog. 2017, 107, 234–242. [Google Scholar] [CrossRef]
- Rewa, O.; Muscedere, J.; Reynolds, S.; Jiang, X.; Heyland, D.K. Coagulase-Negative Staphylococcus, Catheter-Related, Bloodstream Infections and Their Association with Acute Phase Markers of Inflammation in the Intensive Care Unit: An Observational Study. Can. J. Infect. Dis. Med. Microbiol. 2012, 23, 204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chiesa, C.; Natale, F.; Pascone, R.; Osborn, J.F.; Pacifico, L.; Bonci, E.; De Curtis, M. C Reactive Protein and Procalcitonin: Reference Intervals for Preterm and Term Newborns during the Early Neonatal Period. Clin. Chim. Acta 2011, 412, 1053–1059. [Google Scholar] [CrossRef]
- Dritsakou, K.; Liosis, G.; Gioni, M.; Glynou, E.; Avdeliodi, K.; Papagaroufalis, K. CRP Levels in Extremely Low Birth Weight (ELBW) Septic Infants. J. Matern. Fetal. Neonatal Med. 2015, 28, 237–239. [Google Scholar] [CrossRef]
- Mwesigye, P.; Rizwan, F.; Alassaf, N.; Khan, R. The Role and Validity of Diagnostic Biomarkers in Late-Onset Neonatal Sepsis. Cureus 2021, 13, e17065. [Google Scholar] [CrossRef]
- Brown, J.V.E.; Meader, N.; Wright, K.; Cleminson, J.; McGuire, W. Assessment of C-Reactive Protein Diagnostic Test Accuracy for Late-Onset Infection in Newborn Infants: A Systematic Review and Meta-Analysis. JAMA Pediatr. 2020, 174, 260–268. [Google Scholar] [CrossRef]
- Kurul, Ş.; Simons, S.H.P.; Ramakers, C.R.B.; De Rijke, Y.B.; Kornelisse, R.F.; Reiss, I.K.M.; Taal, H.R. Association of Inflammatory Biomarkers with Subsequent Clinical Course in Suspected Late Onset Sepsis in Preterm Neonates. Crit. Care 2021, 25, 12. [Google Scholar] [CrossRef]
- Beltempo, M.; Viel-Thériault, I.; Thibeault, R.; Julien, A.S.; Piedboeuf, B. C-Reactive Protein for Late-Onset Sepsis Diagnosis in Very Low Birth Weight Infants. BMC Pediatr. 2018, 18, 16. [Google Scholar] [CrossRef] [PubMed]
- Hedegaard, S.S.; Wisborg, K.; Hvas, A.M. Diagnostic Utility of Biomarkers for Neonatal Sepsis--a Systematic Review. Infect. Dis. 2015, 47, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, C.; Panero, A.; Rossi, N.; Stegagno, M.; De Giusti, M.; Osborn, J.F.; Pacifico, L. Reliability of Procalcitonin Concentrations for the Diagnosis of Sepsis in Critically Ill Neonates. Clin. Infect. Dis. 1998, 26, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Ruetsch, V.; Barreault, S.; Le Sache, N.; Tissères, P. Procalcitonin Is a Prognosis Biomarker in Very Preterm Neonates with Late Onset Sepsis: A Pilot Study. Eur. J. Pediatr. 2022, 181, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Prentice, S.; Jallow, A.T.; Sinjanka, E.; Jallow, M.W.; Sise, E.A.; Kessler, N.J.; Wegmuller, R.; Cerami, C.; Prentice, A.M. Hepcidin Mediates Hypoferremia and Reduces the Growth Potential of Bacteria in the Immediate Post-Natal Period in Human Neonates. Sci. Rep. 2019, 9, 16596. [Google Scholar] [CrossRef]
- Sherbiny, H.S.; Mostafa, H.A.F.; Sherief, L.M.; Kamal, N.M.; El-Shal, A.S.; Halm, M.M.A.; Khan, H.Y.; Ali, A.S.A. Validity of Serum and Urinary Hepcidin as Biomarkers for Late-Onset in Premature Infants. Ther. Adv. Chronic Dis. 2022, 13, 204062232211225. [Google Scholar] [CrossRef]
- Wu, T.W.; Tabangin, M.; Kusano, R.; Ma, Y.; Ridsdale, R.; Akinbi, H. The Utility of Serum Hepcidin as a Biomarker for Late-Onset Neonatal Sepsis. J. Pediatr. 2013, 162, 67–71. [Google Scholar] [CrossRef]
- Griffin, M.P.; Lake, D.E.; O’Shea, T.M.; Moorman, J.R. Heart Rate Characteristics and Clinical Signs in Neonatal Sepsis. Pediatr. Res. 2007, 61, 222–227. [Google Scholar] [CrossRef]
- Makkar, M.; Pathak, R.; Garg, S.; Gupta, C.; Mahajan, N. Performance Evaluation of Hematologic Scoring System in Early Diagnosis of Neonatal Sepsis. J. Clin. Neonatol. 2013, 2, 25. [Google Scholar] [CrossRef]
- Narasimha, A.; Harendra Kumar, M.L. Significance of Hematological Scoring System (HSS) in Early Diagnosis of Neonatal Sepsis. Indian J. Hematol. Blood Transfus. 2011, 27, 14–17. [Google Scholar] [CrossRef]
- Eichberger, J.; Resch, E.; Resch, B. Diagnosis of Neonatal Sepsis: The Role of Inflammatory Markers. Front. Pediatr. 2022, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Pammi, M.; Flores, A.; Versalovic, J.; Leeflang, M.M.G. Molecular Assays for the Diagnosis of Sepsis in Neonates. Cochrane database Syst. Rev. 2017, 2, CD011926. [Google Scholar] [CrossRef] [PubMed]
- Kothari, A.; Morgan, M.; Haake, D.A. Emerging Technologies for Rapid Identification of Bloodstream Pathogens. Clin. Infect. Dis. 2014, 59, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Van den Brand, M.; Peters, R.P.H.; Catsburg, A.; Rubenjan, A.; Broeke, F.J.; Van den Dungen, F.A.M.; Van Weissenbruch, M.M.; Van Furth, A.M.; Kõressaar, T.; Remm, M.; et al. Development of a Multiplex Real-Time PCR Assay for the Rapid Diagnosis of Neonatal Late Onset Sepsis. J. Microbiol. Methods 2014, 106, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Sinha, M.; Jupe, J.; Mack, H.; Coleman, T.P.; Lawrence, S.M.; Fraley, S.I. Emerging Technologies for Molecular Diagnosis of Sepsis. Clin. Microbiol. Rev. 2018, 31, e00089-17. [Google Scholar] [CrossRef]
- Straub, J.; Paula, H.; Mayr, M.; Kasper, D.; Assadian, O.; Berger, A.; Rittenschober-Böhm, J. Diagnostic Accuracy of the ROCHE Septifast PCR System for the Rapid Detection of Blood Pathogens in Neonatal Sepsis—A Prospective Clinical Trial. PLoS ONE 2017, 12, e0187688. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.Y.I.; Hernandez-Alvarado, N.; Schleiss, M.R.; Hassan-Hanga, F.; Onuchukwu, C.; Umoru, D.; Obaro, S.K. Molecular Detection of Streptococcus Pneumoniae on Dried Blood Spots from Febrile Nigerian Children Compared to Culture. PLoS ONE 2016, 11, e0152253. [Google Scholar] [CrossRef]
- Liu, C.L.; Ai, H.W.; Wang, W.P.; Chen, L.; Hu, H.B.; Ye, T.; Zhu, X.H.; Wang, F.; Liao, Y.L.; Wang, Y.; et al. Comparison of 16S RRNA Gene PCR and Blood Culture for Diagnosis of Neonatal Sepsis. Arch. Pediatr. 2014, 21, 162–169. [Google Scholar] [CrossRef]
- Reier-Nilsen, T.; Farstad, T.; Nakstad, B.; Lauvrak, V.; Steinbakk, M. Comparison of Broad Range 16S RDNA PCR and Conventional Blood Culture for Diagnosis of Sepsis in the Newborn: A Case Control Study. BMC Pediatr. 2009, 9, 5. [Google Scholar] [CrossRef]
- Jordan, J.A.; Durso, M.B.; Butchko, A.R.; Jones, J.G.; Brozanski, B.S. Evaluating the Near-Term Infant for Early Onset Sepsis: Progress and Challenges to Consider with 16S RDNA Polymerase Chain Reaction Testing. J. Mol. Diagn. 2006, 8, 357–363. [Google Scholar] [CrossRef]
- Mancini, N.; Carletti, S.; Ghidoli, N.; Cichero, P.; Burioni, R.; Clementi, M. The Era of Molecular and Other Non-Culture-Based Methods in Diagnosis of Sepsis. Clin. Microbiol. Rev. 2010, 23, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Luethy, P.M.; Johnson, J.K. The Use of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) for the Identification of Pathogens Causing Sepsis. J. Appl. Lab. Med. 2019, 3, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.; Kostrzewa, M. MALDI-TOF MS in the Microbiology Laboratory: Current Trends. Curr. Issues Mol. Biol. 2017, 23, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Tschudin-Sutter, S.; Frei, R.; Dangel, M.; Jakob, M.; Balmelli, C.; Schaefer, D.J.; Weisser, M.; Elzi, L.; Battegay, M.; Widmer, A.F. Validation of a Treatment Algorithm for Orthopaedic Implant-Related Infections with Device-Retention-Results from a Prospective Observational Cohort Study. Clin. Microbiol. Infect. 2016, 22, 457.e1–457.e9. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.Y.; Chiang-Ni, C.; Teng, S.H. Current Status of MALDI-TOF Mass Spectrometry in Clinical Microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.; Sánchez-Juanes, F.; Muñoz-Bellido, J.L.; González-Buitrago, J.M. Rapid Method for Direct Identification of Bacteria in Urine and Blood Culture Samples by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry: Intact Cell vs. Extraction Method. Clin. Microbiol. Infect. 2011, 17, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Prod’Hom, G.; Bizzini, A.; Durussel, C.; Bille, J.; Greub, G. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Direct Bacterial Identification from Positive Blood Culture Pellets. J. Clin. Microbiol. 2010, 48, 1481–1483. [Google Scholar] [CrossRef]
- Verroken, A.; Defourny, L.; Le Polain De Waroux, O.; Belkhir, L.; Laterre, P.F.; Delmée, M.; Glupczynski, Y. Clinical Impact of MALDI-TOF MS Identification and Rapid Susceptibility Testing on Adequate Antimicrobial Treatment in Sepsis with Positive Blood Cultures. PLoS ONE 2016, 11, e0156299. [Google Scholar] [CrossRef]
- Hrabák, J.; Chudác ková, E.; Walková, R. Matrix-Assisted Laser Desorption Ionization-Time of Flight (Maldi-Tof) Mass Spectrometry for Detection of Antibiotic Resistance Mechanisms: From Research to Routine Diagnosis. Clin. Microbiol. Rev. 2013, 26, 103–114. [Google Scholar] [CrossRef]
- Ruiz-Aragón, J.; Ballestero-Téllez, M.; Gutiérrez-Gutiérrez, B.; de Cueto, M.; Rodríguez-Baño, J.; Pascual, Á. Direct Bacterial Identification from Positive Blood Cultures Using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) Mass Spectrometry: A Systematic Review and Meta-Analysis. Enfermedades Infecc. y Microbiol. Clin. (Engl. ed.) 2018, 36, 484–492. [Google Scholar] [CrossRef]
- Hamilton, F.; Evans, R.; MacGowan, A. The Value of MALDI-TOF Failure to Provide an Identification of Staphylococcal Species Direct from Blood Cultures and Rule out Staphylococcus Aureus Bacteraemia: A Post-Hoc Analysis of the RAPIDO Trial. Access Microbiol. 2020, 3, 000192. [Google Scholar] [CrossRef] [PubMed]
- Fink, T.; Wolf, A.; Maurer, F.; Albrecht, F.W.; Heim, N.; Wolf, B.; Hauschild, A.C.; Bödeker, B.; Baumbach, J.I.; Volk, T.; et al. Volatile Organic Compounds during Inflammation and Sepsis in Rats: A Potential Breath Test Using Ion-Mobility Spectrometry. Anesthesiology 2015, 122, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Guamán, A.V.; Carreras, A.; Calvo, D.; Agudo, I.; Navajas, D.; Pardo, A.; Marco, S.; Farré, R. Rapid Detection of Sepsis in Rats through Volatile Organic Compounds in Breath. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2012, 881–882, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Oeschger, T.; McCloskey, D.; Kopparthy, V.; Singh, A.; Erickson, D. Point of Care Technologies for Sepsis Diagnosis and Treatment. Lab Chip 2019, 19, 728–737. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Strunk, T.; Lee, A.H.; Gill, E.E.; Falsafi, R.; Woodman, T.; Hibbert, J.; Hancock, R.E.W.; Currie, A. Whole Blood Transcriptional Responses of Very Preterm Infants during Late-Onset Sepsis. PLoS ONE 2020, 15, e0233841. [Google Scholar] [CrossRef]
- Cernada, M.; Pinilla-González, A.; Kuligowski, J.; Morales, J.M.; Lorente-Pozo, S.; Piñeiro-Ramos, J.D.; Parra-Llorca, A.; Lara-Cantón, I.; Vento, M.; Serna, E. Transcriptome Profiles Discriminate between Gram-Positive and Gram-Negative Sepsis in Preterm Neonates. Pediatr. Res. 2022, 91, 637–645. [Google Scholar] [CrossRef]
- Raymond, S.L.; López, M.C.; Baker, H.V.; Larson, S.D.; Efron, P.A.; Sweeney, T.E.; Khatri, P.; Moldawer, L.L.; Wynn, J.L. Unique Transcriptomic Response to Sepsis Is Observed among Patients of Different Age Groups. PLoS ONE 2017, 12, e0184159. [Google Scholar] [CrossRef]
- Bai, Y.; Zhao, N.; Zhang, Z.; Jia, Y.; Zhang, G.; Dong, G. Identification and Validation of a Novel Four-Gene Diagnostic Model for Neonatal Early-Onset Sepsis with Bacterial Infection. Eur. J. Pediatr. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Miao, H.; Chen, S.; Ding, R. Evaluation of the Molecular Mechanisms of Sepsis Using Proteomics. Front. Immunol. 2021, 12, 733537. [Google Scholar] [CrossRef]
- Stewart, C.J.; Nelson, A.; Treumann, A.; Skeath, T.; Cummings, S.P.; Embleton, N.D.; Berrington, J.E. Metabolomic and Proteomic Analysis of Serum from Preterm Infants with Necrotising Entercolitis and Late-Onset Sepsis. Pediatr. Res. 2016, 79, 425–431. [Google Scholar] [CrossRef]
- Pilar-Orive, F.J.; Astigarraga, I.; Azkargorta, M.; Elortza, F.; Garcia-Obregon, S. A Three-Protein Panel to Support the Diagnosis of Sepsis in Children. J. Clin. Med. 2022, 11, 1563. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Obregon, S.; Azkargorta, M.; Seijas, I.; Pilar-Orive, J.; Borrego, F.; Elortza, F.; Boyano, M.D.; Astigarraga, I. Identification of a Panel of Serum Protein Markers in Early Stage of Sepsis and Its Validation in a Cohort of Patients. J. Microbiol. Immunol. Infect. 2018, 51, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, K.R.; Hummon, A.B. Mass Spectrometry for the Discovery of Biomarkers of Sepsis. Mol. Biosyst. 2017, 13, 648–664. [Google Scholar] [CrossRef] [PubMed]
- Bjerkhaug, A.U.; Granslo, H.N.; Klingenberg, C. Metabolic Responses in Neonatal Sepsis-A Systematic Review of Human Metabolomic Studies. Acta Paediatr. 2021, 110, 2316–2325. [Google Scholar] [CrossRef]
- Tong, P.; Huang, F.R.; Xu, J.; Wu, Z.Q.; Hu, X.; Ling, M.; Wang, D.; Wu, B.F.; Yang, D.J.; Zhang, A.M. Metabolomic Changes of Neonatal Sepsis: An Exploratory Clinical Study. Zhongguo Dang Dai Er Ke Za Zhi 2022, 24, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Mardegan, V.; Giordano, G.; Stocchero, M.; Pirillo, P.; Poloniato, G.; Donadel, E.; Salvadori, S.; Giaquinto, C.; Priante, E.; Baraldi, E. Untargeted and Targeted Metabolomic Profiling of Preterm Newborns with EarlyOnset Sepsis: A Case-Control Study. Metabolites 2021, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.; El-Manzalawy, Y. Machine Learning Based Refined Differential Gene Expression Analysis of Pediatric Sepsis. BMC Med. Genomics 2020, 13. [Google Scholar] [CrossRef]
- Wong, H.R.; Caldwell, J.T.; Cvijanovich, N.Z.; Weiss, S.L.; Fitzgerald, J.C.; Bigham, M.T.; Jain, P.N.; Schwarz, A.; Lutfi, R.; Nowak, J.; et al. Prospective Clinical Testing and Experimental Validation of the Pediatric Sepsis Biomarker Risk Model. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Mangioni, D.; Peri, A.M.; Rossolini, G.M.; Viaggi, B.; Perno, C.F.; Gori, A.; Bandera, A. Toward Rapid Sepsis Diagnosis and Patient Stratification: What’s New From Microbiology and Omics Science. J. Infect. Dis. 2020, 221, 1039–1047. [Google Scholar] [CrossRef]
- Lu, H.; Wen, D.; Wang, X.; Gan, L.; Du, J.; Sun, J.; Zeng, L.; Jiang, J.; Zhang, A. Host Genetic Variants in Sepsis Risk: A Field Synopsis and Meta-Analysis. Crit. Care 2019, 23, 26. [Google Scholar] [CrossRef]
- Wong, H.R.; Reeder, R.W.; Banks, R.; Berg, R.A.; Meert, K.L.; Hall, M.W.; McQuillen, P.S.; Mourani, P.M.; Chima, R.S.; Sorenson, S.; et al. Biomarkers for Estimating Risk of Hospital Mortality and Long-Term Quality-of-Life Morbidity After Surviving Pediatric Septic Shock: A Secondary Analysis of the Life After Pediatric Sepsis Evaluation Investigation. Pediatr. Crit. Care Med. 2021, 22, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Fjalstad, J.W.; Esaiassen, E.; Juvet, L.K.; van den Anker, J.N.; Klingenberg, C. Antibiotic Therapy in Neonates and Impact on Gut Microbiota and Antibiotic Resistance Development: A Systematic Review. J. Antimicrob. Chemother. 2018, 73, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Esaiassen, E.; Fjalstad, J.W.; Juvet, L.K.; van den Anker, J.N.; Klingenberg, C. Antibiotic Exposure in Neonates and Early Adverse Outcomes: A Systematic Review and Meta-Analysis. J. Antimicrob. Chemother. 2017, 72, 1858–1870. [Google Scholar] [CrossRef] [PubMed]
- Kronman, M.P.; Zaoutis, T.E.; Haynes, K.; Feng, R.; Coffin, S.E. Antibiotic Exposure and IBD Development among Children: A Population-Based Cohort Study. Pediatrics 2012, 130. [Google Scholar] [CrossRef]
- Bailey, L.C.; Forrest, C.B.; Zhang, P.; Richards, T.M.; Livshits, A.; DeRusso, P.A. Association of Antibiotics in Infancy with Early Childhood Obesity. JAMA Pediatr. 2014, 168, 1063–1069. [Google Scholar] [CrossRef]
- Ferrer-Espada, R.; Liu, X.; Goh, X.S.; Dai, T. Antimicrobial Blue Light Inactivation of Polymicrobial Biofilms. Front. Microbiol. 2019, 10, 721. [Google Scholar] [CrossRef]
- Halstead, F.D.; Thwaite, J.E.; Burt, R.; Laws, T.R.; Raguse, M.; Moeller, R.; Webber, M.A.; Oppenheim, B.A. Antibacterial Activity of Blue Light against Nosocomial Wound Pathogens Growing Planktonically and as Mature Biofilms. Appl. Environ. Microbiol. 2016, 82, 4006–4016. [Google Scholar] [CrossRef]
- Hu, D.; Li, H.; Wang, B.; Ye, Z.; Lei, W.; Jia, F.; Jin, Q.; Ren, K.F.; Ji, J. Surface-Adaptive Gold Nanoparticles with Effective Adherence and Enhanced Photothermal Ablation of Methicillin-Resistant Staphylococcus Aureus Biofilm. ACS Nano 2017, 11, 9330–9339. [Google Scholar] [CrossRef]
- Kim, W.; Zou, G.; Hari, T.P.A.; Wilt, I.K.; Zhu, W.; Galle, N.; Faizi, H.A.; Hendricks, G.L.; Tori, K.; Pan, W.; et al. A Selective Membrane-Targeting Repurposed Antibiotic with Activity against Persistent Methicillin-Resistant Staphylococcus Aureus. Proc. Natl. Acad. Sci. USA 2019, 116, 16529–16534. [Google Scholar] [CrossRef]
- Kim, W.; Zhu, W.; Hendricks, G.L.; Van Tyne, D.; Steele, A.D.; Keohane, C.E.; Fricke, N.; Conery, A.L.; Shen, S.; Pan, W.; et al. A New Class of Synthetic Retinoid Antibiotics Effective against Bacterial Persisters. Nature 2018, 556, 103–107. [Google Scholar] [CrossRef]
- Sun, F.; Bian, M.; Li, Z.; Lv, B.; Gao, Y.; Wang, Y.; Fu, X. 5-Methylindole Potentiates Aminoglycoside Against Gram-Positive Bacteria Including Staphylococcus Aureus Persisters Under Hypoionic Conditions. Front. Cell. Infect. Microbiol. 2020, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Grassi, L.; Maisetta, G.; Esin, S.; Batoni, G. Combination Strategies to Enhance the Efficacy of Antimicrobial Peptides against Bacterial Biofilms. Front. Microbiol. 2017, 8, 2409. [Google Scholar] [CrossRef] [PubMed]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic Action of Phage and Antibiotics: Parameters to Enhance the Killing Efficacy Against Mono and Dual-Species Biofilms. Antibiotics 2019, 8, 103. [Google Scholar] [CrossRef]
- Bi, Y.; Xia, G.; Shi, C.; Wan, J.; Liu, L.; Chen, Y.; Wu, Y.; Zhang, W.; Zhou, M.; He, H.; et al. Therapeutic Strategies against Bacterial Biofilms. Fundam. Res. 2021, 1, 193–212. [Google Scholar] [CrossRef]
- Bakhuizen, S.E.; De Haan, T.R.; Teune, M.J.; Van Wassenaer-Leemhuis, A.G.; Van Der Heyden, J.L.; Van Der Ham, D.P.; Mol, B.W.J. Meta-Analysis Shows That Infants Who Have Suffered Neonatal Sepsis Face an Increased Risk of Mortality and Severe Complications. Acta Paediatr. 2014, 103, 1211–1218. [Google Scholar] [CrossRef]
- Clark, R.H.; Bloom, B.T.; Spitzer, A.R.; Gerstmann, D.R. Empiric Use of Ampicillin and Cefotaxime, Compared with Ampicillin and Gentamicin, for Neonates at Risk for Sepsis Is Associated with an Increased Risk of Neonatal Death. Pediatrics 2006, 117, 67–74. [Google Scholar] [CrossRef]
- Tzialla, C.; Borghesi, A.; Serra, G.; Stronati, M.; Corsello, G. Antimicrobial Therapy in Neonatal Intensive Care Unit. Ital. J. Pediatr. 2015, 41, 27. [Google Scholar] [CrossRef]
- Cantey, J.B.; Wozniak, P.S.; Sánchez, P.J. Prospective Surveillance of Antibiotic Use in the Neonatal Intensive Care Unit: Results from the SCOUT Study. Pediatr. Infect. Dis. J. 2015, 34, 267–272. [Google Scholar] [CrossRef]
- Rubin, L.G.; Sánchez, P.J.; Siegel, J.; Levine, G.; Saiman, L.; Jarvis, W.R. Evaluation and Treatment of Neonates with Suspected Late-Onset Sepsis: A Survey of Neonatologists’ Practices. Pediatrics 2002, 110, e42. [Google Scholar] [CrossRef]
- Bizzarro, M.J.; Shabanova, V.; Baltimore, R.S.; Dembry, L.M.; Ehrenkranz, R.A.; Gallagher, P.G. Neonatal Sepsis 2004-2013: The Rise and Fall of Coagulase-Negative Staphylococci. J. Pediatr. 2015, 166, 1193–1199. [Google Scholar] [CrossRef]
- Korang, S.K.; Safi, S.; Nava, C.; Greisen, G.; Gupta, M.; Lausten-Thomsen, U.; Jakobsen, J.C. Antibiotic Regimens for Late-Onset Neonatal Sepsis. Cochrane database Syst. Rev. 2021, 5, CD013836. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.X.; Wang, E.H.; Liu, Y.; Luo, E.J. Antibiotic Susceptibility of Coagulase-Negative Staphylococci (CoNS): Emergence of Teicoplanin-Non-Susceptible CoNS Strains with Inducible Resistance to Vancomycin. J. Med. Microbiol. 2011, 60, 1661–1668. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Daley, A.J.; Istivan, T.S.; Garland, S.M.; Deighton, M.A. Antibiotic Susceptibility of Coagulase-Negative Staphylococci Isolated from Very Low Birth Weight Babies: Comprehensive Comparisons of Bacteria at Different Stages of Biofilm Formation. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Lopes, N.; Freitas, A.I.; Ramos, H.; Vasconcelos, C.S. Epidermidis Isolates from a Tertiary Care Portuguese Hospital Show Very High Antibiotic Non-Susceptible Rates and Significant Ability to Form Biofilms. Appl. Microbiol. 2021, 1, 150–161. [Google Scholar] [CrossRef]
- Mehta, G.; Kumari, S. Multi-Resistant Staphylococcus Haemolyticus in a Neonatal Unit in New Delhi. Ann. Trop. Paediatr. 1997, 17, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Westberg, R.; Stegger, M.; Söderquist, B. Molecular Epidemiology of Neonatal-Associated Staphylococcus Haemolyticus Reveals Endemic Outbreak. Microbiol. Spectr. 2022, 10, e0245222. [Google Scholar] [CrossRef]
- Klingenberg, C.; Rønnestad, A.; Anderson, A.S.; Abrahamsen, T.G.; Zorman, J.; Villaruz, A.; Flægstad, T.; Otto, M.; Sollid, E.J. Persistent Strains of Coagulase-Negative Staphylococci in a Neonatal Intensive Care Unit: Virulence Factors and Invasiveness. Clin. Microbiol. Infect. 2007, 13, 1100–1111. [Google Scholar] [CrossRef]
- Van Der Zwet, W.C.; Debets-Ossenkopp, Y.J.; Reinders, E.; Kapi, M.; Savelkoul, P.H.M.; Van Elburg, R.M.; Hiramatsu, K.; Vandenbroucke-Grauls, C.M.J.E. Nosocomial Spread of a Staphylococcus Capitis Strain with Heteroresistance to Vancomycin in a Neonatal Intensive Care Unit. J. Clin. Microbiol. 2002, 40, 2520–2525. [Google Scholar] [CrossRef]
- Ng, P.C.; Chow, V.C.Y.; Lee, C.H.; Ling, J.M.L.; Wong, H.L.; Chan, R.C.Y. Persistent Staphylococcus Capitis Septicemia in a Preterm Infant. Pediatr. Infect. Dis. J. 2006, 25, 652–654. [Google Scholar] [CrossRef]
- Laurent, F.; Butin, M. Staphylococcus Capitis and NRCS-A Clone: The Story of an Unrecognized Pathogen in Neonatal Intensive Care Units. Clin. Microbiol. Infect. 2019, 25, 1081–1085. [Google Scholar] [CrossRef]
- Asfour, S.S.; Asfour, R.S.; Khalil, T.M.; Al-Mouqdad, M.M. The Use of Daptomycin in the Treatment of Persistent Coagulase-Negative Staphylococcal Sepsis in Premature Infants: A Case Series. J. Pediatr. Pharmacol. Ther. JPPT 2021, 26, 92. [Google Scholar] [CrossRef]
- Principi, N.; Caironi, M.; Venturini, F.; Pani, L.; Esposito, S. Daptomycin in Paediatrics: Current Knowledge and the Need for Future Research. J. Antimicrob. Chemother. 2015, 70, 643–648. [Google Scholar] [CrossRef]
- Karageorgos, S.A.; Miligkos, M.; Dakoutrou, M.; Tsioutis, C. Clinical Effectiveness, Safety Profile, and Pharmacokinetics of Daptomycin in Pediatric Patients: A Systematic Review. J. Pediatric Infect. Dis. Soc. 2016, 5, 446–457. [Google Scholar] [CrossRef]
- Kocher, S.; Müller, W.; Resch, B. Linezolid Treatment of Nosocomial Bacterial Infection with Multiresistant Gram-Positive Pathogens in Preterm Infants: A Systematic Review. Int. J. Antimicrob. Agents 2010, 36, 106–110. [Google Scholar] [CrossRef]
- Butin, M.; Martins-Simões, P.; Picaud, J.C.; Kearns, A.; Claris, O.; Vandenesch, F.; Laurent, F.; Rasigade, J.P. Adaptation to Vancomycin Pressure of Multiresistant Staphylococcus Capitis NRCS-A Involved in Neonatal Sepsis. J. Antimicrob. Chemother. 2015, 70, 3027–3031. [Google Scholar] [CrossRef]
- Deville, J.G.; Adler, S.; Azimi, P.H.; Jantausch, B.A.; Morfin, M.R.; Beltran, S.; Edge-Padbury, B.; Naberhuis-Stehouwer, S.; Bruss, J.B. Linezolid versus Vancomycin in the Treatment of Known or Suspected Resistant Gram-Positive Infections in Neonates. Pediatr. Infect. Dis. J. 2003, 22, S158–S163. [Google Scholar] [CrossRef]
- Trampuz, A.; Zimmerli, W. Antimicrobial Agents in Orthopaedic Surgery: Prophylaxis and Treatment. Drugs 2006, 66, 1089–1106. [Google Scholar] [CrossRef]
- Monzón, M.; Oteiza, C.; Leiva, J.; Lamata, M.; Amorena, B. Biofilm Testing of Staphylococcus Epidermidis Clinical Isolates: Low Performance of Vancomycin in Relation to Other Antibiotics. Diagn. Microbiol. Infect. Dis. 2002, 44, 319–324. [Google Scholar] [CrossRef]
- Lee, J.Y.H.; Monk, I.R.; Gonçalves da Silva, A.; Seemann, T.; Chua, K.Y.L.; Kearns, A.; Hill, R.; Woodford, N.; Bartels, M.D.; Strommenger, B.; et al. Global Spread of Three Multidrug-Resistant Lineages of Staphylococcus Epidermidis. Nat. Microbiol. 2018, 3, 1175–1185. [Google Scholar] [CrossRef]
- Tan, T.Q.; Mason, E.O.; Ou, C.N.; Kaplan, S.L. Use of Intravenous Rifampin in Neonates with Persistent Staphylococcal Bacteremia. Antimicrob. Agents Chemother. 1993, 37, 2401. [Google Scholar] [CrossRef]
- van der Lugt, N.M.; Steggerda, S.J.; Walther, F.J. Use of Rifampin in Persistent Coagulase Negative Staphylococcal Bacteremia in Neonates. BMC Pediatr. 2010, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shama, A.; Patole, S.K.; Whitehall, J.S. Intravenous Rifampicin in Neonates with Persistent Staphylococcal Bacteraemia. Acta Paediatr. 2002, 91, 670–673. [Google Scholar] [CrossRef] [PubMed]
- Dall, G.F.; Tsang, S.T.J.; Gwynne, P.J.; MacKenzie, S.P.; Simpson, A.H.R.W.; Breusch, S.J.; Gallagher, M.P. Unexpected Synergistic and Antagonistic Antibiotic Activity against Staphylococcus Biofilms. J. Antimicrob. Chemother. 2018, 73, 1830–1840. [Google Scholar] [CrossRef] [PubMed]
- Rybak, M.J.; Lomaestro, B.M.; Rotschafer, J.C.; Moellering, R.C.; Craig, W.A.; Billeter, M.; Dalovisio, J.R.; Levine, D.P. Therapeutic Monitoring of Vancomycin in Adults Summary of Consensus Recommendations from the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, and the Society of Infectious Diseases Pharmacists. Pharmacotherapy 2009, 29, 1275–1279. [Google Scholar] [CrossRef]
- Hailemeskel, B.; Namanny, M.; Wutoh, A. Frequency of Nephrotoxicity with Vancomycin and Aminoglycoside Antibiotic Therapy. Hosp. Pharm. 1999, 34, 1417–1420. [Google Scholar] [CrossRef]
- Sorrell, T.C.; Collignon, P.J. A Prospective Study of Adverse Reactions Associated with Vancomycin Therapy. J. Antimicrob. Chemother. 1985, 16, 235–241. [Google Scholar] [CrossRef]
- Ozkaya-Parlakay, A.; Kara, A.; Celik, M.; Ozsurekci, Y.; Oncel, E.K.; Ceyhan, M.; Cengiz, A.B. Early Lactic Acidosis Associated with Linezolid Therapy in Paediatric Patients. Int. J. Antimicrob. Agents 2014, 44, 334–336. [Google Scholar] [CrossRef]
- Alsayyed, B. Rifampin. Pediatr. Rev. 2004, 25, 216–217. [Google Scholar] [CrossRef]
- Arnold, C.J.; Ericson, J.; Kohman, J.; Corey, K.L.; Oh, M.; Onabanjo, J.; Hornik, C.P.; Clark, R.H.; Benjamin, D.K.; Smith, P.B.; et al. Rifampin Use and Safety in Hospitalized Infants. Am. J. Perinatol. 2015, 32, 565–570. [Google Scholar] [CrossRef]
- Brian Smith, P.; Michael Cotten, C.; Hudak, M.L.; Sullivan, J.E.; Poindexter, B.B.; Cohen-Wolkowiez, M.; Boakye-Agyeman, F.; Lewandowski, A.; Anand, R.; Benjamin, D.K.; et al. Rifampin Pharmacokinetics and Safety in Preterm and Term Infants. Antimicrob. Agents Chemother. 2019, 63, e00284-19. [Google Scholar] [CrossRef]
- Filioti, J.; Spiroglou, K.; Roilides, E. Invasive Candidiasis in Pediatric Intensive Care Patients: Epidemiology, Risk Factors, Management, and Outcome. Intensive Care Med. 2007, 33, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Spiliopoulou, A.; Dimitriou, G.; Jelastopulu, E.; Giannakopoulos, I.; Anastassiou, E.D.; Christofidou, M. Neonatal Intensive Care Unit Candidemia: Epidemiology, Risk Factors, Outcome, and Critical Review of Published Case Series. Mycopathologia 2012, 173, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Antonoplis, A.; Zang, X.; Huttner, M.A.; Chong, K.K.L.; Lee, Y.B.; Co, J.Y.; Amieva, M.R.; Kline, K.A.; Wender, P.A.; Cegelski, L. A Dual-Function Antibiotic-Transporter Conjugate Exhibits Superior Activity in Sterilizing MRSA Biofilms and Killing Persister Cells. J. Am. Chem. Soc. 2018, 140, 16140–16151. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.W.; Deshayes, S.; Hawker, S.; Blacker, A.; Kasko, A.M.; Wong, G.C.L. Engineering Persister-Specific Antibiotics with Synergistic Antimicrobial Functions. Deshayes Contrib. Equal. 2014, 8, 8786–8793. [Google Scholar] [CrossRef]
- Joo, H.S.; Otto, M. Mechanisms of Resistance to Antimicrobial Peptides in Staphylococci. Biochim. Biophys. Acta 2015, 1848, 3055–3061. [Google Scholar] [CrossRef]
- Peschel, A.; Sahl, H.G. The Co-Evolution of Host Cationic Antimicrobial Peptides and Microbial Resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef]
- Zhu, C.; Zhao, Y.; Zhao, X.; Liu, S.; Xia, X.; Zhang, S.; Wang, Y.; Zhang, H.; Xu, Y.; Chen, S.; et al. The Antimicrobial Peptide MPX Can Kill Staphylococcus Aureus, Reduce Biofilm Formation, and Effectively Treat Bacterial Skin Infections in Mice. Front. Vet. Sci. 2022, 9, 327. [Google Scholar] [CrossRef]
- Fleming, D.; Rumbaugh, K. The Consequences of Biofilm Dispersal on the Host. Sci. Rep. 2018, 8, 10738. [Google Scholar] [CrossRef]
- Donelli, G.; Francolini, I.; Romoli, D.; Guaglianone, E.; Piozzi, A.; Ragunath, C.; Kaplan, J.B. Synergistic Activity of Dispersin B and Cefamandole Nafate in Inhibition of Staphylococcal Biofilm Growth on Polyurethanes. Antimicrob. Agents Chemother. 2007, 51, 2733–2740. [Google Scholar] [CrossRef]
- França, A.; Pérez-Cabezas, B.; Correia, A.; Pier, G.B.; Cerca, N.; Vilanova, M. Staphylococcus Epidermidis Biofilm-Released Cells Induce a Prompt and More Marked in Vivo Inflammatory-Type Response than Planktonic or Biofilm Cells. Front. Microbiol. 2016, 7, 1530. [Google Scholar] [CrossRef]
- Izano, E.A.; Amarante, M.A.; Kher, W.B.; Kaplan, J.B. Differential Roles of Poly-N-Acetylglucosamine Surface Polysaccharide and Extracellular DNA in Staphylococcus Aureus and Staphylococcus Epidermidis Biofilms. Appl. Environ. Microbiol. 2008, 74, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Chaignon, P.; Sadovskaya, I.; Ragunah, C.; Ramasubbu, N.; Kaplan, J.B.; Jabbouri, S. Susceptibility of Staphylococcal Biofilms to Enzymatic Treatments Depends on Their Chemical Composition. Appl. Microbiol. Biotechnol. 2007, 75, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Ramasubbu, N.; Thomas, L.M.; Ragunath, C.; Kaplan, J.B. Structural Analysis of Dispersin B, a Biofilm-Releasing Glycoside Hydrolase from the Periodontopathogen Actinobacillus Actinomycetemcomitans. J. Mol. Biol. 2005, 349, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.A.; Kusuma, C.; Mond, J.J.; Kokai-Kun, J.F. Lysostaphin Disrupts Staphylococcus Aureus and Staphylococcus Epidermidis Biofilms on Artificial Surfaces. Antimicrob. Agents Chemother. 2003, 47, 3407–3414. [Google Scholar] [CrossRef] [PubMed]
- Sieprawska-Lupa, M.; Mydel, P.; Krawczyk, K.; Wójcik, K.; Puklo, M.; Lupa, B.; Suder, P.; Silberring, J.; Reed, M.; Pohl, J.; et al. Degradation of Human Antimicrobial Peptide LL-37 by Staphylococcus Aureus-Derived Proteinases. Antimicrob. Agents Chemother. 2004, 48, 4673–4679. [Google Scholar] [CrossRef]
- Prokešová, L.; Potuẑníková, B.; Potempa, J.; Zikán, J.; Radl, J.; Hachová, L.; Baran, K.; Porwit-Bbr, Z.; John, C. Cleavage of Human Immunoglobulins by Serine Proteinase from Staphylococcus Aureus. Immunol. Lett. 1992, 31, 259–265. [Google Scholar] [CrossRef]
- Pires, D.P.; Melo, L.D.R.; Vilas Boas, D.; Sillankorva, S.; Azeredo, J. Phage Therapy as an Alternative or Complementary Strategy to Prevent and Control Biofilm-Related Infections. Curr. Opin. Microbiol. 2017, 39, 48–56. [Google Scholar] [CrossRef]
- Melo, L.D.R.; Oliveira, H.; Pires, D.P.; Dabrowska, K.; Azeredo, J. Phage Therapy Efficacy: A Review of the Last 10 Years of Preclinical Studies. Crit. Rev. Microbiol. 2020, 46, 78–99. [Google Scholar] [CrossRef]
- Rohde, C.; Wittmann, J.; Kutter, E. Bacteriophages: A Therapy Concept against Multi-Drug-Resistant Bacteria. Surg. Infect. 2018, 19, 737–744. [Google Scholar] [CrossRef]
- González, S.; Fernández, L.; Campelo, A.B.; Gutiérrez, D.; Martínez, B.; Rodríguez, A.; García, P. The Behavior of Staphylococcus Aureus Dual-Species Biofilms Treated with Bacteriophage PhiIPLA-RODI Depends on the Accompanying Microorganism. Appl. Environ. Microbiol. 2017, 83, e02821-16. [Google Scholar] [CrossRef]
- Cerca, N.; Oliveira, R.; Azeredo, J. Susceptibility of Staphylococcus Epidermidis Planktonic Cells and Biofilms to the Lytic Action of Staphylococcus Bacteriophage K. Lett. Appl. Microbiol. 2007, 45, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.R.; Gaudion, A.; Bean, J.E.; Perez Esteban, P.; Arnot, T.C.; Harper, D.R.; Kot, W.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. Combined Use of Bacteriophage K and a Novel Bacteriophage to Reduce Staphylococcus Aureus Biofilm Formation. Appl. Environ. Microbiol. 2014, 80, 6694–6703. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.R.; Pinto, G.; Oliveira, F.; Vilas-Boas, D.; Almeida, C.; Sillankorva, S.; Cerca, N.; Azeredo, J. The Protective Effect of Staphylococcus Epidermidis Biofilm Matrix against Phage Predation. Viruses 2020, 12, 1076. [Google Scholar] [CrossRef]
- Melo, L.D.R.; França, A.; Brandão, A.; Sillankorva, S.; Cerca, N.; Azeredo, J. Assessment of Sep1virus Interaction with Stationary Cultures by Transcriptional and Flow Cytometry Studies. FEMS Microbiol. Ecol. 2018, 94, fiy143. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Vandenheuvel, D.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Two Phages, PhiIPLA-RODI and PhiIPLA-C1C, Lyse Mono- and Dual-Species Staphylococcal Biofilms. Appl. Environ. Microbiol. 2015, 81, 3336–3348. [Google Scholar] [CrossRef]
- Fernández, L.; González, S.; Campelo, A.B.; Martínez, B.; Rodríguez, A.; García, P. Low-Level Predation by Lytic Phage PhiIPLA-RODI Promotes Biofilm Formation and Triggers the Stringent Response in Staphylococcus Aureus. Sci. Rep. 2017, 7, 40965. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Tufenkji, N.; van de Ven, T.G.M. Formation of Biofilms under Phage Predation: Considerations Concerning a Biofilm Increase. Biofouling 2013, 29, 457–468. [Google Scholar] [CrossRef]
- García, P.; Martínez, B.; Rodríguez, L.; Rodríguez, A. Synergy between the Phage Endolysin LysH5 and Nisin to Kill Staphylococcus Aureus in Pasteurized Milk. Int. J. Food Microbiol. 2010, 141, 151–155. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Ruas-Madiedo, P.; Martínez, B.; Rodríguez, A.; García, P. Effective Removal of Staphylococcal Biofilms by the Endolysin LysH5. PLoS ONE 2014, 9, e107307. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, M.; Zhang, H.; Dai, J.; Guo, Z.; Li, X.; Ji, Y.; Cai, R.; Xi, H.; Wang, X.; et al. Antibacterial Effects of Phage Lysin LysGH15 on Planktonic Cells and Biofilms of Diverse Staphylococci. Appl. Environ. Microbiol. 2018, 84, e00886-18. [Google Scholar] [CrossRef]
- Schuch, R.; Khan, B.K.; Raz, A.; Rotolo, J.A.; Wittekind, M. Bacteriophage Lysin CF-301, a Potent Antistaphylococcal Biofilm Agent. Antimicrob. Agents Chemother. 2017, 61, e02666-16. [Google Scholar] [CrossRef] [PubMed]
- Dickey, J.; Perrot, V. Adjunct Phage Treatment Enhances the Effectiveness of Low Antibiotic Concentration against Staphylococcus Aureus Biofilms in Vitro. PLoS ONE 2019, 14, e0209390. [Google Scholar] [CrossRef] [PubMed]
- Tkhilaishvili, T.; Lombardi, L.; Klatt, A.B.; Trampuz, A.; Di Luca, M. Bacteriophage Sb-1 Enhances Antibiotic Activity against Biofilm, Degrades Exopolysaccharide Matrix and Targets Persisters of Staphylococcus Aureus. Int. J. Antimicrob. Agents 2018, 52, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Cobb, L.H.; Park, J.Y.; Swanson, E.A.; Beard, M.C.; McCabe, E.M.; Rourke, A.S.; Seo, K.S.; Olivier, A.K.; Priddy, L.B. CRISPR-Cas9 Modified Bacteriophage for Treatment of Staphylococcus Aureus Induced Osteomyelitis and Soft Tissue Infection. PLoS ONE 2019, 14, e0220421. [Google Scholar] [CrossRef]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable Repression and Activation of Bacterial Gene Expression Using an Engineered CRISPR-Cas System. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef]
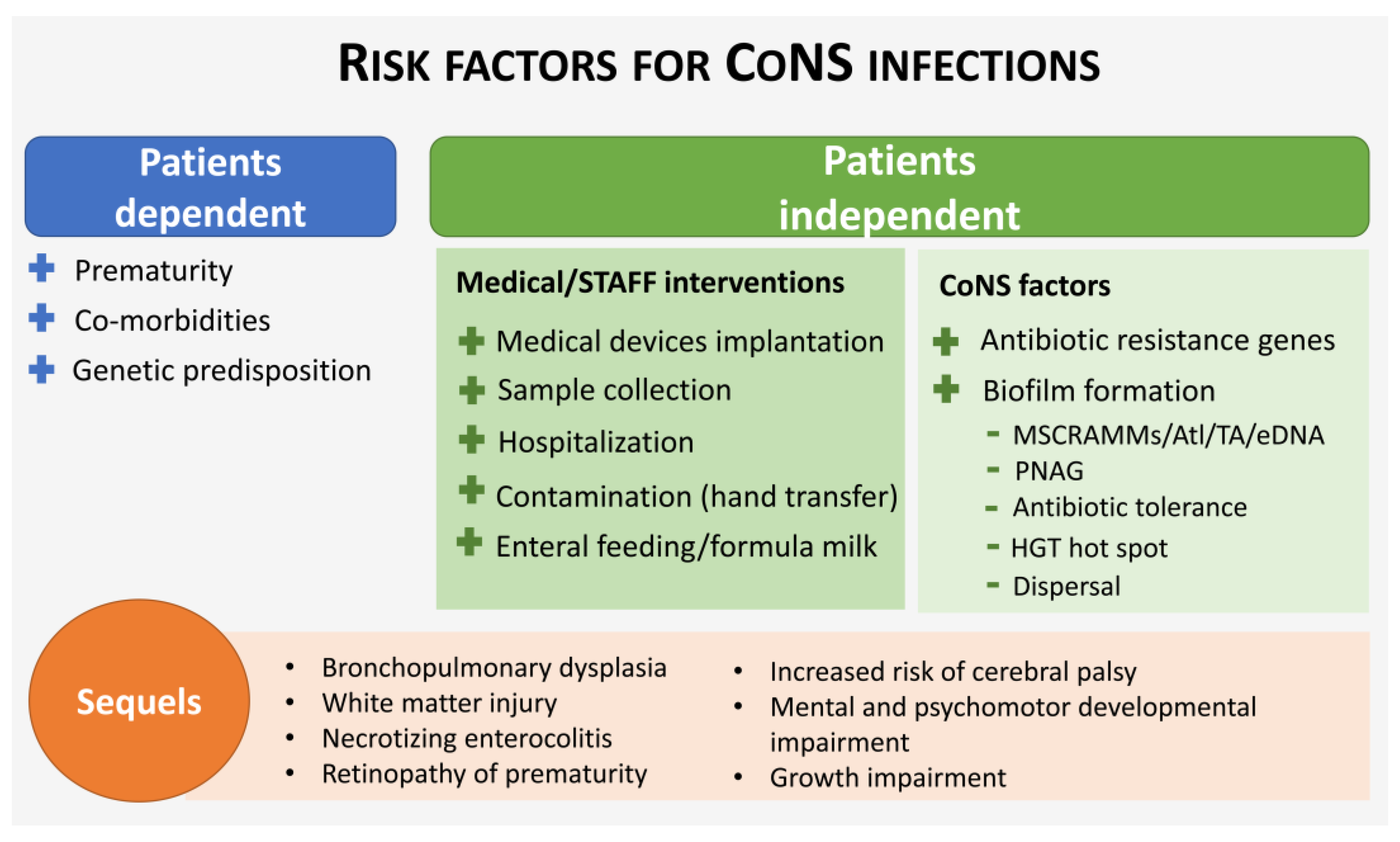
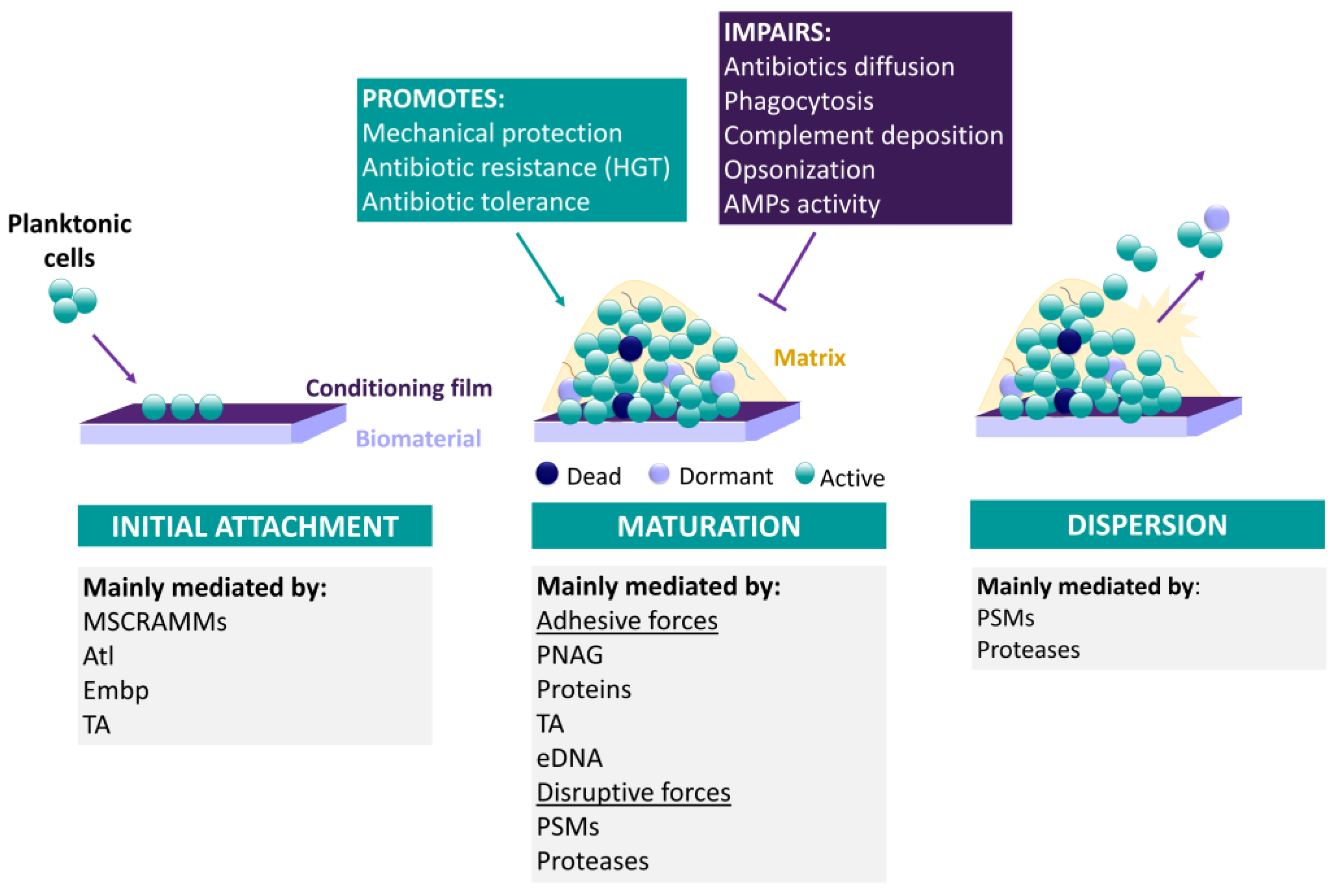
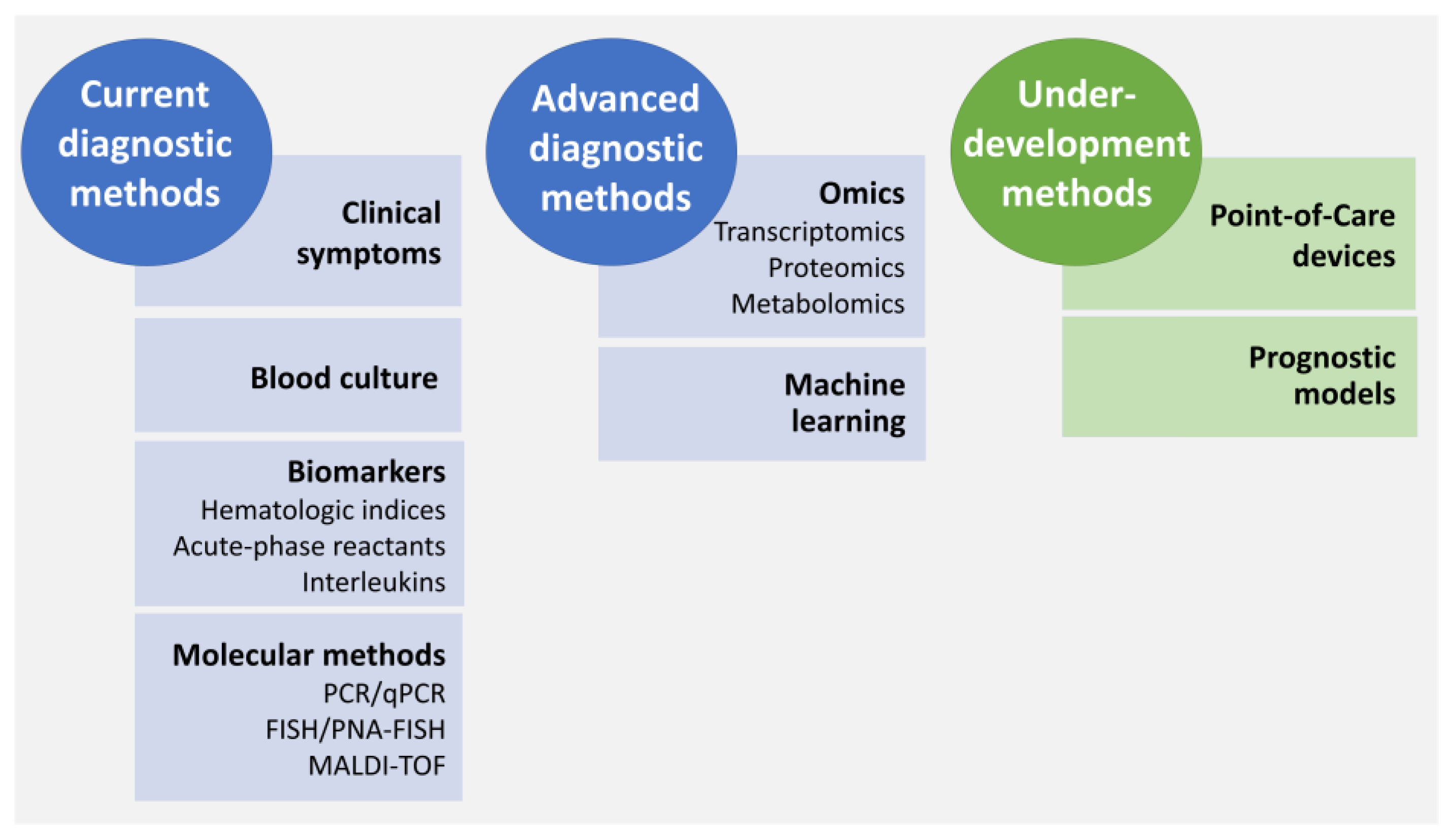
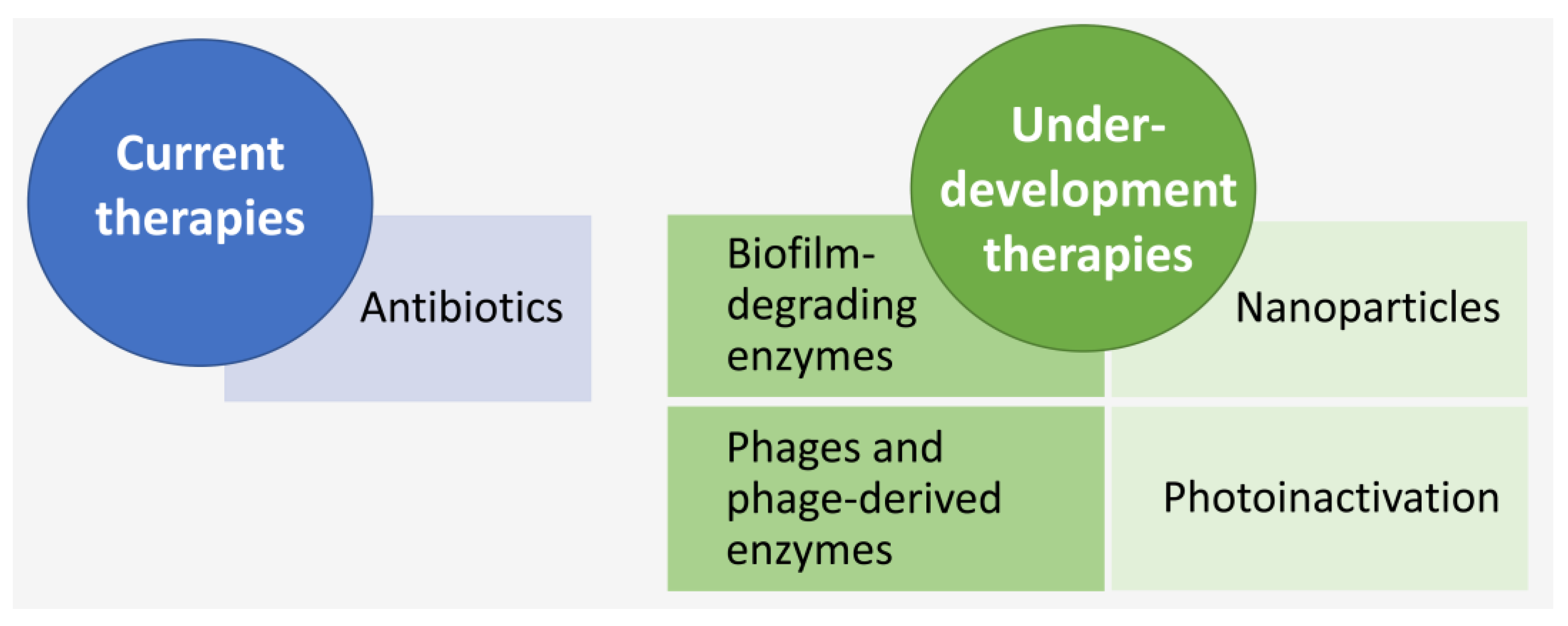
| Characteristics | EOS | LOS |
|---|---|---|
| Onset | 0–3 days | 3–28 days |
| Transmission | Vertical (female genitourinary system) | Horizontal (neonatal/community environments) |
| Frequent causative agents | Group B Streptococcus (GBS) | CoNS |
| Escherichia coli | Staphylococcus aureus | |
| Haemophilus influenza | Escherichia coli | |
| Listeria monocytogenes (dietary intake) | Klebsiella pneumonia | |
| Acinetobacter baumannii | ||
| Risk factors |
|
|
|
| |
|
| |
|
| |
|
| |
| ||
| ||
| Mortality | 3–16% | 36–52% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
França, A. The Role of Coagulase-Negative Staphylococci Biofilms on Late-Onset Sepsis: Current Challenges and Emerging Diagnostics and Therapies. Antibiotics 2023, 12, 554. https://doi.org/10.3390/antibiotics12030554
França A. The Role of Coagulase-Negative Staphylococci Biofilms on Late-Onset Sepsis: Current Challenges and Emerging Diagnostics and Therapies. Antibiotics. 2023; 12(3):554. https://doi.org/10.3390/antibiotics12030554
Chicago/Turabian StyleFrança, Angela. 2023. "The Role of Coagulase-Negative Staphylococci Biofilms on Late-Onset Sepsis: Current Challenges and Emerging Diagnostics and Therapies" Antibiotics 12, no. 3: 554. https://doi.org/10.3390/antibiotics12030554
APA StyleFrança, A. (2023). The Role of Coagulase-Negative Staphylococci Biofilms on Late-Onset Sepsis: Current Challenges and Emerging Diagnostics and Therapies. Antibiotics, 12(3), 554. https://doi.org/10.3390/antibiotics12030554






