Mechanisms of Antimicrobial Peptides from Bagasse against Human Pathogenic Bacteria
Abstract
1. Introduction
2. Results
2.1. Antibacterial Efficiency of Lower than 3 kDa Protein Hydrolysates/Peptides
2.2. Antibacterial Activity of Purified Peptides from Each Purification Step
2.3. Antibacterial Analysis of Synthetic Peptides
2.4. Study of Peptide-Microbe Interaction Mechanisms
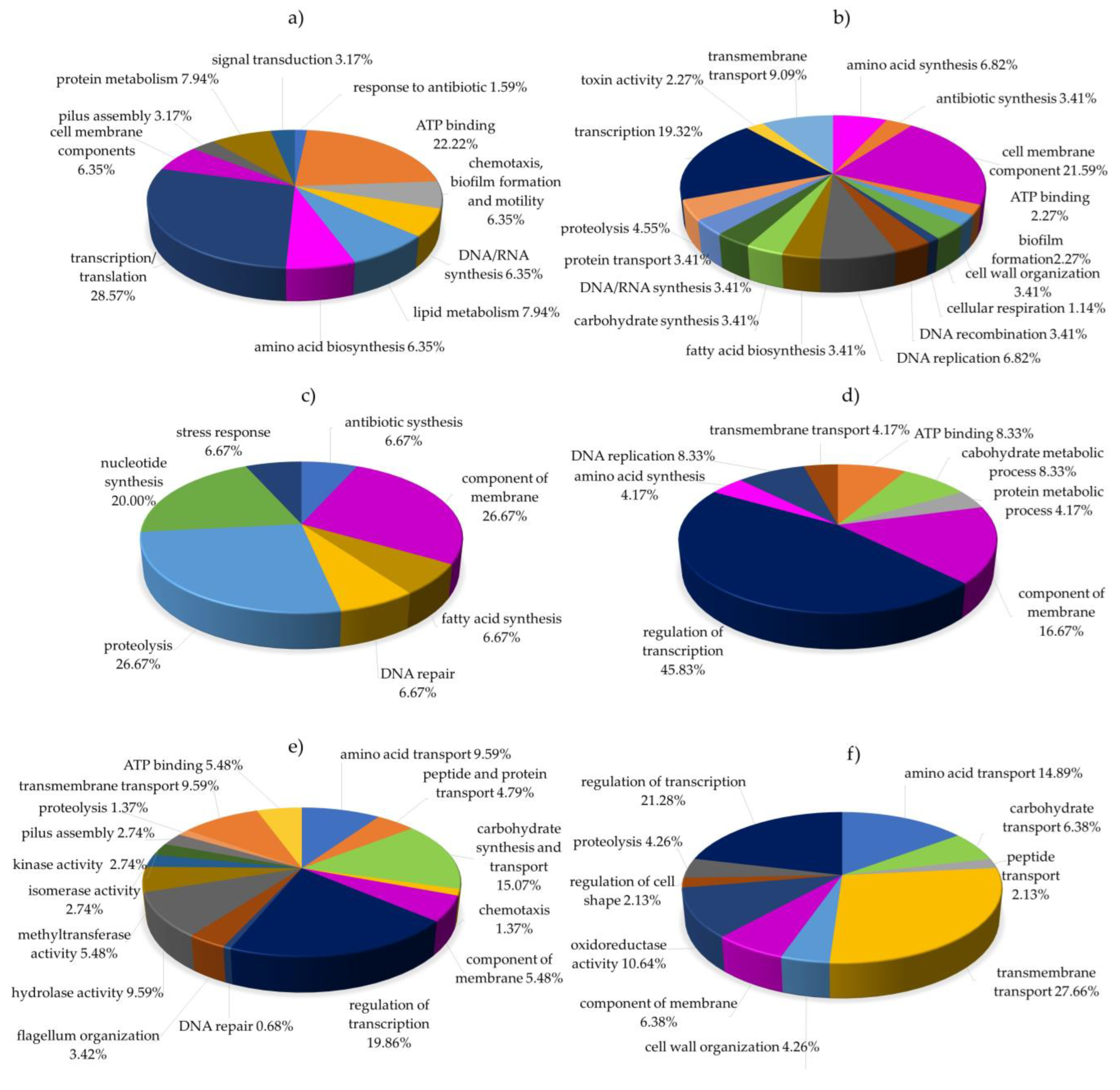
2.5. Functional Classification of Differentially Expressed Proteins
3. Discussion
3.1. Protein Hydrolysate Preparation and Antibacterial Activity Screening
3.2. Protein Hydrolysate and Peptide Purification
3.3. Peptide-Microbe Interaction Mechanisms
4. Materials and Methods
4.1. Time and Place of Research
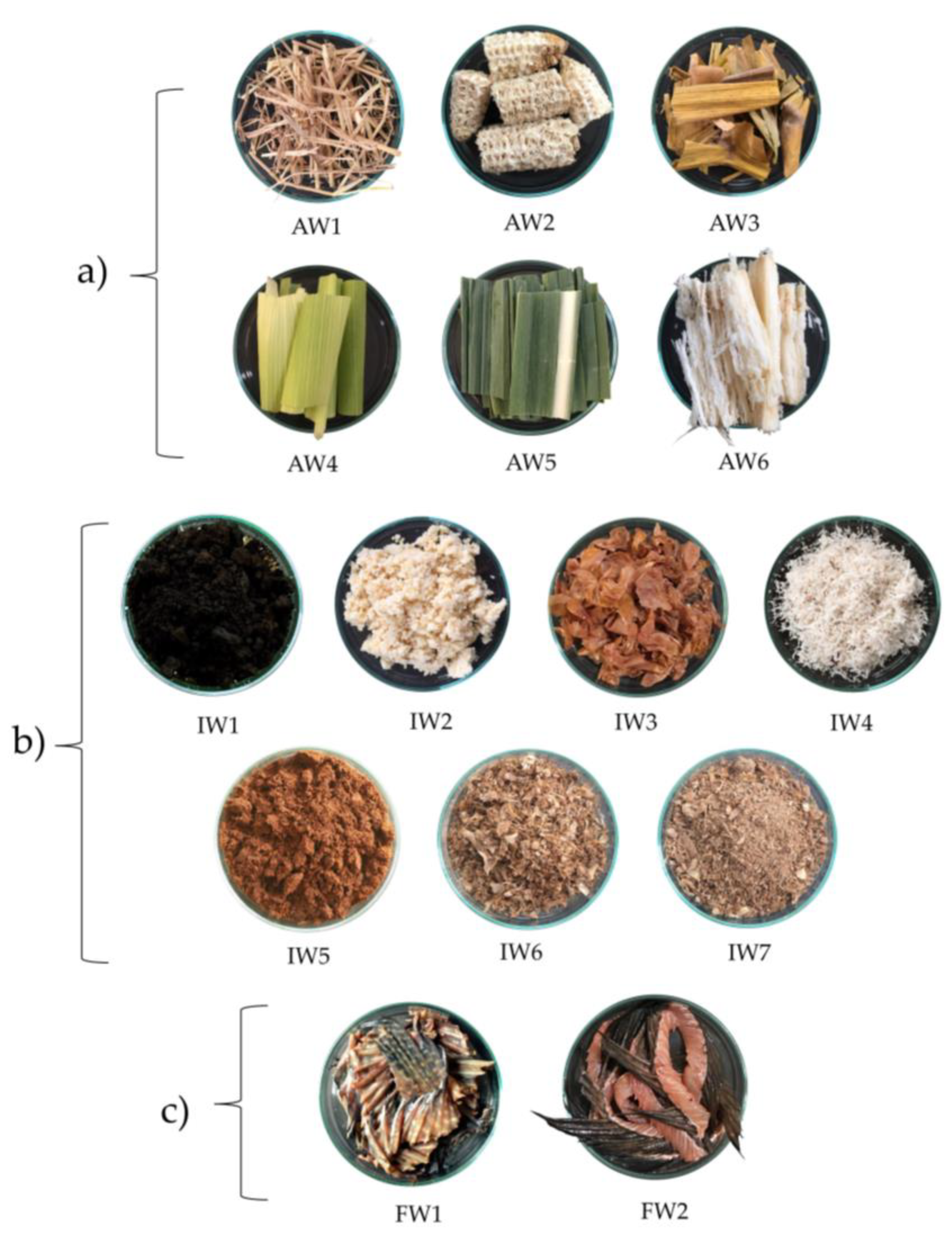
4.2. Sample Collection
4.3. Preparation of Lower than 3 kDa Protein Hydrolysates
4.4. Human Pathogenic Bacteria and Antimicrobial Activity Assay
4.5. Experimental and Statistical Design
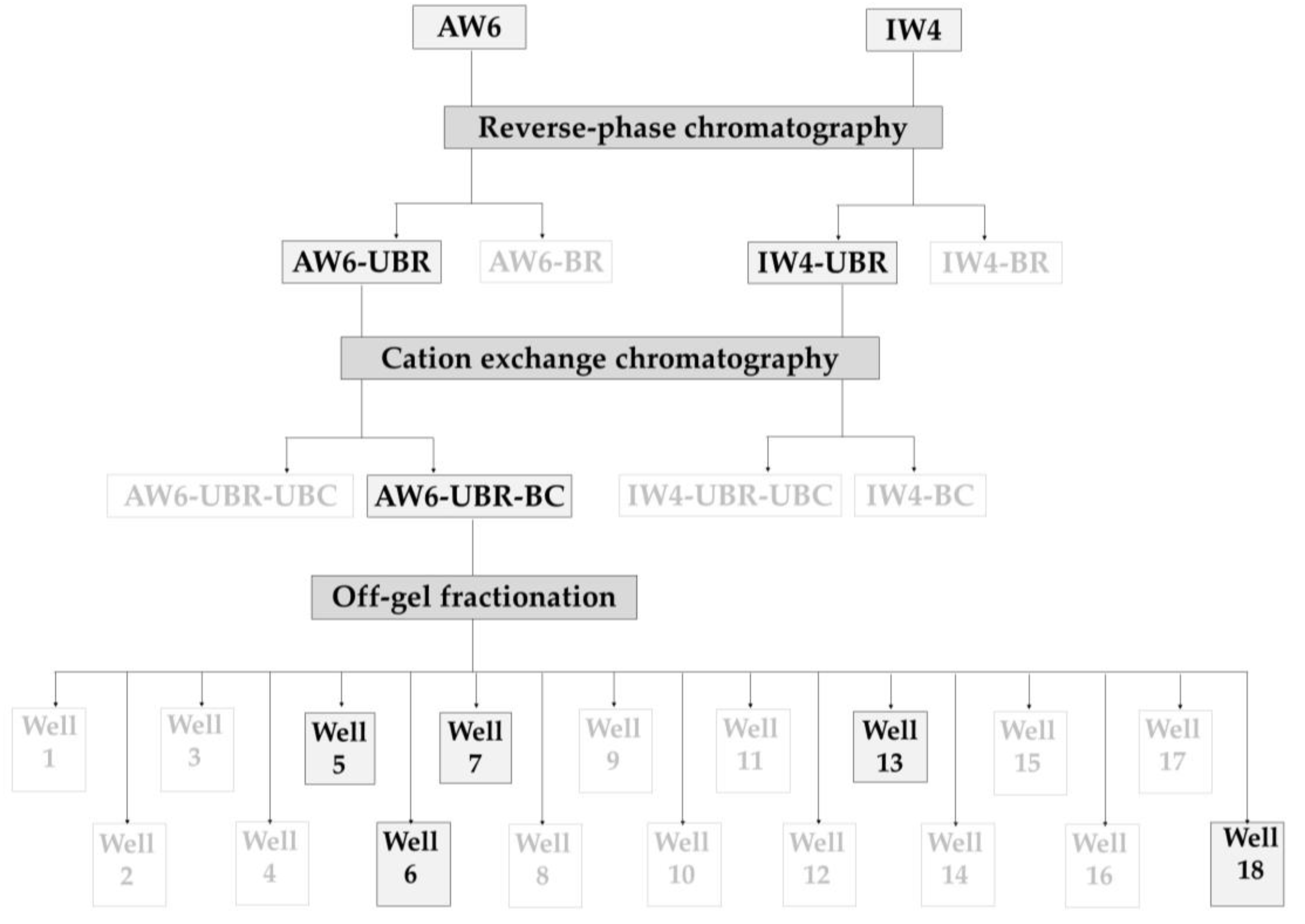
4.6. Reversed-Phase Chromatography (1st Step of Peptide Purification)
4.7. Cation Exchange Chromatography (2nd Step of Peptide Purification)
4.8. Off-Gel Fractionation (3rd Step of Peptide Purification)
4.9. Peptide Synthesis
4.10. Study of Peptide-Microbe Interaction Mechanisms Using Proteomics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kadri, S.S. Key Takeaways From the U.S. Cdc’s 2019 Antibiotic Resistance Threats Report for Frontline Providers. Crit. Care Med. 2020, 48, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to Manage Pseudomonas aeruginosa Infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef]
- Maraolo, A.E.; Cascella, M.; Corcione, S.; Cuomo, A.; Nappa, S.; Borgia, G.; De Rosa, F.G.; Gentile, I. Management of Multidrug-Resistant Pseudomonas aeruginosa in the Intensive Care Unit: State of the Art. Expert Rev. Anti Infect. Ther. 2017, 15, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Adimpong, D.B.; Sørensen, K.I.; Thorsen, L.; Stuer-Lauridsen, B.; Abdelgadir, W.S.; Nielsen, D.S.; Derkx, P.M.; Jespersen, L. Antimicrobial susceptibility of Bacillus strains isolated from primary starters for African traditional bread production and characterization of the bacitracin operon and bacitracin biosynthesis. Appl. Environ. Microbiol. 2012, 78, 7903–7914. [Google Scholar] [CrossRef]
- Mazza, P.; Zani, F.; Martelli, P. Studies on the antibiotic resistance of Bacillus subtilis strains used in oral bacteriotherapy. Boll. Chim. Farm. 1992, 131, 401–408. [Google Scholar] [PubMed]
- Eberl, L.; Tümmler, B. Pseudomonas aeruginosa and Burkholderia cepacia in cystic fibrosis: Genome evolution, interactions and adaptation. Int. J. Med. Microbiol. 2004, 294, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Cho, J.H.; Kim, K.S.; Kim, Y.B.; Kim, M.S.; Kim, S.C. Helix stability confers salt resistance upon helical antimicrobial peptides. J. Biol. Chem. 2004, 279, 13896–13901. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, H.; Lu, H.; Li, G.; Huang, Q. Lamp: A database linking antimicrobial peptides. PLoS ONE 2013, 8, e66557. [Google Scholar] [CrossRef]
- Zhang, L.J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Ben Hamida, J.; Vergoten, G.; Fliss, I. PhytAMP: A database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009, 37, D963–D968. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Hall, K.N.; Aguilar, M.I. Antimicrobial peptide structure and mechanism of action: A focus on the role of membrane structure. Curr. Top. Med. Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.P.S.; Rosek, A.; Hancock, R.E.W. Structure activity relationship for the β-hairpin cationic antimicrobial peptide polyphemusin I. Biochim. Biophys. Acta Proteins Proteom. 2004, 1698, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Adejumo, I.O.; Adebiyi, O.A. Agricultural Solid Wastes: Causes, Effects, and Effective Management. Strategies of Sustainable Solid Waste Management; IntechOpen Limited: London, UK, 2021; pp. 1–2. [Google Scholar] [CrossRef]
- Varma, V.S.; Yadav, J.; Das, S.; Kalamdhad, A.S. Potential of waste carbide sludge addition on earthworm growth and organic matter degradation during vermicomposting of agricultural wastes. Ecol. Eng. 2015, 83, 90–95. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- Pickardt, C.; Neidhart, S.; Griesbach, C.; Dube, M.; Knauf, U.; Kammerer, D.R.; Carle, R. Optimisation of mild-acidic protein extraction from defatted sunflower (Helianthus annuus L.) meal. Food Hydrocoll. 2009, 23, 1966–1973. [Google Scholar] [CrossRef]
- Lay, F.T.; Brugliera, F.; Anderson, M.A. Isolation and Properties of Floral Defensins from Ornamental Tobacco and Petunia. Plant Physiol. 2003, 131, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Kameda, M.; Namae, T.; Ochiai, A.; Saitoh, E.; Tanaka, T. Identification and characterization of multifunctional cationic peptides derived from peptic hydrolysates of rice bran protein. J. Funct. Foods 2017, 34, 287–296. [Google Scholar] [CrossRef]
- Lay, F.T.; Anderson, M.A. Defensins—Components of the Innate Immune System in Plants. Curr. Protein Pept. Sci. 2005, 6, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Cammue, B.P.A.; De Bolle, M.F.C.; Thevissen, K.; De Samblanx, G.W.; Osborn, R.W. Antimicrobial peptides from plants. Crit. Rev. Plant Sci. 1997, 16, 297–323. [Google Scholar] [CrossRef]
- Gordon, Y.J.; Romanowski, E.G.; McDermott, A.M. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005, 30, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Martinez, V.; Valles-Rosales, D.; Rodriguez-Uribe, L.; Holguin, O.; Quintero-Quiroz, J.; Reyes-Jaquez, D.; Rodriguez-Borbon, M.I.; Villagrán-Villegas, L.Y.; Delgado, E. Antimicrobial, shelf-life stability, and effect of maltodextrin and gum arabic on the encapsulation efficiency of sugarcane bagasse bioactive compounds. Foods 2021, 10, 116. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.M.; Dey, S.; Mandal, M.; Sarkar, S.; Maria-Neto, S.; Franco, O.L. Identification and structural insights of three novel antimicrobial peptides isolated from green coconut water. Peptides 2009, 30, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.; Peter, F.; Sander, J. Biomass in the manufacture of industrial products-the use of proteins and amino acids. Appl. Microbiol. Biotechnol. 2007, 75, 751–762. [Google Scholar] [CrossRef]
- Otvos, L. Antibacterial peptides and proteins with multiple cellular targets. J. Pept. Sci. 2005, 11, 697–706. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Casteels, P.; Ampe, C.; Jacobs, F.; Vaeck, M.; Tempst, P. Apidaecins: Antibacterial peptides from honeybees. EMBO J. 1989, 8, 2387–2391. [Google Scholar] [CrossRef]
- Cudic, M.; Otvos, J.L. Intracellular targets of antibacterial peptides. Curr. Drug Targets 2002, 3, 101–106. [Google Scholar] [CrossRef]
- Sani, M.A.; Separovic, F. How Membrane-Active Peptides Get into Lipid Membranes. Acc. Chem. Res. 2017, 49, 1130–1138. [Google Scholar] [CrossRef]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.H.; Chen, C.; Jou, M.L.; Lee, A.Y.; Lin, Y.C.; Yu, Y.P.; Huang, W.T.; Wu, S.H. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: Evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 2005, 33, 4053–4064. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrel, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Hansen, P.R.; Oddo, A. Fmoc Solid-Phase Peptide Synthesis. Methods Mol. Biol. 2015, 1348, 33–50. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
| Antibacterial Activity Ranking | Inhibitory Percentage of Target Human Pathogenic Bacteria * | |||||
|---|---|---|---|---|---|---|
| Pseudomonas aeruginosa | Bacillus subtilis | Burkholderia cepacia | ||||
| 1 | 79.49 ± 0.49 a | AW6 | 52.73 ± 4.58 a | AW6 | 85.66 ± 0.91 a | AW6 |
| 2 | 56.33 ± 8.79 b | IW4 | 23.71 ± 4.28 b | IW4 | 82.13 ± 0.97 ab | IW4 |
| 3 | 15.03 ± 19.42 c | IW1 | 11.13 ± 1.73 c | IW1 | 79.57 ± 1.97 ab | IW3 |
| 4 | 15.00 ± 11.23 c | AW1 | 10.89 ± 0.95 c | IW5 | 75.40 ± 1.32 abc | AW1 |
| 5 | 10.24 ± 3.86 cd | AW3 | 6.10 ± 0.95 cd | AW1 | 71.71 ± 2.57 bc | AW5 |
| 6 | 8.70 ± 7.83 cd | AW4 | 3.10 ± 1.05 cde | AW3 | 71.55 ± 3.85 bc | AW4 |
| 7 | 3.91 ± 6.58 cde | IW7 | 0.10 ± 0.94 def | IW2 | 67.39 ± 7.32 c | IW7 |
| 8 | 3.21 ± 8.27 cde | AW5 | −0.44 ± 2.03 def | AW5 | 66.75 ± 2.18 c | FW2 |
| 9 | 2.08 ± 14.40 cdef | IW3 | −2.18 ± 0.75 def | AW2 | 66.59 ± 2.60 c | AW3 |
| 10 | 2.05 ± 7.41 cdef | AW2 | −2.90 ± 2.55 def | IW6 | 64.34 ± 2.04 c | IW5 |
| 11 | −0.95 ± 2.41 cdefg | FW2 | −3.97 ± 0.92 ef | AW4 | 51.60 ± 2.47 d | FW1 |
| 12 | −2.46 ± 8.56 defg | IW5 | −5.90 ± 3.83 ef | IW7 | 47.04 ± 14.65 d | IW6 |
| 13 | −11.50 ± 5.05 efg | IW2 | −8.66 ± 12.68 fg | IW3 | 46.47 ± 9.94 d | IW2 |
| 14 | −14.49 ± 1.99 fg | FW1 | −15.34 ± 11.78 g | FW2 | 41.03 ± 11.67 d | AW2 |
| 15 | −15.03 ± 5.91 g | IW6 | −17.56 ± 4.71 g | FW1 | 24.60 ± 9.02 e | IW1 |
| kanamycin | 39.00 ± 2.81 | 77.60 ± 0.85 | 87.02 ± 0.42 | |||
| ampicillin | 31.98 ± 1.59 | 85.20 ± 10.57 | 32.77 ± 4.18 | |||
| Peptide No. | Accession No. | Protein Name | Peptide Sequence | Inhibitory Percentage | ||
|---|---|---|---|---|---|---|
| P. aeruginosa | Bacillus subtilis | Burkholderia cepacia | ||||
| 1 | A0A3P3YVC5 | Photosystem II CP47 reaction center protein (PSII 47 kDa protein) (Protein CP-47) | GAFHVTGL | 21.74 ± 0.96 | 3.82 ± 0.09 | 9.09 ± 0.04 |
| 2 | A0A059PYU1 | RNA helicase (EC 3.6.4.13) | VLSSWGDESTL | 23.61 ± 0.12 | 4.45 ± 0.03 | 14.92 ± 0.72 |
| 3 | A0A678TPX2 | LRR receptor-like serine/threonine-protein kinase GSO2 | NLWSNEINQDMAEF | 24.87 ± 0.81 | 11.33 ± 0.29 | 23.32 ± 0.68 |
| 4 | A0A059Q010 | 3-ketoacyl-CoA synthase (EC 2.3.1.) | VSNCL | 24.70 ± 0.81 | 10.39 ± 0.33 | 20.56 ± 0.28 |
| 5 | A0A059PZS2 | Chitinase | WDTDNLSPDAVAAIKAAHPNVAVMAGL | 30.34 ± 0.95 | 5.97 ± 0.06 | 20.51 ± 0.48 |
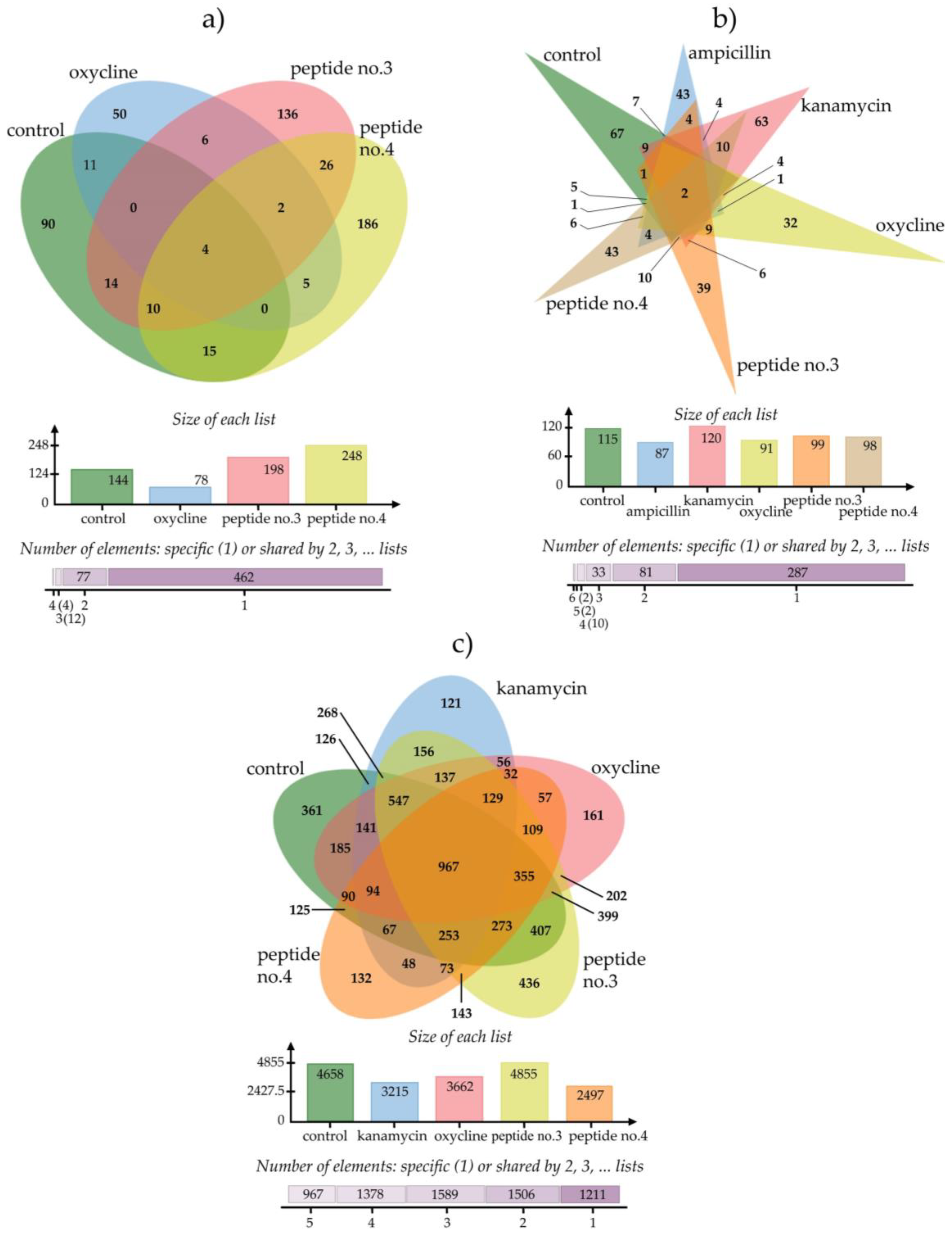
| Human Pathogen | Total Proteins Differentially Expressed | Uniqueness and Commonality of Differentially Expressed Proteins | |||
|---|---|---|---|---|---|
| Unique | Shared with Ampicillin | Shared with Kanamycin | Shared with Oxycline | ||
| Peptide no.3 | |||||
| P. aeruginosa | 198 | 136 | - | - | 8 |
| Bacillus subtilis | 99 | 39 | 4 | 6 | 9 |
| Burkholderia cepacia | 4855 | 436 | - | 422 | 375 |
| Peptide no.4 | |||||
| P. aeruginosa | 248 | 186 | - | - | 7 |
| Bacillus subtilis | 98 | 43 | 4 | 10 | 6 |
| Burkholderia cepacia | 2947 | 132 | - | 209 | 327 |
| Code | Waste Samples | Source of Waste Sample | Location (Latitude, Longitude) |
|---|---|---|---|
| AW1 | Rice straw | rice farm | Chachoengsao (13.6690° N, 101.0891° E) |
| AW2 | Corn cobs | corn farm | Sakaeo (13.5035° N, 102.2872° E) |
| AW3 | Corn leaves | corn farm | Sakaeo (13.5035° N, 102.2872° E) |
| AW4 | Corn husks | corn farm | Sakaeo (13.5035° N, 102.2872° E) |
| AW5 | Sugarcane leaves | sugarcane farm | Sakaeo (13.50181° N, 102.2875° E) |
| AW6 | Bagasse | sugarcane farm | Sakaeo (13.50181° N, 102.2875° E) |
| IW1 | Fermented soybeans | light soy sauce productions | Chachoengsao (13.7489° N, 100.9518° E) |
| IW2 | Soybean pellets | soybean milk productions | Chachoengsao (13.6924° N, 101.0807° E) |
| IW3 | Peanut seed coats | peanut-based snack productions | Bangkok (13.6557° N, 100.4305° E) |
| IW4 | Coconut residue | coconut milk productions | Chachoengsao (13.6924° N, 101.0807° E) |
| IW5 | Coffee grounds | Arabica grounds, mainly coffee industry | Chachoengsao (13.6701° N, 101.0562° E) |
| IW6 | Fish residue | fish sauce productions | Bangkok (13.5790° N, 100.4418° E) |
| IW7 | Fish residue (water rinsed) | fish sauce productions | Bangkok (13.5790° N, 100.4418° E) |
| FW1 | Nile tilapia fish fin | fresh market | Chachoengsao (13.6623° N, 101.0343° E) |
| FW2 | Clarias sp. Fish fin | fresh market | Chachoengsao (13.6623° N, 101.0343° E) |
| Step | Voltage Mode | Voltage (V) | Time (h:min) | kVh |
|---|---|---|---|---|
| 1 | Step and hold | 500 | 1:00 (8:00) | 0.5 |
| 2 | Gradient | 1000 | 1:00 | 0.8 |
| 3a | Gradient | 8000 | 3:00 | 13.5 |
| 4a | Step and hold | 8000 | 0:46–1:30 | 6.2–12.2 |
| 3b | Gradient | 10,000 | 3:00 | 16.5 |
| 4b | Step and hold | 10,000 | 0:20–0:55 | 3.2–9.2 |
| Total | 21.0–27.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ditsawanon, T.; Phaonakrob, N.; Roytrakul, S. Mechanisms of Antimicrobial Peptides from Bagasse against Human Pathogenic Bacteria. Antibiotics 2023, 12, 448. https://doi.org/10.3390/antibiotics12030448
Ditsawanon T, Phaonakrob N, Roytrakul S. Mechanisms of Antimicrobial Peptides from Bagasse against Human Pathogenic Bacteria. Antibiotics. 2023; 12(3):448. https://doi.org/10.3390/antibiotics12030448
Chicago/Turabian StyleDitsawanon, Thitiporn, Narumon Phaonakrob, and Sittiruk Roytrakul. 2023. "Mechanisms of Antimicrobial Peptides from Bagasse against Human Pathogenic Bacteria" Antibiotics 12, no. 3: 448. https://doi.org/10.3390/antibiotics12030448
APA StyleDitsawanon, T., Phaonakrob, N., & Roytrakul, S. (2023). Mechanisms of Antimicrobial Peptides from Bagasse against Human Pathogenic Bacteria. Antibiotics, 12(3), 448. https://doi.org/10.3390/antibiotics12030448






