Identification of mcr-1 Genes and Characterization of Resistance Mechanisms to Colistin in Escherichia coli Isolates from Colombian Hospitals
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Susceptibility
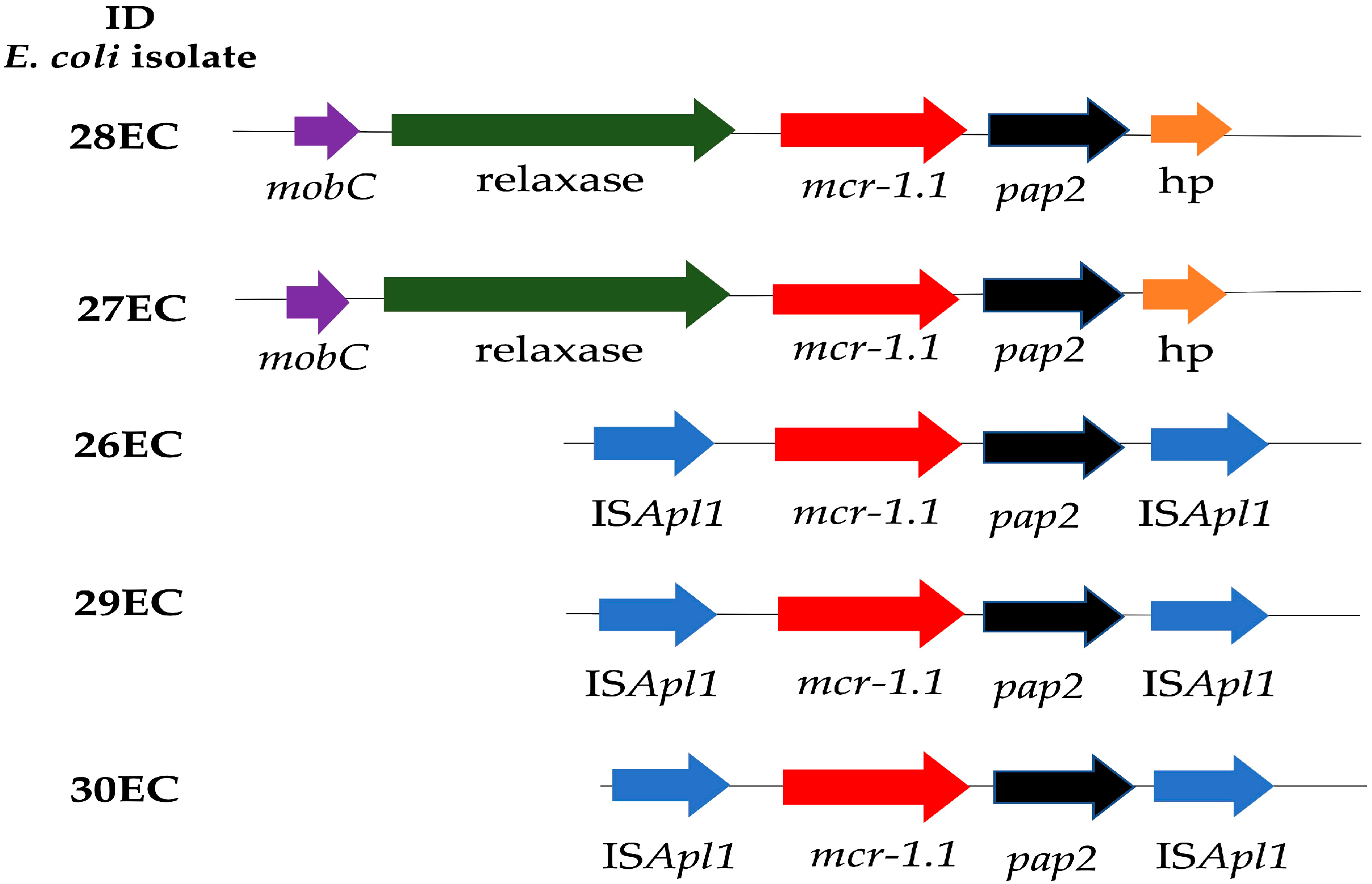
2.2. Whole Genome Sequencing
3. Discussion
4. Materials and Methods
4.1. Antimicrobial Susceptibility Testing
4.2. Whole Genome Sequencing (WGS)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, S.H.; Yang, K.Y.; Sheu, C.C.; Lin, Y.C.; Chan, M.C.; Feng, J.Y.; Chen, C.M.; Chen, C.Y.; Zheng, Z.R.; Chou, Y.C.; et al. The prevalence, presentation and outcome of colistin susceptible-only Acinetobacter Baumannii-associated pneumonia in intensive care unit: A multicenter observational study. Sci. Rep. 2023, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Bialvaei, A.Z.; Samadi Kafil, H. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Stefaniuk, E.M.; Tyski, S. Colistin resistance in Enterobacterales strains—A current view. Pol. J. Microbiol. 2019, 68, 417–427. [Google Scholar] [CrossRef]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef]
- Jayol, A.; Nordmann, P.; Brink, A.; Poirel, L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 2015, 59, 2780–2784. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Stojanoski, V.; Sankaran, B.; Prasad, B.V.V.; Poirel, L.; Nordmann, P.; Palzkill, T. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol. 2016, 14, 81. [Google Scholar] [CrossRef]
- Payne, M.; Croxen, M.A.; Lee, T.D.; Mayson, B.; Champagne, S.; Leung, V.; Bariso, S.; Hoang, L.; Lowe, C. Mcr-1–positive colistin-resistant escherichia coli in traveler returning to Canada from China. Emerg. Infect. Dis. 2016, 22, 1673–1675. [Google Scholar] [CrossRef]
- Macesic, N.; Green, D.; Wang, Z.; Sullivan, S.B.; Shim, K.; Park, S.; Whittier, S.; Furuya, E.Y.; Gomez-Simmonds, A.; Uhlemann, A.-C. Detection of mcr-1-Carrying Escherichia coli Causing Bloodstream Infection in a New York City Hospital: Avian Origins, Human Concerns? Open Forum Infect. Dis. 2017, 4, 2–5. [Google Scholar] [CrossRef]
- Xu, L.; Wan, F.; Fu, H.; Tang, B.; Ruan, Z.; Xiao, Y.; Luo, Q. Emergence of Colistin Resistance Gene mcr—10 in Enterobacterales Isolates Recovered from Fecal Samples of Chickens, Slaughterhouse Workers, and a Nearby Resident. Microbiol. Spectr. 2022, 10, e00418-22. [Google Scholar] [CrossRef] [PubMed]
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.M. Epidemiology of mobile colistin resistance (mcr) genes in aquatic environments. J. Glob. Antimicrob. Resist. 2021, 27, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, S.Y.; Diaz, L.; Wiesner, M.; Correa, A.; Alejandra Arévalo, S.; Reyes, J.; Hidalgo, A.M.; De La Cadena, E.; Perenguez, M.; Montaño, L.A.; et al. Genomic and molecular characterization of clinical isolates of Enterobacteriaceae harboring mcr-1 in Colombia, 2002 to 2016. Antimicrob. Agents Chemother. 2017, 61, e00841-17. [Google Scholar] [CrossRef] [PubMed]
- Wise, M.G.; Estabrook, M.A.; Sahm, D.F.; Stone, G.G.; Kazmierczak, K.M. Prevalence of mcr-type genes among colistinresistant Enterobacteriaceae collected in 2014-2016 as part of the INFORM global surveillance program. PLoS ONE 2018, 13, e0195281. [Google Scholar] [CrossRef]
- Kieffer, N.; Nordmann, P.; Moreno, A.M.; Moreno, L.Z.; Chaby, R.; Breton, A.; Tissières, P.; Poirel, L. Genetic and functional characterization of an MCR-3-like enzyme-producing Escherichia coli isolate recovered from Swine in Brazil. Antimicrob. Agents Chemother. 2018, 62, e00278-18. [Google Scholar] [CrossRef]
- Rada, A.M.; de la Cadena, E.; Agudelo, C.; Capataz, C.; Orozco, N.; Pallares, C.; Dinh, A.Q.; Panesso, D.; Ríos, R.; Diaz, L.; et al. Dynamics of blaKPC-2 Dissemination from Non-CG258 Klebsiella pneumoniae to Other Enterobacterales via IncN Plasmids in an Area of High Endemicity. Antimicrob. Agents Chemother. 2020, 64, e01743-20. [Google Scholar] [CrossRef]
- Mendes Oliveira, V.R.; Paiva, M.C.; Lima, W.G. Plasmid-mediated colistin resistance in Latin America and Caribbean: A systematic review. Travel Med. Infect. Dis. 2019, 31, 101459. [Google Scholar] [CrossRef]
- Yang, R.; Feng, Y.; Lv, X.; Duan, J.; Chen, J.; Fang, L.; Xia, J.; Liao, X.; Sun, J.; Liu, Y. ST648 and ST156 from a Single Muscovy Duck (Cairina moschata). Antimicrob. Agents Chemother. 2016, 60, 6899–6902. [Google Scholar] [CrossRef]
- Wu, R.; Yi, L.X.; Yu, L.F.; Wang, J.; Liu, Y.; Chen, X.; Lv, L.; Yang, J.; Liu, J.H. Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 2018, 9, 331. [Google Scholar] [CrossRef]
- Monte, D.F.; Mem, A.; Fernandes, M.R.; Cerdeira, L.; Esposito, F.; Galvão, J.A.; Franco, B.D.G.M.; Lincopan, N.; Landgraf, M. Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 2017, 61, 2–5. [Google Scholar] [CrossRef]
- Sato, T.; Shiraishi, T.; Hiyama, Y.; Honda, H.; Shinagawa, M.; Usui, M.; Kuronuma, K.; Masumori, N.; Takahashi, S.; Tamura, Y.; et al. Contribution of novel amino acid alterations in PmrA or PmrB to colistin resistance in mcr-negative Escherichia coli clinical isolates, including major multidrug-resistant lineages O25b:H4-ST131-H30Rx and Non-x. Antimicrob. Agents Chemother. 2018, 62, e00864-18. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Devendec, L.L.; Jouy, E.; Fach, P.; Drider, D.; Kempf, I. Characterization of colistin-resistant escherichia coli isolated from diseased pigs in France. Front. Microbiol. 2017, 8, 2278. [Google Scholar] [CrossRef]
- Rossi, F.; Girardello, R.; Morais, C.; Cury, A.P.; Martins, L.F.; Da Silva, A.M.; Abdala, E.; Setubal, J.C.; Da Silva Duarte, A.J. Plasmid-mediated mcr-1 in carbapenem-susceptible escherichia coli ST156 causing a blood infection: An unnoticeable spread of colistin resistance in Brazil? Clinics 2017, 72, 642–644. [Google Scholar] [CrossRef]
- Organización Panamericana de la Salud/Organización Mundial de la Salud. Alerta Epidemiológica: Enterobacterias con Resistencia Transferible a Colistina, Implicaciones para la Salud Publica en las Américas, 10 de Junio de 2016, Washington, DC. OPS/OMS. 2016. Available online: https://www.paho.org/es/documentos/10-junio-2016-enterobacterias-con-resistencia-transferible-colistina-implicaciones-para (accessed on 24 January 2023).
- Ling, Z.; Yin, W.; Shen, Z.; Wang, Y.; Shen, J.; Walsh, T.R. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 2020, 75, 3087–3095. [Google Scholar] [CrossRef] [PubMed]
- Rada, A.M.; Hernández-Gómez, C.; Restrepo, E.; Villegas, M.V. Distribución y caracterización molecular de betalactamasas en bacterias Gram negativas en Colombia, 2001–2016. Biomédica 2019, 39, 199–220. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Pillonetto, M.; Mazzetti, A.; Becker, G.N.; Siebra, C.A.; Arend, L.N.V.S.; Barth, A.L. Low level of polymyxin resistance among nonclonal mcr-1–positive Escherichia coli from human sources in Brazil. Diagn. Microbiol. Infect. Dis. 2019, 93, 140–142. [Google Scholar] [CrossRef]
- Castanheira, M.; Griffin, M.A.; Deshpande, L.M.; Mendes, R.E.; Jones, R.N.; Flamm, R.K. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob. Agents Chemother. 2016, 60, 5623–5624. [Google Scholar] [CrossRef]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an ‘old’ class of antibiotics. Futur. Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef]
- Chew, K.L.; La, M.V.; Lin, R.T.P.; Teo, J.W.P. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive enterobacteriaceae: Comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J. Clin. Microbiol. 2017, 55, 2609–2616. [Google Scholar] [CrossRef]
- Dalmolin, T.V.; Martins, A.F.; Zavascki, A.P.; Daiana de Lima-Morales, A.L.B. Acquisition of the mcr-1 gene by a high-risk clone of KPC-2-producing Klebsiella pneumoniae ST437/CC258, Brazil. Diagn. Microbiol. Infect. Dis. 2018, 90, 132–133. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Hu, Y.; Sun, Q.; Hu, F.; Zhou, H.; Shu, L.; Ma, T.; Shen, Y.; Wang, Y.; Li, J.; et al. Emerging Carriage of NDM-5 and MCR-1 in Escherichia coli From Healthy People in Multiple Regions in China: A Cross Sectional Observational Study. EClinicalMedicine 2018, 6, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mediavilla, J.R.; Patrawalla, A.; Chen, L.; Chavda, K.D.; Mathema, B.; Vinnard, C.; Dever, L.L.; Kreiswirth, B.N. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio 2016, 7, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.Q.; Ong, R.T.; Xia, E.; Koh, T.; Khor, C.; Lee, S.J.; Lim, T. Enterobacteriaceae Isolates in Singapore. Antimicrob. Agents Chemother. 2016, 60, 6435–6437. [Google Scholar] [CrossRef]
- Ruiz, S.J.; Montealegre, M.C.; Ruiz-Garbajosa, P.; Correa, A.; Briceño, D.F.; Martinez, E.; Rosso, F.; Muñoz, M.; Quinn, J.P.; Cantón, R.; et al. First characterization of CTX-M-15-producing Escherichia coli ST131 and ST405 clones causing community-onset infections in South America. J. Clin. Microbiol. 2011, 49, 1993–1996. [Google Scholar] [CrossRef]
- Cadena, E.D.L.; Mojica, M.F.; Castillo, N.; Correa, A.; Appel, T.M.; García-Betancur, J.C.; Pallares, C.J.; Villegas, M.V. Genomic analysis of ctx-m-group-1-producing extraintestinal pathogenic e. Coli (expec) from patients with urinary tract infections (uti) from colombia. Antibiotics 2020, 9, 899. [Google Scholar] [CrossRef]
- Li, R.; Lu, X.; Munir, A.; Abdullah, S.; Liu, Y.; Xiao, X.; Wang, Z.; Mohsin, M. Widespread prevalence and molecular epidemiology of tet(X4) and mcr-1 harboring Escherichia coli isolated from chickens in Pakistan. Sci. Total Environ. 2022, 806, 150689. [Google Scholar] [CrossRef]
- Wu, C.; Wang, Y.; Shi, X.; Wang, S.; Ren, H.; Shen, Z.; Wang, Y.; Lin, J.; Wang, S. Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008–2014. Emerg. Microbes Infect. 2018, 7, 1–10. [Google Scholar] [CrossRef]
- Nakayama, T.; Le Thi, H.; Thanh, P.N.; Minh, D.T.N.; Hoang, O.N.; Hoai, P.H.; Yamaguchi, T.; Jinnai, M.; Do, P.N.; Van, C.D.; et al. Abundance of colistin-resistant Escherichia coli harbouring mcr-1 and extended-spectrum β-lactamase-producing E. coli co-harbouring bla CTX-M-55 or -65 with bla TEM isolates from chicken meat in Vietnam. Arch. Microbiol. 2022, 204, 137. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D.D. The role of epidemic resistance plasmids and international high- risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef]
- Loucif, L.; Chelaghma, W.; Bendjama, E.; Cherak, Z.; Khellaf, M.; Khemri, A.; Rolain, J.M. Detection of blaOXA-48 and mcr-1 Genes in Escherichia coli Isolates from Pigeon (Columba livia) in Algeria. Microorganisms 2022, 10, 975. [Google Scholar] [CrossRef] [PubMed]
- Sacramento, A.G.; Fernandes, M.R.; Sellera, F.P.; Muñoz, M.E.; Vivas, R.; Dolabella, S.S.; Lincopan, N. Genomic analysis of MCR-1 and CTX-M-8 co-producing Escherichia coli ST58 isolated from a polluted mangrove ecosystem in Brazil. J. Glob. Antimicrob. Resist. 2018, 15, 288–289. [Google Scholar] [CrossRef] [PubMed]
- Snesrud, E.; He, S.; Chandler, M.; Dekker, J.P.; Hickman, A.B.; McGann, P.; Dyda, F. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 2016, 60, 6973–6976. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef]
- Petrillo, M.; Angers-Loustau, A.; Kreysa, J. Possible genetic events producing colistin resistance gene mcr-1. Lancet Infect. Dis. 2016, 16, 280. [Google Scholar] [CrossRef]
- Matamoros, S.; Van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; Van Genderen, P.J.; et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 2017, 7, 15364. [Google Scholar] [CrossRef]
- Paiva, Y.; Nagano, D.S.; Cotia, A.L.F.; Guimarães, T.; Martins, R.C.R.; Neto, L.V.P.; Côrtes, M.F.; Marchi, A.P.; Corscadden, L.; Machado, A.S.; et al. Colistin-resistant Escherichia coli belonging to different sequence types: Genetic characterization of isolates responsible for colonization, community- and healthcare-acquired infections. Rev. Inst. Med. Trop. São Paulo 2021, 63, e38. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing M100; 32th Informational Supplement; CLSI: Wayne, PA, USA, 2022. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
| ID | City | Isolation Date | MIC (µg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRO | CTX | CAZ | FEP | TZP | ETP | CIP | FOS | COL | |||
| 27EC | Bogotá | 2018 | <=0.5 | <=0.5 | <=2 | <=2 | <=2/4 | <=0.25 | 0.5 | <=8 | >32 |
| 28EC | Bogotá | 2018 | <=0.5 | <=0.5 | <=2 | <=2 | <=2/4 | <=0.25 | ≥64 | <=8 | >32 |
| 29EC | Cali | 2017 | <=0.5 | <=0.5 | <=2 | <=2 | <=2/4 | <=0.25 | 0.25 | <=8 | >32 |
| 26EC | Ibagué | 2017 | >4 | 4 | 8 | 8 | <=2/4 | <=0.25 | <0.25 | 64 | >32 |
| 30EC | Pereira | 2017 | <=0.5 | <=0.5 | <=2 | <=2 | 32/4 | <=0.25 | 1 | <=8 | >32 |
| ID | MLST | Phylogroup | mcr-Variant | Resistance Genes | Plasmid Incompatibility Types | Virulence Factors |
|---|---|---|---|---|---|---|
| 26EC | 14315 | B1 | mcr-1.1 | CTX-M-55, TEM-1B, aph(4)-Ia, aac(3)-IV, aadA1, mdf(A), drfA1, fosA, tet(B), floR | Col, IncFIA, IncFIB, IncFII, IncN, IncI1 | cia, cvaC, etsC, hlyF, iroN, iss, iucC, iutA, IpfA, ompT, tsh, sitA, terC, traT |
| 27EC | 58 | B1 | mcr-1.1 | TEM-1B, qnrS1, Sul3, drfA15, tet(A), mdf(A), cm1A1 | Col, IncFIB, IncFIC (II), IncI2 | cea, cib, cma, cvaC, fyuA, gad, hlyF, iroN, irp2, iss, iucC, iutA, ipfA, ompT, sitA, terC, tratT |
| 28EC | 46 | A | mcr-1.1 | TEM-1B, sull2, sul3, aac(3)-Iid, aph(3′)-Ia, aadA1, tet(A), tet(M), dfrA12, mdf(A) | Col, IncFIB, IncI2, IncX4 | celb, fyuA, gad, irp2, ompT, sitA, terC, traT |
| 29EC | 393 | D | mcr-1.1 | qnrB19, sul2, aadA1, aadA5, mdf(A), drfA17, tet(A), floR | IncFIA, IncFII, IncHI1A, IncHI1B | chuA, eilA, fyuA, gad, hra, iha, irp2, iss, iucC, iutA, kpsE, kpsMII, lpfA, mcmA, ompT, papA, papC, sat, sitA, terC, traT |
| 30EC | 58 | B1 | mcr-1.1 | TEM-1B, qnrB19, aph(6)-Id, aadA12, aph(3″)-IIb, drfA5, sul2, floR | InFIB, IncFII, IncI1-1, IncQ1, IncX1 | cia, cvaC, etsC, fyuA, gad, hlyF, iroN, irp2, iss, iucC, iutA, ipfA, mchF, ompT, sitA, terC, traT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De La Cadena, E.; Mahecha, M.; Velandia, A.M.; García-Betancur, J.C.; Rojas, L.J.; Porras, J.; Pallares, C.; Villegas, M.V. Identification of mcr-1 Genes and Characterization of Resistance Mechanisms to Colistin in Escherichia coli Isolates from Colombian Hospitals. Antibiotics 2023, 12, 488. https://doi.org/10.3390/antibiotics12030488
De La Cadena E, Mahecha M, Velandia AM, García-Betancur JC, Rojas LJ, Porras J, Pallares C, Villegas MV. Identification of mcr-1 Genes and Characterization of Resistance Mechanisms to Colistin in Escherichia coli Isolates from Colombian Hospitals. Antibiotics. 2023; 12(3):488. https://doi.org/10.3390/antibiotics12030488
Chicago/Turabian StyleDe La Cadena, Elsa, Mateo Mahecha, Ana María Velandia, Juan Carlos García-Betancur, Laura J. Rojas, Jessica Porras, Christian Pallares, and María Virginia Villegas. 2023. "Identification of mcr-1 Genes and Characterization of Resistance Mechanisms to Colistin in Escherichia coli Isolates from Colombian Hospitals" Antibiotics 12, no. 3: 488. https://doi.org/10.3390/antibiotics12030488
APA StyleDe La Cadena, E., Mahecha, M., Velandia, A. M., García-Betancur, J. C., Rojas, L. J., Porras, J., Pallares, C., & Villegas, M. V. (2023). Identification of mcr-1 Genes and Characterization of Resistance Mechanisms to Colistin in Escherichia coli Isolates from Colombian Hospitals. Antibiotics, 12(3), 488. https://doi.org/10.3390/antibiotics12030488





