Abstract
Rifampin resistance (RIF-R) in Staphylococcus aureus (S. aureus) with rpoB mutations as one of its resistance mechanisms has raised concern about clinical treatment and infection prevention strategies. Data on the prevalence and molecular epidemiology of RIF-R S. aureus blood isolates in South Korea are scarce. We used broth microdilution to investigate RIF-R prevalence and analyzed the rpoB gene mutation in 1615 S. aureus blood isolates (772 methicillin-susceptible and 843 methicillin-resistant S. aureus (MRSA)) from patients with bacteremia, between 2008 and 2017. RIF-R prevalence and antimicrobial susceptibility were determined. Multilocus sequence typing was used to characterize the isolate’s molecular epidemiology; Staphylococcus protein A (spa), staphylococcal cassette chromosome mec (SCCmec), and rpoB gene mutations were detected by PCR. Among 52 RIF-R MRSA isolates out of 57 RIF-R S. aureus blood isolates (57/1615, 0.4%; 5 methicillin-susceptible and 52 MRSA), ST5 (44/52, 84.6%), SCCmec IIb (40/52, 76.9%), and spa t2460 (27/52, 51.9%) were predominant. rpoB gene mutations with amino acid substitutions showed that A477D (17/48, 35.4%) frequently conferred high-level RIF resistance (MIC > 128 mg/L), followed by H481Y (4/48, 8.3%). RIF-R S. aureus blood isolates in South Korea have unique molecular characteristics and are closely associated with rpoB gene mutations. RIF-R surveillance through S. aureus–blood isolate epidemiology could enable effective therapeutic management.
1. Introduction
Staphylococcus aureus (S. aureus) is one of the most notorious and clinically significant pathogens in both community-acquired and nosocomial infections that causes various illnesses, which range from relatively minor local infections to life-threatening systemic infections [1]. Widespread antibiotic use has contributed to the emergence of multidrug-resistant S. aureus strains, particularly those that are resistant to methicillin, and this has become a global concern [2]. Rifampin is an antimicrobial agent with activity against multidrug-resistant S. aureus, including methicillin-resistant S. aureus (MRSA). Thus, rifampin is currently indicated in combination therapy for S. aureus bacteremia, prosthetic joint infections, and prosthetic valve endocarditis [3,4,5]. However, in MRSA, rifampin resistance (RIF-R) has emerged and is attributed to rpoB gene mutations [6]. RIF-R prevalence has rapidly increased to create a high RIF-R resistance rate [7,8]. The molecular characteristics of RIF-R and rpoB mutations in clinical MRSA [8,9] and those of S. aureus bloodstream isolates [10,11,12] have been previously evaluated. However, in South Korea, an accurate understanding of these characteristics is lacking, as there is limited information on the molecular characteristics of RIP-R and rpoB mutations in S. aureus infections. A recent study identified rifamycin resistance and rpoB gene mutations in Staphylococcus species isolates from prosthetic joint infections [13]; nonetheless, there is a paucity of data on the molecular epidemiology and prevalence of RIF-R and rpoB mutations in S. aureus isolates from patients with bacteremia in South Korea. With regard to treatment and infection control, the dissemination of RIF-R isolates in S. aureus bacteremia may pose a serious threat, and this highlights the importance of understanding the molecular epidemiology of S. aureus isolates.
Therefore, we aimed to investigate the prevalence of RIF-R and analyze the rpoB gene mutation in S. aureus isolates obtained from patients with bacteremia.
2. Results
2.1. Characteristics of Study Isolates and Profile of rpoB Gene Mutations
The study sample comprised 1615 S. aureus, 843 MRSA, and 772 MSSA isolates obtained from patients with bacteremia. Among these, 57 (57/1615, 0.4%) were resistant to RIF. Antibiotic susceptibility testing using the broth microdilution method revealed that 52 RIF-R S. aureus isolates (52/57, 91.2%) were MRSA, and five RIF-R S. aureus isolates (5/57, 8.8%) were methicillin-susceptible S. aureus (MSSA). RIF resistance was more common in MRSA than in MSSA (p < 0.001) isolates.
Among the 52 RIF-R MRSA isolates, 51 isolates (51/52, 98.1%) showed high-level rifampin resistance (MIC ≥ 8 mg/L), and 1 isolate (1/52, 1.9%) showed low-level rifampin resistance (MIC 4 mg/L) (Figure 1). No mutations in the rifampin resistance-determining region (RRDR) of the rpoB gene were detected in 4 of the 52 RIP-R MRSA isolates (4/52, 7.7%), though these isolates were phenotypically resistant to RIF. Amplification of the 714-bp RRDR region of the rpoB gene processed for DNA sequence analysis and the MIC distributions for RIP related to rpoB mutations are shown in Table 1. rpoB gene mutations were detected in 41 out of 48 (41/48, 85.4%) isolates compared with the rpoB gene sequence of the reference strain. We identified 19 different types of mutations in rpoB gene mutation analysis, with 33 single mutations (33/48, 68.8%) and 15 multiple mutations (15/48, 31.3%). Of the 48 isolates, 17 (17/48, 35.4%) showed a mutation at codon 477 (gct to gat) and presented the amino acid substitution A477D, which was associated with high-level RIF resistance (MIC > 128 mg/L). We found several amino acid substitutions of H481Y and S468P that were common mutations of the rpoB proteins and some relatively rare mutations of R484H, S486L, and D471G that some previous studies have reported [2,8]. In addition, some isolates in our study carried rare mutations in RRDR, such as Q486L, Q486R, H481R, E568K, and R197L. However, when we sequenced 50 RIF-S MRSA strains as a negative control, we found that there was no detection of amino acid substitutions in any of the 50 RIF-S S. aureus isolates.
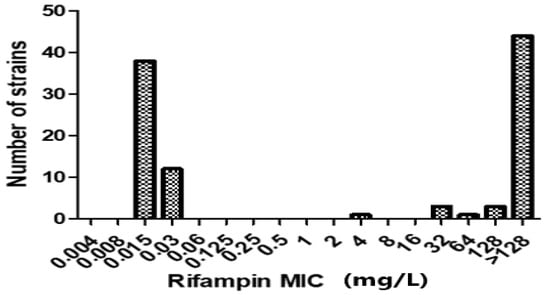
Figure 1.
Distribution of rifampin (RIF) minimum inhibitory concentrations (MICs) for 52 RIF-R MRSA isolates. Of the 52 isolates, 51 (98.1%) showed high-level RIF resistance (MIC ≥ 8 mg/L), and only 1 (1.9%) had low-level RIF resistance (MIC 4 mg/L). Additionally, the broth microdilution method was used to determine the MIC distributions of 50 RIF-susceptible MRSA isolates.

Table 1.
Correlation of rpoB gene mutations and the level of resistance to rifampin in MRSA isolates.
2.2. Antimicrobial Resistance Profiles of RIF-R MRSA Isolates
The antibiotic resistance profiles of the 52 RIF-R MRSA isolates are shown in Figure 2. Our results revealed that the resistance levels of 52 RIF-R MRSA isolates were as follows: 100% to ampicillin, 94.2% to clindamycin, 92.3% to ciprofloxacin, 94.2% to erythromycin, 76.9% to fusidic acid, 73.1% to gentamicin, 100% to oxacillin, 100% to penicillin, 71.1% to tetracycline, 0% to quinupristin, and 7.7% to trimethoprim–sulfamethoxazole (TMP-SMX).
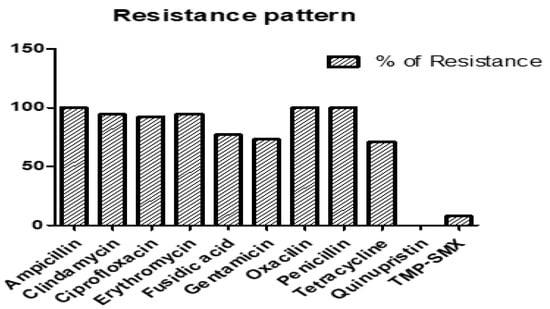
Figure 2.
Percentage of antibiotic resistance in 52 rifampin (RIF)-R MRSA isolates.
2.3. Molecular Typing
Among the total of 1615 S. aureus isolates (843 MRSA and 772 MSSA), ST5 was the predominant ST (543/1615, 33.6%), followed by ST72 (403/1615, 25.0%). Of the 57 RIF-R S. aureus isolates (52 MRSA and 5 MSSA), ST5 was predominant (44/57, 77.2%), and there were 6 non-ST5 isolates (1 ST72, 2 ST254, 1 ST1, 1 ST6, 1 ST101, and 1 ST45). In 1558 RIF-S S. aureus isolates (791 MRSA and 767 MSSA), 463 ST5 (29.7%) in MRSA and 137 ST72 (17.9%) in MSSA were the predominant STs.
Staphylococcal chromosomal cassette mec (SCCmec) typing, MLST, and spa typing were performed on the 52 RIF-R MRSA strains (Table 2). SCCmec typing revealed that the most common type was SCCmec type IIb (40/52, 76.9%), followed by IVa (7/52, 13.5%), Ⅰc (2/52, 3.8%), II (2/52, 3.8%), and IIa (1/52, 1.9%). The results of spa typing revealed a diverse distribution of the 52 RIF-R MRSA strains. The most predominant spa type was t2460 (27/52, 51.9%), followed by t002 (4/52, 7.7%) and t324 (4/52, 7.7%). Four spa-type isolates remained undetermined.

Table 2.
Molecular characteristics of the 52 rifampin-resistant MRSA isolates.
We found that the predominant RIF-R MRSA was the ST5-Ⅱb-t2460 (26/52, 50%) molecular type, which had high resistance to RIF. Among the MSSA isolates, ST72-I-t126 (2/5, 40%) isolates from persistent carriers were resistant to RIF.
2.4. δ-Hemolysin Activities As Genotypic Characteristics of S. aureus Isolates by Rifampin
Table 3 shows the differences in the genotypic characteristics of S. aureus isolates with RIF susceptibility and resistance. The agr functionality test was performed on 1615 S. aureus isolates. Fourteen (24.6%) functional agr (hemolytic strains) were resistant to RIF, and 835 (53.6%) agr (hemolytic strains) were susceptible to RIF. There were 43 (75.4%) dysfunctional agr (nonhemolytic strains) resistant to RIF and 723 (46.4%) dysfunctional agr (nonhemolytic strains) susceptible to RFP.

Table 3.
Genotypic characteristics of the S. aureus isolates stratified by their rifampin-resistance status.
3. Discussion
In this study, we evaluated the molecular characteristics and prevalence of RIF resistance in S. aureus isolates obtained from patients with bacteremia. Our results suggested that RIF resistance was detected in 0.4% (57/1615) of S. aureus isolates associated with bloodstream infections, and ST5-SCCmec Ⅱb-spat 2460 was the most common type of RIF-R S. aureus isolate. Amino acid A477D substitution was most common in RIF-R MRSA isolates that harbored multiple mutations, which conferred high-level RIF resistance (MIC > 128 mg/L). Notably, the RIF-R MRSA isolates showed high rates of resistance to various antibiotics.
Several recent studies have reported the geographically diverse molecular epidemiology of prevalent MRSA clones in bacteremia, with a high prevalence of ST5 and ST59 (People’s Republic of China) [14,15,16], ST 239 (Turkey) [12], ST239 (Iran and Kuwait) [11,17], and ST1011 (Mexico) [10]. However, limited information is available regarding the molecular epidemiology of S. aureus in patients with bacteremia in South Korea. ST5-SCCmecII and ST239-SCCmecIII clones were predominant among a relatively small number of blood isolates (n, 45–96) [18,19]. Community-associated (CA) MRSA strains of ST72-SCCmec IV accounted for a major proportion of MRSA blood isolates (from 76 to 83) in South Korea [20,21,22]. A key strength of our study was the evaluation of a large number of S. aureus blood isolates (n = 1615); ST5-SCCmecII (543/1615, 33.6%) and ST72-SCCmecIV (403/1615, 25.0%) were the major clones in South Korea, which reinforced the results of previous molecular epidemiology studies. To the best of our knowledge, this is the first study to evaluate the molecular characteristics of RIF-R S. aureus blood isolates in South Korea, and we identified molecular characteristics distinct from those reported in previous studies on RIF-R S. aureus clinical isolates [8,9,13,23].
Mutations conferring RIF-R are almost exclusively confined to the rpoB gene in most microorganisms [24], and rpoB mutations can be associated with high-level RIF resistance [25]. In our study, we amplified and sequenced portions of rpoB from RIF-R S. aureus isolates. High-level RIF resistance may be attributed to additional mutations within the rpoB gene, as previously described [26]. Though sequencing of the RRDR successfully identified mutations in RIF-R MRSA isolates, four RIF-R MRSA isolates (4/52, 7.7%) lacked any mutation in the RRDR region of the rpoB gene despite phenotypic resistance to RIF. This difference might persist because of genotypic variations that are prevailing worldwide, and the presence of mutations in the rpoB gene other than in the RRDR region could not be excluded [27]. The lack of RRDR mutations in RIF-R MRSA isolates may be due to the presence of other rare rpoB mutations or another mechanism of RIF-R [27], which is supported by previous reports from several countries [28,29,30]. We posit that the four isolates (7.7%) likely possessed additional mutations outside the RRDR because they exhibited higher RIF-R levels than those conferred by the mutations in their RRDR alleles. On the contrary, 15 multiple mutations (15/48, 31.3%) were identified in rpoB gene mutation analysis in our study, all of which were associated with high-level RIF-R (MIC ≥ 8 mg/L). Although most rpoB gene mutations were correlated with high-level resistance [6], it is difficult to identify how much each mutation contributed to the increased MIC. Further molecular analysis of rpoB gene mutations in RIF-R S. aureus isolates to identify the correlation between high-level MIC and mutation at specific codons will provide higher robustness on isolates’ ability to survive and affect clinical practice in the region of endemicity.
The amino acid substitution A477D was the most prevalent in our study, which was inconsistent with findings reported from previous studies that showed the H481Y MRSA clone was predominant [8,9,13,31]. Furthermore, the transformation of A477D-mutated rpoB into wild-type S. aureus strains resulted in a RIF-R phenotype, indicating that these mutations contribute to rifampicin resistance in S. aureus. In the A477D mutant, this substitution places a negatively charged carboxylate unit in close proximity to an existing carboxylate from H481, which is situated near the protein–DNA interface and thereby increases the negative charge on the surface of the protein and destabilizes the enzyme–DNA interaction, owing to electrostatic repulsion [32]. Even if the abovementioned substitution does not face the RIF target region, it can induce a conformational change that indirectly prevents antibiotic binding to the target site, which determines the RIF-R [8]. Another interesting finding of our study was that the analyzed isolates lacked the L466S mutation, which was one of the most commonly found mutations in the clinical isolates of other studies [7,9]. We posit that there should be regional differences in the mutation codon of RIF-R blood S. aureus isolates, given that only limited information is available regarding the molecular characteristics of rpoB mutations of blood S. aureus isolates in multiple countries, including South Korea. Future multi-center studies using isolates of national or global pathogen resource networks to further characterize the mutations associated with RIF-F and investigate new mutations are essential.
rpoB mutations are associated not only with resistance to RIF but also resistance to other last-line antibiotics for MRSA, including vancomycin and daptomycin [33,34]. In particular, the reduced susceptibility to vancomycin that is linked to rpoB mutations may result in poor clinical outcomes and persistent MRSA infections [35]. We posit that the potentially decreased susceptibility to vancomycin promoted persistent infection which was associated with the rpoB-A477D mutation; this aspect may be a critical concern in the treatment of MRSA bacteremia because the A477D mutant was most prevalent in our 52 RIF-R MRSA isolates from patients with bacteremia [2,33,34]. In addition, given that RIF exposure can be the main selective pressure for decreased susceptibility to RIF and vancomycin and that a higher prevalence of RIF resistance in ST5 was prevalent in previous large-scale S. aureus genome studies [2], judicious use of RIF and preventive infection-control measures against the spread of RIF-R S. aureus clones should be prioritized based on large-scale genomic surveillance in the fight against S. aureus bacteremia. Further research is required to understand whether cross-resistance to other antibiotics develops during prolonged RIF exposure or whether resistance rapidly evolves during exposure to antibiotics [36].
Our study has certain limitations. First, this study was conducted at a single center. Second, we analyzed the clinical outcomes and risk factors of RIF-R S. aureus isolates obtained from patients with bacteremia. Third, we did not evaluate the molecular characteristics of the RIF-R MSSA isolates. We posit that our study could be strengthened further if we had sequenced more sufficient RIF-R and RIF-S S. aureus isolates, and it has limitations in being able to acquire strong confirmation that RIF-R in S. aureus isolates was caused by a rpoB mutation. However, our study included a relatively large number of S. aureus isolates from bacteremia (n = 1615) with RIF-R MRSA isolates (n = 52) and was conducted in a 2700-bed tertiary-care teaching hospital where patients from across the country were admitted during a 10-year period. Therefore, our study has important clinical implications for antibiotic use and provides useful information for preventive infection-control measures based on the molecular epidemiology of RIF-R S. aureus blood isolates in South Korea.
4. Materials and Methods
4.1. Collection of S. aureus Isolates
The study sample consisted of S. aureus strain isolates obtained from Asan Medical Center, Seoul, Republic of Korea, from 2008 to 2017. The S. aureus samples were plated on a blood agar plate. This sterile medium was streaked with a cotton swab, and the plates were incubated overnight at 37 °C. The isolate was grown to screen for and analyze S. aureus. The strains were stored in 20% glycerol-tryptic soy broth at −80 °C (Becton Dickinson, Sparks, MD, USA). We determined the methicillin resistance of S. aureus isolates based on the oxacillin MIC and the presence of the mecA gene.
4.2. Agr Functionality Test
We used δ-hemolysin activity to determine agr functionality by cross-streaking vertically to RN4220 and a test strain on a sheep blood agar plate (BAP). The β-hemolysin produced by RN4220 enables detection of δ-hemolysin [37]. δ-hemolytic activity was indicated by an enhanced area of hemolysis at the intersection of the streaks.
4.3. Antimicrobial Susceptibility Tests
The antimicrobial susceptibility profiles of the S. aureus isolates were determined using the broth microdilution method according to the Clinical and Laboratory Standard Institute (CLSI) guidelines [38]. For the broth microdilution method, serial two-fold dilutions were carried out in cation-adjusted Mueller–Hinton II broth (Becton Dickinson, Sparks, MD, USA) in microtiter plates according to standard criteria [38]. The MIC was determined using the broth microdilution method, and each spot was inoculated with 106 CFU. After incubation at 37 °C for 16–20 h, the MIC value was considered when the bacteria did not grow at the minimum antibiotic concentration. The reference strain from the American Type Culture Collection 29,213 was used for quality control. The MIC for rifampin (Sigma, Darmstadt, Germany) was determined using the broth microdilution method, following standard criteria [38]. Based on the CLSI guidelines, the isolates were classified as RIF susceptible (MIC ≤ 1 mg/L) or RIF resistant (MIC ≤ 4 mg/L).
4.4. Molecular Typing
4.4.1. Detection of the mecA Gene
The mecA gene sequence (532 bp) of all the MRSA isolates was amplified by PCR. The amplification of the mecA gene was achieved using mecA1 (5′ AAA ATC GAT GGT AAA GGT TGG C 3′) and mecA2 (3′ AGT TCT GCA GTA CCG GAT TTG C 5′) primers and sequence analysis. The PCR conditions involved an initial denaturation for 3 min, followed by 30 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C 30 s and a final extension at 72 °C for 4 min. The PCR products (10 μL) were separated on a 1% agarose gel in 0.5 × Tris-borate-EDTA buffer at 100 V and visualized using RedSafe.
4.4.2. Multilocus Sequence Typing
Multilocus sequence typing (MLST) of the isolates was conducted by amplifying internal fragments of seven housekeeping genes of S. aureus, as described previously [39]. The fragments were amplified using the following primers (reference): carbamate kinase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glpF), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL). Following the purification and sequencing of these genes, allele quantification and sequence typing were performed using a well-characterized online database (https://pubmlst.org/, accessed on 10 August 2023).
4.4.3. SCCmec Typing
The SCCmec typing of the MRSA isolates was performed using the multiplex PCR method described by Oliveira and de Lencastre [40]. The eight loci (A through H) and specific pairs of primers for SCCmec types and subtypes I, II, III, and IV have been described previously [41]. The multiplex PCR conditions included an initial denaturation at 4 min, followed by 30 cycles at 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 1 min as well as a final extension at 72 °C for 4 min. The PCR products (10 μL) were separated on a 1.8% agarose gel in 0.5 × Tris-borate-EDTA buffer at 135 V and visualized using RedSafe.
4.4.4. Spa Typing
The Staphylococcus protein A (spa) variable repeat region from each MRSA isolate was amplified using simplex PCR oligonucleotide primers, as described previously [40,42]. The purified spa PCR products were sequenced, and the typing of spa was performed using the public spa database website (http://spa.ridom.de/, accessed on 10 August 2023) for all S. aureus isolates.
4.5. PCR Detection of Rifampin Resistance-Associated Mutations
Template DNA for PCR was obtained using a WizPrep gDNA Mini Kit (Seongnam, Republic of Korea). Total DNA from S. aureus was purified and used as a template for PCR amplification. The rifampin resistance-determining region (RRDR) of the rpoB gene sequence, measuring 690 bp (nucleotides 1307–2020), was amplified using PCR. Amplification of rpoB was performed using RRDR1 (5′ TTC AAG ATA CTG AGT CTA TCA CAC C 3′) and rpoB RRDR2 (3′ GCA CG T GAT TCT GGT GCA GCT ATT A 5′) primers, followed by sequence analysis. Amplification was performed for 52 RIF-R and 50 RIF-S strains as previously described [43], and the 50 RIF-S strains were sequenced as a negative control: 5 oligonucleotide primers rpoB1-F (5′ ATG GTA TTT AGC TAA AAG CGG 3′), rpoB1-R (3′ GCA CTG AAA ACA CTG AAC AA 5′), rpoB2-F (5′ ATT AGG TTT CTC AAG TGA CC 3′), rpoB2-R (3′ CCA TTA GCT GAG TTA ACG CAT 5′), rpoB3-F (5′ AAG CAG TGC CTT TGA ATC C 3′), rpoB3-R (3′ CCT AAA GGT GTA ACT GAG TT 5′), rpoB4-F (5′ TTG GTG CAG AAG TAA AAG ATG G 3′), rpoB4-F (3′ GGT GTA ATG TAC ATG TTG AA 5′), rpoB5-F (5′ AAT CTT GGT ATT CAC GTT GC 3′), and rpoB5-R (3′ GCT GAA TTT TAT TGA TT 5′) were identified using for mutation. The PCR products were purified and analyzed using DNA sequencing. PCR was performed in a DNA thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA). The rpoB PCR cycling programs consisted of an initial denaturation (4 min at 94 °C) followed by 35 cycles of denaturation (30 s at 94 °C), annealing (45 s at 53 °C), and extension (45 s at 72 °C), with a final extension (3 min at 72 °C). The except RRDR PCR cycling programs consisted of an initial denaturation (4 min at 94 °C) followed by 35 cycles of denaturation (45 s at 94 °C), annealing (45 s at 50 °C), and extension (1 min at 72 °C), with a final extension (10 min at 72 °C). DNA sequencing was performed by COSMO Genetech (Seoul, Republic of Korea). The nucleotide sequences obtained were compared to the rpoB wild-type sequence from S. aureus subsp. aureus (GenBank accession number: N315) using clustalw software (https://www.genome.jp/tools-bin/clustalw, accessed on 10 August 2023).
4.6. Statistical Analysis
The statistical distribution and clinical characteristics of the patients were compared using cross-analysis and the chi-square test, respectively. Statistical significance was set at p < 0.05. All statistical analyses were performed using SPSS version 24.0 (SPSS Inc., Chicago, IL, USA).
5. Conclusions
We showed that 0.4% of S. aureus blood isolates (57/1615) demonstrated RIF resistance in a tertiary care hospital in South Korea during a 10-year period. The RIF-R S. aureus blood isolates had unique molecular characteristics, revealing that RIF-R in S. aureus was highly related to mutations in rpoB and ST5-SCCmec Ⅱb-spat 2460, with high-level RIF resistance being the most predominant type. A surveillance system based on the molecular epidemiology of S. aureus blood isolates should be implemented in hospitals to improve clinical treatment and infection prevention strategies.
Author Contributions
Conceptualization, Y.S.K.; methodology, Y.E. and Y.S.K.; software, E.K. and Y.E.; validation, Y.K.K. and Y.S.K.; formal analysis, Y.K.K. and Y.S.K.; investigation, Y.K.K.; resources, Y.E., E.K., E.C., S.B., J.J., M.J.K., Y.P.C., S.-H.K., S.-H.C. and S.-O.L.; data curation, Y.K.K. and Y.E.; writing—original draft preparation, Y.K.K. and Y.E.; writing—review and editing, Y.K.K. and Y.S.K.; visualization, Y.E.; supervision, Y.S.K.; project administration, Y.S.K.; funding acquisition, Y.S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Yang Soo Kim, with a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (grant number HV22C2029).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the Asan Medical Center.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Guérillot, R.; da Silva, A.G.; Monk, I.; Giulieri, S.; Tomita, T.; Alison, E.; Porter, J.; Pidot, S.; Gao, W.; Peleg, A.Y.; et al. Convergent evolution driven by rifampin exacerbates the global burden of drug-resistant Staphylococcus aureus. mSphere 2018, 3, e00550-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Bayer, A.; Cosgrove, S.E.; Daum, R.S.; Fridkin, S.K.; Gorwitz, R.J.; Kaplan, S.L.; Karchmer, A.W.; Levine, D.P.; Murray, B.E.; et al. Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 2011, 52, e18–e55. [Google Scholar] [CrossRef] [PubMed]
- Osmon, D.R.; Berbari, E.F.; Berendt, A.R.; Lew, D.; Zimmerli, W.; Steckelberg, J.M.; Rao, N.; Hanssen, A.; Wilson, W.R. Diagnosis and management of prosthetic joint infection: Clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 2013, 56, e1–e25. [Google Scholar] [CrossRef] [PubMed]
- Habib, G.; Lancellotti, P.; Antunes, M.J.; Bongiorni, M.G.; Casalta, J.P.; Del Zotti, F.; Dulgheru, R.; El Khoury, G.; Erba, P.A.; Iung, B.; et al. 2015 ESC Guidelines for the management of infective endocarditis: The Task Force for the Management of Infective Endocarditis of the European Society of Cardiology (ESC). Endorsed by: European Association for Cardio-Thoracic Surgery (EACTS), the European Association of Nuclear Medicine (EANM). Eur. Heart J. 2015, 36, 3075–3128. [Google Scholar] [CrossRef] [PubMed]
- Aubry-Damon, H.; Soussy, C.J.; Courvalin, P. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1998, 42, 2590–2594. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Shan, W.; Ma, X.; Chang, W.; Zhou, X.; Lu, H.; Dai, Y. Molecular characterization of rifampicin-resistant Staphylococcus aureus isolates in a Chinese teaching hospital from Anhui, China. BMC Microbiol. 2012, 12, 240. [Google Scholar] [CrossRef] [PubMed]
- Bongiorno, D.; Mongelli, G.; Stefani, S.; Campanile, F. Burden of rifampicin- and methicillin-resistant Staphylococcus aureus in Italy. Microb. Drug Resist. 2018, 24, 732–738. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, B.; Rao, L.; Wang, X.; Zhao, H.; Li, M.; Yu, F. Molecular characteristics of rifampin-sensitive and -resistant isolates and characteristics of rpoB gene mutations in methicillin-resistant Staphylococcus aureus. Infect. Drug Resist. 2021, 14, 4591–4600. [Google Scholar] [CrossRef]
- Vazquez-Rosas, G.J.; Merida-Vieyra, J.; Aparicio-Ozores, G.; Lara-Hernandez, A.; De Colsa, A.; Aquino-Andrade, A. Molecular characterization of Staphylococcus aureus obtained from blood cultures of paediatric patients treated in a tertiary care hospital in Mexico. Infect. Drug Resist. 2021, 14, 1545–1556. [Google Scholar] [CrossRef]
- Goudarzi, M.; Seyedjavadi, S.S.; Nasiri, M.J.; Goudarzi, H.; Nia, R.S.; Dabiri, H. Molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from patients with bacteremia based on MLST, SCCmec, spa, and agr locus types analysis. Microb. Pathog. 2017, 104, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Tekeli, A.; Ocal, D.N.; Ozmen, B.B.; Karahan, Z.C.; Dolapci, I. Molecular characterization of methicillin-resistant Staphylococcus aureus bloodstream isolates in a Turkish university hospital between 2002 and 2012. Microb. Drug Resist. 2016, 22, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, S.S.; Choi, S.M.; Bae, C.J.; Oh, T.H.; Kim, S.E.; Kim, U.J.; Kang, S.J.; Jung, S.I.; Park, K.H. Rifamycin resistance, rpoB gene mutation and clinical outcomes of Staphylococcus species isolates from prosthetic joint infections in Republic of Korea. J. Glob. Antimicrob. Resist. 2022, 28, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Wang, X.; Wang, X.; Du, B.; Xu, K.; Zhang, F.; Jiang, C.; Zhao, Y.; Zhu, Y. Molecular characterization and virulence gene profiling of methicillin-resistant Staphylococcus aureus associated with bloodstream infections in southern China. Front. Microbiol. 2022, 13, 1008052. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, D.; Huang, Z.; Zhang, J.; Xie, W.; Liu, P.; Jing, H.; Wang, J. Clonality, virulence genes, and antibiotic resistance of Staphylococcus aureus isolated from blood in Shandong, China. BMC Microbiol. 2021, 21, 281. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; He, W.; Xiao, S.; Wang, S.; Li, X.; Zeng, Q.; Ni, Y.; Han, L. Antimicrobial resistance and molecular epidemiology of Staphylococcus aureus causing bloodstream infections at Ruijin Hospital in Shanghai from 2013 to 2018. Sci. Rep. 2020, 10, 6019. [Google Scholar] [CrossRef] [PubMed]
- Alfouzan, W.; Udo, E.E.; Modhaffer, A.; Alosaimi, A. Molecular characterization of methicillin- resistant Staphylococcus aureus in a tertiary care hospital in Kuwait. Sci. Rep. 2019, 9, 18527. [Google Scholar] [CrossRef]
- Soo Ko, K.; Kim, Y.S.; Song, J.H.; Yeom, J.S.; Lee, H.; Jung, S.I.; Jeong, D.R.; Kim, S.W.; Chang, H.H.; Ki, H.K.; et al. Genotypic diversity of methicillin-resistant Staphylococcus aureus isolates in Korean hospitals. Antimicrob. Agents Chemother. 2005, 49, 3583–3585. [Google Scholar] [CrossRef]
- Cha, H.Y.; Moon, D.C.; Choi, C.H.; Oh, J.Y.; Jeong, Y.S.; Lee, Y.C.; Seol, S.Y.; Cho, D.T.; Chang, H.H.; Kim, S.W.; et al. Prevalence of the ST239 clone of methicillin-resistant Staphylococcus aureus and differences in antimicrobial susceptibilities of ST239 and ST5 clones identified in a Korean hospital. J. Clin. Microbiol. 2005, 43, 3610–3614. [Google Scholar] [CrossRef]
- Kwon, J.C.; Kim, S.H.; Park, S.H.; Choi, S.M.; Lee, D.G.; Choi, J.H.; Park, C.; Shin, N.Y.; Yoo, J.H. Molecular epidemiologic analysis of methicillin-resistant Staphylococcus aureus isolates from bacteremia and nasal colonization at 10 intensive care units: Multicenter prospective study in Korea. J. Korean Med. Sci. 2011, 26, 604–611. [Google Scholar] [CrossRef]
- Park, S.H.; Park, C.; Yoo, J.H.; Choi, S.M.; Choi, J.H.; Shin, H.H.; Lee, D.G.; Lee, S.; Kim, J.; Choi, S.E.; et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated bloodstream infections in Korea. Infect. Control Hosp. Epidemiol. 2009, 30, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.Y.; Lee, H.J.; Lee, M.S. Molecular characteristics of methicillin-resistant Staphylococcus aureus blood isolates: Clonal spread of staphylococcal cassette chromosome mec type IVA between the community and the hospital. Microb. Drug Resist. 2010, 16, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Fang, R.; Wang, C.; Tian, X.; Lin, J.; Zeng, W.; Zhou, T.; Xu, C. Resistance profiles and biological characteristics of rifampicin-resistant Staphylococcus aureus small-colony variants. Infect. Drug Resist. 2021, 14, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Qazi, O.; Rahman, H.; Tahir, Z.; Qasim, M.; Khan, S.; Ahmad Anjum, A.; Yaqub, T.; Tayyab, M.; Ali, N.; Firyal, S. Mutation pattern in rifampicin resistance determining region of rpoB gene in multidrug-resistant Mycobacterium tuberculosis isolates from Pakistan. Int. J. Mycobacteriol. 2014, 3, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Adikaram, C.P.; Perera, J.; Wijesundera, S.S. Geographical profile of rpoB gene mutations in rifampicin resistant Mycobacterium tuberculosis isolates in Sri Lanka. Microb. Drug Resist. 2012, 18, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Frénay, H.M.; Bunschoten, A.E.; Schouls, L.M.; van Leeuwen, W.J.; Vandenbroucke-Grauls, C.M.; Verhoef, J.; Mooi, F.R. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 1996, 15, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Heep, M.; Brandstätter, B.; Rieger, U.; Lehn, N.; Richter, E.; Rüsch-Gerdes, S.; Niemann, S. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 2001, 39, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.P.; Crawford, J.T.; Shinnick, T.M. The rpoB gene of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1994, 38, 805–811. [Google Scholar] [CrossRef]
- Kapur, V.; Li, L.L.; Iordanescu, S.; Hamrick, M.R.; Wanger, A.; Kreiswirth, B.N.; Musser, J.M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 1994, 32, 1095–1098. [Google Scholar] [CrossRef]
- Musser, J.M. Antimicrobial agent resistance in mycobacteria: Molecular genetic insights. Clin. Microbiol. Rev. 1995, 8, 496–514. [Google Scholar] [CrossRef]
- Chuang, Y.Y.; Huang, Y.C. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect. Dis. 2013, 13, 698–708. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, A.J.; Huovinen, T.; Fishwick, C.W.; Chopra, I. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob. Agents Chemother. 2006, 50, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Cui, L.; Katayama, Y.; Kozue, K.; Hiramatsu, K. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol. 2011, 49, 2680–2684. [Google Scholar] [CrossRef] [PubMed]
- Bæk, K.T.; Thøgersen, L.; Mogenssen, R.G.; Mellergaard, M.; Thomsen, L.E.; Petersen, A.; Skov, S.; Cameron, D.R.; Peleg, A.Y.; Frees, D. Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob. Agents Chemother. 2015, 59, 6983–6991. [Google Scholar] [CrossRef]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Liao, C.H.; Chen, S.Y.; Yang, C.J.; Hsu, H.S.; Teng, L.J.; Hsueh, P.R. Characterization of rifampin-resistant Staphylococcus aureus nasal carriage in patients receiving rifampin-containing regimens for tuberculosis. Infect. Drug Resist. 2018, 11, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Traber, K.E.; Lee, E.; Benson, S.; Corrigan, R.; Cantera, M.; Shopsin, B.; Novick, R.P. agr function in clinical Staphylococcus aureus isolates. Microbiology 2008, 154, 2265–2274. [Google Scholar] [CrossRef]
- Wayne, P.A. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Six Informational Supplement M100-S26; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Koreen, L.; Ramaswamy, S.V.; Graviss, E.A.; Naidich, S.; Musser, J.M.; Kreiswirth, B.N. spa typing method for discriminating among Staphylococcus aureus isolates: Implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 2004, 42, 792–799. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 5026–5033. [Google Scholar] [CrossRef]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef]
- Mick, V.; Domínguez, M.A.; Tubau, F.; Liñares, J.; Pujol, M.; Martín, R. Molecular characterization of resistance to Rifampicin in an emerging hospital-associated Methicillin-resistant Staphylococcus aureus clone ST228, Spain. BMC Microbiol. 2010, 10, 68. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).