Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review
Abstract
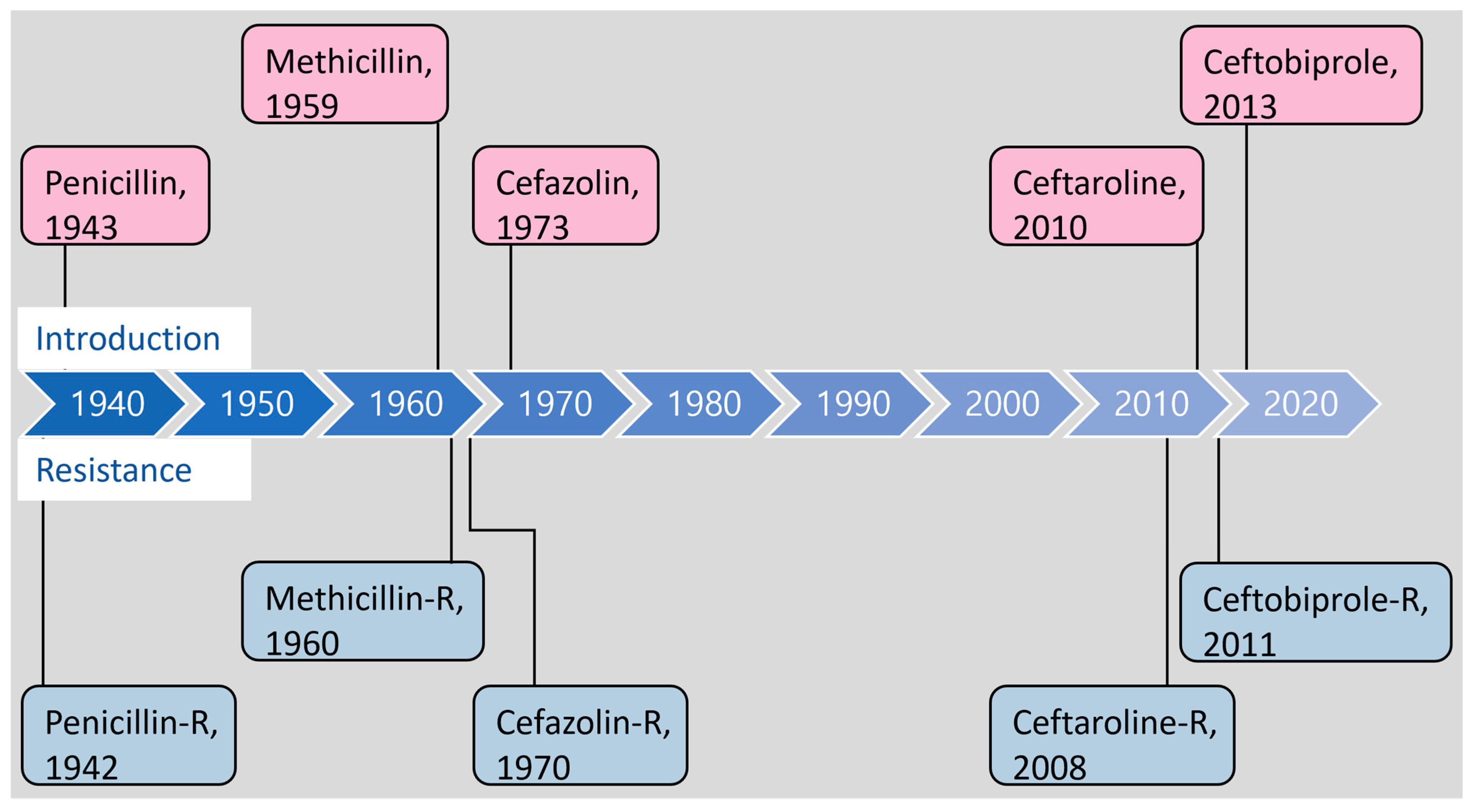
1. Introduction
2. SCCmec as a Carrier of Methicillin Resistance
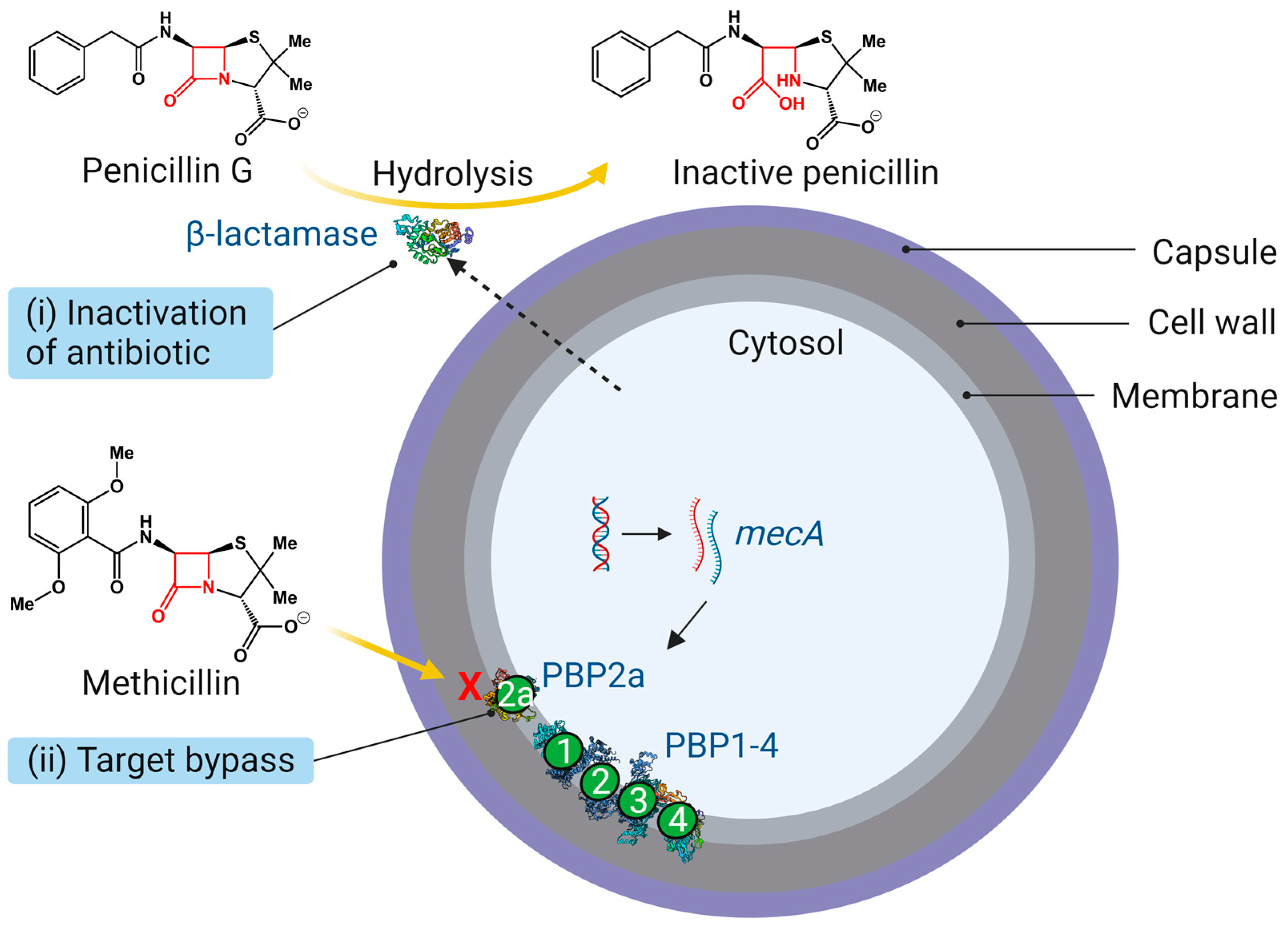
3. Molecular Mechanisms of β-Lactam Resistance
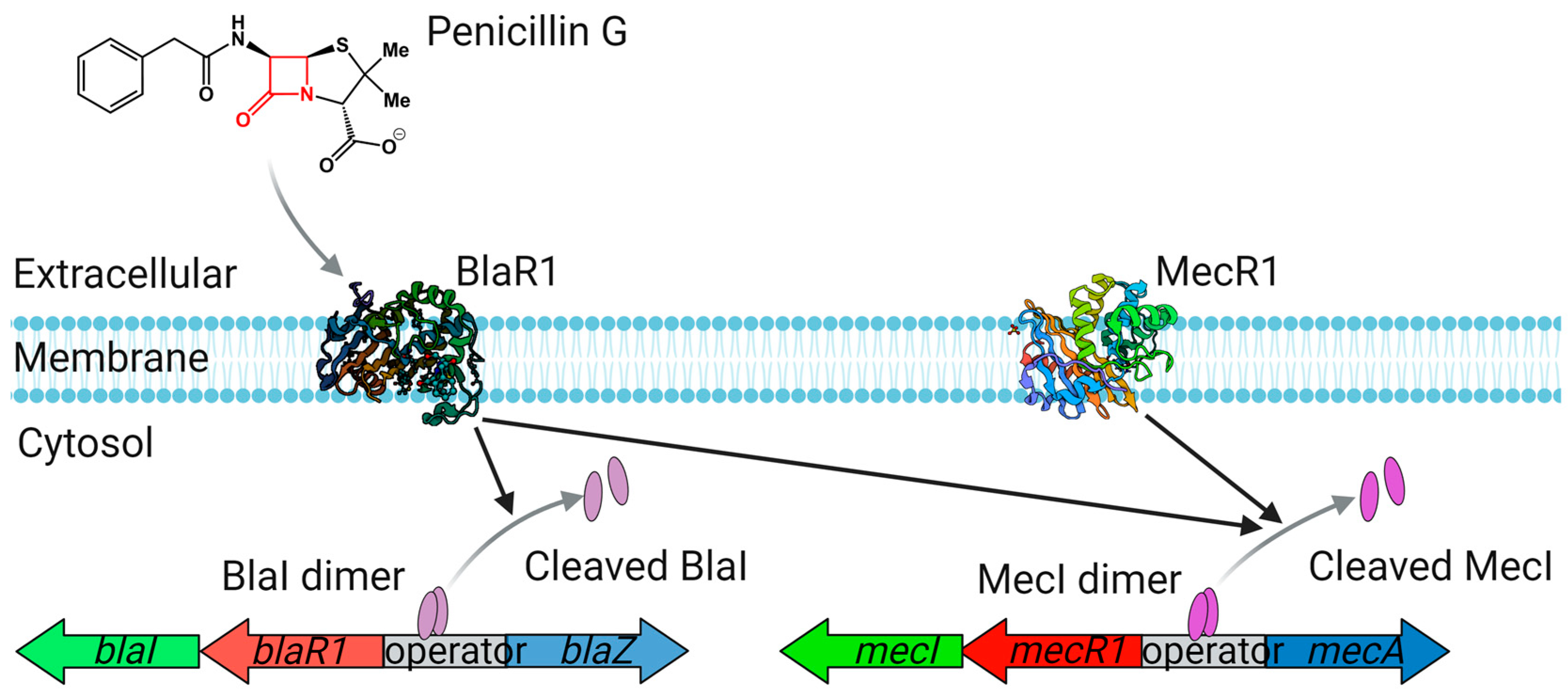
3.1. β-Lactamases
3.2. Penicillin-Binding Proteins (PBP1-4 and PBP2a)
3.3. PBP2a Mutation and New-Generation Cephalosporin Resistance
| β-Lactam Antibiotics | Year of Discovery/ Introduced or Approved | Year of Resistance Reported | Resistance Determinant/Gene Product |
|---|---|---|---|
| Penicillin G | 1928 [2]/1943 | 1942 [6] | blaZ/BlaZ [7] (β-lactamase hydrolyzes the peptide bond in the β-lactam ring) |
| Methicillin # | 1959 [9]/-- | 1960 [10] | mecA/PBP2a [12,13] (PBP2a has a low affinity for methicillin) |
| Oxacillin | 1960/U.S. FDA 1971 | mecA/PBP2a | |
| Cefoxitin * | 1974 [94]/U.S. FDA 1978 | mecA/PBP2a | |
| Cefazolin | 1970 [95]/U.S. FDA 1973 | 1970 [96] | mecA/PBP2a [97] |
| Ceftaroline | 2003 (Ceftaroline fosamil) [98]/U.S. FDA 2010, E.U. EMA 2012 | 2013 [90] | mecA/PBP2a Mutations in mecA [88,90], pbp2, pbp4 [93,99,100], clpX, and gdpP [93,101] |
| Ceftobiprole | 1998 [102]/E.U. EMA 2013 | 2008 [103] | mecA/PBP2a Mutations in mecA [103], pbp2, pbp4 [93,104], clpX, and gdpP [93,101] |
4. Genetic Factors Modulating β-Lactam Resistance
4.1. ClpXP System
4.2. Cyclic-di-AMP Phosphodiesterase (GdpP)
4.3. RNA Polymerase (RpoB/RpoC)
4.4. Accessory Gene Regulator (Agr) System
4.5. Staphylococcal Accessory Regulator A (SarA)
4.6. Serine/Threonine Kinase and Phosphatase (Stk1/Stp1)
4.7. FtsH Protease
4.8. AuxA and AuxB
4.9. PrsA
4.10. PBP4
4.11. Filamentous Temperature-Sensitive Protein Z (FtsZ)
4.12. D-alanyl Carrier Protein Ligase DltA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Lowy, F.D. Staphylococcus aureus Infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Abraham, D.J. Burger’s Medicinal Chemistry, Drug Discovery and Development; Wiley: Hoboken, NJ, USA, 2021; ISBN 9781119821489. [Google Scholar]
- Brown, N.M.; Goodman, A.L.; Horner, C.; Jenkins, A.; Brown, E.M. Treatment of methicillin-resistant Staphylococcus aureus (MRSA): Updated guidelines from the UK. JAC-Antimicrob. Resist. 2021, 3, dlaa114. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Bayer, A.; Cosgrove, S.E.; Daum, R.S.; Fridkin, S.K.; Gorwitz, R.J.; Kaplan, S.L.; Karchmer, A.W.; Levine, D.P.; Murray, B.E.; et al. Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children. Clin. Infect. Dis. 2011, 52, e18–e55. [Google Scholar] [CrossRef] [PubMed]
- Rammelkamp, C.H.; Maxon, T. Resistance of Staphylococcus aureus to the Action of Penicillin. Exp. Biol. Med. 1942, 51, 386–389. [Google Scholar] [CrossRef]
- Kirby, W.M.M. Extraction of a highly potent penicillin inactivator from penicillin resistant staphylococci. Science 1944, 99, 452–453. [Google Scholar] [CrossRef]
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. Nature 1940, 146, 837. [Google Scholar] [CrossRef]
- Knox, R. A New Penicillin (BRL 1241) Active Against Penicillin-resistant Staphylococci. BMJ 1960, 2, 690–693. [Google Scholar] [CrossRef]
- Jevons, M.P. “Celbenin”—resistant Staphylococci. Br. Med. J. 1961, 1, 124–125. [Google Scholar] [CrossRef]
- Harkins, C.P.; Pichon, B.; Doumith, M.; Parkhill, J.; Westh, H.; Tomasz, A.; de Lencastre, H.; Bentley, S.D.; Kearns, A.M.; Holden, M.T.G. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 2017, 18, 130. [Google Scholar] [CrossRef]
- Hartman, B.J.; Tomasz, A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 1984, 158, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Utsui, Y.; Yokota, T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1985, 28, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Matsuhashi, M.; Song, M.D.; Ishino, F.; Wachi, M.; Doi, M.; Inoue, M.; Ubukata, K.; Yamashita, N.; Konno, M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 1986, 167, 975–980. [Google Scholar] [CrossRef]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. Antibiotic-Resistant Bugs in the 21st Century—A Clinical Super-Challenge. N. Engl. J. Med. 2009, 360, 439–443. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef]
- Panlilio, A.L.; Culver, D.H.; Gaynes, R.P.; Banerjee, S.; Henderson, T.S.; Tolson, J.S.; Martone, W.J.; System, N.N.I.S. Methicillin-Resistant Staphylococcus aureus in U.S. Hospitals, 1975–1991. Infect. Control Hosp. Epidemiol. 1992, 13, 582–586. [Google Scholar]
- Dantes, R. National Burden of Invasive Methicillin-Resistant Staphylococcus aureus Infections, United States, 2011. JAMA Intern. Med. 2013, 173, 1970–1978. [Google Scholar]
- Klein, E.; Smith, D.L.; Laxminarayan, R. Hospitalizations and Deaths Caused by Methicillin-Resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 2007, 13, 1840–1846. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Assessing the Health Burden of Infections with Antibiotic-Resistant Bacteria in the EU/EEA, 2016–2020; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2022.
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention: Atlanta, Georgia, 2019.
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-Resistant Bacterial Infections in U.S. Hospitalized Patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; Centers for Disease Control and Prevention: Atlanta, Georgia, 2022.
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Mwangi, M.; Chung, M.; Milheirço, C.; de Lencastre, H.; Tomasz, A. The Mechanism of Heterogeneous Beta-Lactam Resistance in MRSA: Key Role of the Stringent Stress Response. PLoS ONE 2013, 8, e82814. [Google Scholar] [CrossRef] [PubMed]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Ryffel, C.; Strässle, A.; Kayser, F.H.; Berger-Bächi, B. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1994, 38, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Finan, J.E.; Rosato, A.E.; Dickinson, T.M.; Ko, D.; Archer, G.L. Conversion of Oxacillin-Resistant Staphylococci from Heterotypic to Homotypic Resistance Expression. Antimicrob. Agents Chemother. 2002, 46, 24–30. [Google Scholar] [CrossRef]
- Gallagher, L.A.; Coughlan, S.; Black, N.S.; Lalor, P.; Waters, E.M.; Wee, B.; Watson, M.; Downing, T.; Fitzgerald, J.R.; Fleming, G.T.A.; et al. Tandem Amplification of the Staphylococcal Cassette Chromosome mec Element Can Drive High-Level Methicillin Resistance in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2017, 61, e0086917. [Google Scholar] [CrossRef]
- Katayama, Y.; Ito, T.; Hiramatsu, K. A New Class of Genetic Element, Staphylococcus Cassette Chromosome mec, Encodes Methicillin Resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000, 44, 1549–1555. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Asada, K.; Suzuki, E.; Okonogi, K.; Yokota, T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 1992, 298, 133–136. [Google Scholar] [CrossRef]
- Boundy, S.; Safo, M.K.; Wang, L.; Musayev, F.N.; O’Farrell, H.C.; Rife, J.P.; Archer, G.L. Characterization of the Staphylococcus aureus rRNA Methyltransferase Encoded by orfX, the Gene Containing the Staphylococcal Chromosome Cassette mec (SCCmec) Insertion Site. J. Biol. Chem. 2013, 288, 132–140. [Google Scholar] [CrossRef]
- Ito, T.; Katayama, Y.; Hiramatsu, K. Cloning and Nucleotide Sequence Determination of the Entire mec DNA of Pre-Methicillin-Resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 1999, 43, 1449–1458. [Google Scholar] [CrossRef]
- García-Álvarez, L.; Holden, M.T.G.; Lindsay, H.; Webb, C.R.; Brown, D.F.J.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Ballhausen, B.; Kriegeskorte, A.; Schleimer, N.; Peters, G.; Becker, K. The mecA Homolog mecC Confers Resistance against β-Lactams in Staphylococcus aureus Irrespective of the Genetic Strain Background. Antimicrob. Agents Chemother. 2014, 58, 3791–3798. [Google Scholar] [CrossRef] [PubMed]
- Tsubakishita, S.; Kuwahara-Arai, K.; Sasaki, T.; Hiramatsu, K. Origin and Molecular Evolution of the Determinant of Methicillin Resistance in Staphylococci. Antimicrob. Agents Chemother. 2010, 54, 4352–4359. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Antignac, A.; Wu, S.W.; Tomasz, A. Penicillin-Binding Proteins and Cell Wall Composition in β-Lactam-Sensitive and -Resistant Strains of Staphylococcus sciuri. J. Bacteriol. 2008, 190, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Couto, I.; de Lencastre, H.; Severina, E.; Kloos, W.; Webster, J.A.; Hubner, R.J.; Sanches, I.S.; Tomasz, A. Ubiquitous Presence of a mecA Homologue in Natural Isolates of Staphylococcus sciuri. Microb. Drug Resist. 1996, 2, 377–391. [Google Scholar] [CrossRef]
- Hanssen, A.-M.; Ericson Sollid, J.U. SCC mec in staphylococci: Genes on the move. FEMS Immunol. Med. Microbiol. 2006, 46, 8–20. [Google Scholar] [CrossRef]
- Hiramatsu, K. Molecular Evolution of MRSA. Microbiol. Immunol. 1995, 39, 531–543. [Google Scholar] [CrossRef]
- Ito, T.; Hiramatsu, K.; Oliveira, D.C.; De Lencastre, H.; Zhang, K.; Westh, H.; O’Brien, F.; Giffard, P.M.; Coleman, D.; Tenover, F.C.; et al. Classification of staphylococcal cassette chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 2009, 53, 4961–4967. [Google Scholar]
- Uehara, Y. Current Status of Staphylococcal Cassette Chromosome mec (SCCmec). Antibiotics 2022, 11, 86. [Google Scholar] [CrossRef]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef]
- Ito, T.; Katayama, Y.; Asada, K.; Mori, N.; Tsutsumimoto, K.; Tiensasitorn, C.; Hiramatsu, K. Structural Comparison of Three Types of Staphylococcal Cassette Chromosome mec Integrated in the Chromosome in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2001, 45, 1323–1336. [Google Scholar] [CrossRef] [PubMed]
- Wielders, C.; Vriens, M.; Brisse, S.; de Graaf-Miltenburg, L.; Troelstra, A.; Fleer, A.; Schmitz, F.; Verhoef, J.; Fluit, A. Evidence for in-vivo transfer of mecA DNA between strains of Staphylococcus aureus. Lancet 2001, 357, 1674–1675. [Google Scholar] [CrossRef] [PubMed]
- Bloemendaal, A.L.A.; Brouwer, E.C.; Fluit, A.C. Methicillin Resistance Transfer from Staphylocccus epidermidis to Methicillin-Susceptible Staphylococcus aureus in a Patient during Antibiotic Therapy. PLoS ONE 2010, 5, e11841. [Google Scholar] [CrossRef]
- Jansen, W.T.M.; Beitsma, M.M.; Koeman, C.J.; Van Wamel, W.J.B.; Verhoef, J.; Fluit, A.C. Novel mobile variants of staphylococcal cassette chromosome mec in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 2072–2078. [Google Scholar] [CrossRef]
- Becker, K.; van Alen, S.; Idelevich, E.A.; Schleimer, N.; Seggewiß, J.; Mellmann, A.; Kaspar, U.; Peters, G. Plasmid-Encoded Transferable mecB -Mediated Methicillin Resistance in Staphylococcus aureus. Emerg. Infect. Dis. 2018, 24, 242–248. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.B.K.; Lampen, J.O. Membrane-bound penicillinases in Gram-positive bacteria. J. Biol. Chem. 1982, 257, 4490–4495. [Google Scholar] [CrossRef]
- Fisher, J.F.; Mobashery, S. β-Lactams against the Fortress of the Gram-Positive Staphylococcus aureus Bacterium. Chem. Rev. 2021, 121, 3412–3463. [Google Scholar] [CrossRef]
- Holden, M.T.G.; Feil, E.J.; Lindsay, J.A.; Peacock, S.J.; Day, N.P.J.; Enright, M.C.; Foster, T.J.; Moore, C.E.; Hurst, L.; Atkin, R.; et al. Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 2004, 101, 9786–9791. [Google Scholar] [CrossRef]
- Wright, G.D. Q&A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 123. [Google Scholar]
- Hackbarth, C.J.; Chambers, H.F. blaI and blaR1 regulate beta-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1993, 37, 1144–1149. [Google Scholar] [CrossRef]
- Zhang, H.Z.; Hackbarth, C.J.; Chansky, K.M.; Chambers, H.F. A Proteolytic Transmembrane Signaling Pathway and Resistance to β-Lactams in Staphylococci. Science 2001, 291, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.D.; Lewis, R.A.; Curnock, S.P.; Dyke, K.G.H. Studies of the repressor (BlaI) of β-lactamase synthesis in Staphylococcus aureus. Mol. Microbiol. 1997, 24, 1025–1037. [Google Scholar] [CrossRef]
- Blázquez, B.; Llarrull, L.I.; Luque-Ortega, J.R.; Alfonso, C.; Boggess, B.; Mobashery, S. Regulation of the expression of the β-lactam antibiotic-resistance determinants in methicillin-resistant Staphylococcus aureus (MRSA). Biochemistry 2014, 53, 1548–1550. [Google Scholar] [CrossRef] [PubMed]
- Herzberg, O.; Moult, J. Bacterial Resistance to β-Lactam Antibiotics: Crystal Structure of β-Lactamase from Staphylococcus aureus PC1 at 2.5 Å Resolution. Science 1987, 236, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.A.N.; Worrall, L.J.; Hu, J.; Vuckovic, M.; Satishkumar, N.; Poon, R.; Sobhanifar, S.; Rosell, F.I.; Jenkins, J.; Chiang, D.; et al. Structural basis of broad-spectrum β-lactam resistance in Staphylococcus aureus. Nature 2023, 613, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Richmond, M.H. Wild-type variants of exopenicillinase from Staphylococcus aureus. Biochem. J. 1965, 94, 584–593. [Google Scholar] [CrossRef]
- Zygmunt, D.J.; Stratton, C.W.; Kernodle, D.S. Characterization of four beta-lactamases produced by Staphylococcus aureus. Antimicrob. Agents Chemother. 1992, 36, 440–445. [Google Scholar] [CrossRef]
- Kernodle, D.S.; Stratton, C.W.; Mcmurray, L.W.; Chipley, J.R.; Mcgraw, P.A. Differentiation of -Lactamase Variants of Staphylococcus aureus by Substrate Hydrolysis Profiles. J. Infect. Dis. 1989, 159, 103–108. [Google Scholar] [CrossRef]
- Kernodle, D.S.; McGraw, P.A.; Stratton, C.W.; Kaiser, A.B. Use of extracts versus whole-cell bacterial suspensions in the identification of Staphylococcus aureus beta-lactamase variants. Antimicrob. Agents Chemother. 1990, 34, 420–425. [Google Scholar] [CrossRef]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Pasquina-Lemonche, L.; Burns, J.; Turner, R.D.; Kumar, S.; Tank, R.; Mullin, N.; Wilson, J.S.; Chakrabarti, B.; Bullough, P.A.; Foster, S.J.; et al. The architecture of the Gram-positive bacterial cell wall. Nature 2020, 582, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Sahl, H.-G. An oldie but a goodie—cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol. 2010, 300, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; Kjos, M.; Veening, J.-W. How to get (a)round: Mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Microbiol. 2013, 11, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.F.F.; Henriques, A.O.; Pinho, M.G.; de Lencastre, H.; Tomasz, A. Role of PBP1 in Cell Division of Staphylococcus aureus. J. Bacteriol. 2007, 189, 3525–3531. [Google Scholar] [CrossRef] [PubMed]
- Atilano, M.L.; Pereira, P.M.; Yates, J.; Reed, P.; Veiga, H.; Pinho, M.G.; Filipe, S.R. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2010, 107, 18991–18996. [Google Scholar] [CrossRef]
- Pinho, M.G.; Errington, J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 2003, 50, 871–881. [Google Scholar] [CrossRef]
- Lovering, A.L.; de Castro, L.H.; Lim, D.; Strynadka, N.C.J. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 2007, 315, 1402–1405. [Google Scholar] [CrossRef]
- Reed, P.; Veiga, H.; Jorge, A.M.; Terrak, M.; Pinho, M.G. Monofunctional Transglycosylases Are Not Essential for Staphylococcus aureus Cell Wall Synthesis. J. Bacteriol. 2011, 193, 2549–2556. [Google Scholar] [CrossRef]
- Wacnik, K.; Rao, V.A.; Chen, X.; Lafage, L.; Pazos, M.; Booth, S.; Vollmer, W.; Hobbs, J.K.; Lewis, R.J.; Foster, S.J. Penicillin-Binding Protein 1 (PBP1) of Staphylococcus aureus Has Multiple Essential Functions in Cell Division. MBio 2022, 13, e0066922. [Google Scholar] [CrossRef]
- Łęski, T.A.; Tomasz, A. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: Evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 2005, 187, 1815–1824. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; Errington, J. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 2004, 55, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; Filipe, S.R.; de Lencastre, H.; Tomasz, A. Complementation of the Essential Peptidoglycan Transpeptidase Function of Penicillin-Binding Protein 2 (PBP2) by the Drug Resistance Protein PBP2A in Staphylococcus aureus. J. Bacteriol. 2001, 183, 6525–6531. [Google Scholar] [CrossRef] [PubMed]
- Barbuti, M.D.; Myrbråten, I.S.; Angeles, D.M.; Kjos, M. The cell cycle of Staphylococcus aureus: An updated review. Microbiologyopen 2023, 12, e1338. [Google Scholar] [CrossRef]
- Reichmann, N.T.; Tavares, A.C.; Saraiva, B.M.; Jousselin, A.; Reed, P.; Pereira, A.R.; Monteiro, J.M.; Sobral, R.G.; VanNieuwenhze, M.S.; Fernandes, F.; et al. SEDS–bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat. Microbiol. 2019, 4, 1368–1377. [Google Scholar] [CrossRef]
- Loskill, P.; Pereira, P.M.; Jung, P.; Bischoff, M.; Herrmann, M.; Pinho, M.G.; Jacobs, K. Reduction of the Peptidoglycan Crosslinking Causes a Decrease in Stiffness of the Staphylococcus aureus Cell Envelope. Biophys. J. 2014, 107, 1082–1089. [Google Scholar] [CrossRef]
- Pinho, M.G.; de Lencastre, H.; Tomasz, A. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J. Bacteriol. 2000, 182, 1074–1079. [Google Scholar] [CrossRef]
- Pinho, M.G.; de Lencastre, H.; Tomasz, A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 2001, 98, 10886–10891. [Google Scholar] [CrossRef]
- Bilyk, B.L.; Panchal, V.V.; Tinajero-Trejo, M.; Hobbs, J.K.; Foster, S.J. An Interplay of Multiple Positive and Negative Factors Governs Methicillin Resistance in Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 2022, 86, e0015921. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Llarrull, L.I.; Fisher, J.F.; Mobashery, S. Molecular Basis and Phenotype of Methicillin Resistance in Staphylococcus aureus and Insights into New β-Lactams That Meet the Challenge. Antimicrob. Agents Chemother. 2009, 53, 4051–4063. [Google Scholar] [CrossRef] [PubMed]
- Pucci, M.J.; Dougherty, T.J. Direct Quantitation of the Numbers of Individual Penicillin-Binding Proteins per Cell in Staphylococcus aureus. J. Bacteriol. 2002, 184, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N.; Mendes, R.E.; Sader, H.S. Ceftaroline activity against pathogens associated with complicated skin and skin structure infections: Results from an international surveillance study. J. Antimicrob. Chemother. 2010, 65, iv17–iv31. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kelley, W.L.; Jousselin, A.; Barras, C.; Lelong, E.; Renzoni, A. Missense mutations in PBP2A affecting ceftaroline susceptibility detected in epidemic hospital-acquired Methicillin-resistant Staphylococcus aureus clonotypes ST228 and ST247 in Western Switzerland archived since 1998. Antimicrob. Agents Chemother. 2015, 59, 1922–1930. [Google Scholar] [CrossRef]
- McNeil, J.C.; Sommer, L.M.; Vallejo, J.G.; Hulten, K.G.; Kaplan, S.L.; Flores, A.R. Reduced Ceftaroline Susceptibility among Invasive MRSA Infections in Children: A Clinical and Genomic Investigation. Antimicrob. Agents Chemother. 2022, 66, e0074522. [Google Scholar] [CrossRef]
- Long, S.W.; Olsen, R.J.; Mehta, S.C.; Palzkill, T.; Cernoch, P.L.; Perez, K.K.; Musick, W.L.; Rosato, A.E.; Musser, J.M. PBP2a Mutations Causing High-Level Ceftaroline Resistance in Clinical Methicillin-Resistant Staphylococcus aureus Isolates. Antimicrob. Agents Chemother. 2014, 58, 6668–6674. [Google Scholar] [CrossRef]
- Lahiri, S.D.; McLaughlin, R.E.; Whiteaker, J.D.; Ambler, J.E.; Alm, R.A. Molecular characterization of MRSA isolates bracketing the current EUCAST ceftaroline-susceptible breakpoint for Staphylococcus aureus: The role of PBP2a in the activity of ceftaroline. J. Antimicrob. Chemother. 2015, 70, 2488–2498. [Google Scholar] [CrossRef]
- Lee, H.; Yoon, E.-J.; Kim, D.; Kim, J.W.; Lee, K.-J.; Kim, H.S.; Kim, Y.R.; Shin, J.H.; Shin, J.H.; Shin, K.S.; et al. Ceftaroline Resistance by Clone-Specific Polymorphism in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2018, 62, e00485-18. [Google Scholar] [CrossRef]
- Chan, L.C.; Basuino, L.; Diep, B.; Hamilton, S.; Chatterjee, S.S.; Chambers, H.F. Ceftobiprole- and ceftaroline-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 2960–2963. [Google Scholar] [CrossRef]
- Wallick, H.; Hendlin, D. Cefoxitin, a Semisynthetic Cephamycin Antibiotic: Susceptibility Studies. Antimicrob. Agents Chemother. 1974, 5, 25–32. [Google Scholar] [CrossRef][Green Version]
- Kariyone, K.; Harada, H.; Kurita, M.; Takano, T. Cefazolin, a new semisynthetic cephalosporin antibiotic. I. Synthesis and chemical properties of cefazolin. J. Antibiot. 1970, 23, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Nishida, M.; Matsubara, T.; Murakawa, T.; Mine, Y.; Yokota, Y.; Goto, S.; Kuwahara, S. Cefazolin, a new semisynthetic cephalosporin antibiotic. II. In vitro and in vivo antimicrobial activity. J. Antibiot. 1970, 23, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Yotsuji, A.; Mitsuyama, J.; Hori, R.; Yasuda, T.; Saikawa, I.; Inoue, M.; Mitsuhashi, S. Mechanism of action of cephalosporins and resistance caused by decreased affinity for penicillin-binding proteins in Bacteroides fragilis. Antimicrob. Agents Chemother. 1988, 32, 1848–1853. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Matsunaga, N.; Tawada, H.; Kuroda, N.; Nakayama, Y.; Ishibashi, Y.; Tomimoto, M.; Ikeda, Y.; Tagawa, Y.; Iizawa, Y. TAK-599, a novel N-Phosphono type prodrug of anti-MRSA cephalosporin T-91825: Synthesis, physicochemical and pharmacological properties. Bioorg. Med. Chem. 2003, 11, 2427–2437. [Google Scholar] [CrossRef]
- Alexander, J.A.N.; Chatterjee, S.S.; Hamilton, S.M.; Eltis, L.D.; Chambers, H.F.; Strynadka, N.C.J. Structural and kinetic analyses of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance in Staphylococcus aureus. J. Biol. Chem. 2018, 293, 19854–19865. [Google Scholar] [CrossRef]
- Hamilton, S.M.; Alexander, J.A.N.; Choo, E.J.; Basuino, L.; Da Costa, T.M.; Severin, A.; Chung, M.; Aedo, S.; Strynadka, N.C.J.; Tomasz, A.; et al. High-level resistance of Staphylococcus aureus to β-Lactam antibiotics mediated by penicillin-binding protein 4 (PBP4). Antimicrob. Agents Chemother. 2017, 61, e02727-16. [Google Scholar] [CrossRef]
- Greninger, A.L.; Chatterjee, S.S.; Chan, L.C.; Hamilton, S.M.; Chambers, H.F.; Chiu, C.Y. Whole-genome sequencing of methicillin-resistant Staphylococcus aureus resistant to fifth-generation cephalosporins reveals potential non-meca mechanisms of resistance. PLoS ONE 2016, 11, e0149541. [Google Scholar] [CrossRef]
- Jones, R.N. In vitro evaluation of BAL9141, a novel parenteral cephalosporin active against oxacillin-resistant staphylococci. J. Antimicrob. Chemother. 2002, 50, 915–932. [Google Scholar] [CrossRef]
- Banerjee, R.; Gretes, M.; Basuino, L.; Strynadka, N.; Chambers, H.F. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2008, 52, 2089–2096. [Google Scholar] [CrossRef]
- Morroni, G.; Brenciani, A.; Brescini, L.; Fioriti, S.; Simoni, S.; Pocognoli, A.; Mingoia, M.; Giovanetti, E.; Barchiesi, F.; Giacometti, A.; et al. High Rate of Ceftobiprole Resistance among Clinical Methicillin-Resistant Staphylococcus aureus Isolates from a Hospital in Central Italy. Antimicrob. Agents Chemother. 2018, 62, e01663-18. [Google Scholar] [CrossRef]
- Salamaga, B.; Kong, L.; Pasquina-Lemonche, L.; Lafage, L.; von und zur Muhlen, M.; Gibson, J.F.; Grybchuk, D.; Tooke, A.K.; Panchal, V.; Culp, E.J.; et al. Demonstration of the role of cell wall homeostasis in Staphylococcus aureus growth and the action of bactericidal antibiotics. Proc. Natl. Acad. Sci. USA 2021, 118, e2106022118. [Google Scholar] [CrossRef] [PubMed]
- Roemer, T.; Schneider, T.; Pinho, M.G. Auxiliary factors: A chink in the armor of MRSA resistance to β-lactam antibiotics. Curr. Opin. Microbiol. 2013, 16, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Berger-Bächi, B.; Rohrer, S. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 2002, 178, 165–171. [Google Scholar] [CrossRef] [PubMed]
- De Lencastre, H.; Tomasz, A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1994, 38, 2590–2598. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.K.; Milheiriço, C.; de Lencastre, H.; Tomasz, A. Antibiotic Resistance as a Stress Response: Recovery of High-Level Oxacillin Resistance in Methicillin-Resistant Staphylococcus aureus “Auxiliary” (fem) Mutants by Induction of the Stringent Stress Response. Antimicrob. Agents Chemother. 2017, 61, e00313-17. [Google Scholar] [CrossRef]
- Dordel, J.; Kim, C.; Chung, M.; Pardos de la Gándara, M.; Holden, M.T.J.; Parkhill, J.; de Lencastre, H.; Bentley, S.D.; Tomasz, A. Novel Determinants of Antibiotic Resistance: Identification of Mutated Loci in Highly Methicillin-Resistant Subpopulations of Methicillin-Resistant Staphylococcus aureus. MBio 2014, 5, e01000-13. [Google Scholar] [CrossRef] [PubMed]
- Boonsiri, T.; Watanabe, S.; Tan, X.-E.; Thitiananpakorn, K.; Narimatsu, R.; Sasaki, K.; Takenouchi, R.; Sato’o, Y.; Aiba, Y.; Kiga, K.; et al. Identification and characterization of mutations responsible for the β-lactam resistance in oxacillin-susceptible mecA-positive Staphylococcus aureus. Sci. Rep. 2020, 10, 16907. [Google Scholar] [CrossRef]
- Berger-Bächi, B.; Strässle, A.; Gustafson, J.E.; Kayser, F.H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1992, 36, 1367–1373. [Google Scholar] [CrossRef]
- De Lencastre, H.; Wu, S.W.; Pinho, M.G.; Ludovice, A.M.; Filipe, S.; Gardete, S.; Sobral, R.; Gill, S.; Chung, M.; Tomasz, A. Antibiotic resistance as a stress response: Complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 1999, 5, 163–175. [Google Scholar] [CrossRef]
- Baker, T.A.; Sauer, R.T. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta–Mol. Cell Res. 2012, 1823, 15–28. [Google Scholar] [CrossRef]
- Jensen, C.; Bæk, K.T.; Gallay, C.; Thalsø-Madsen, I.; Xu, L.; Jousselin, A.; Ruiz Torrubia, F.; Paulander, W.; Pereira, A.R.; Veening, J.-W.; et al. The ClpX chaperone controls autolytic splitting of Staphylococcus aureus daughter cells, but is bypassed by β-lactam antibiotics or inhibitors of WTA biosynthesis. PLoS Pathog. 2019, 15, e1008044. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.T.; Bowman, L.; Millership, C.; Søgaard, M.D.; Kaever, V.; Siljamäki, P.; Savijoki, K.; Varmanen, P.; Nyman, T.A.; Gründling, A.; et al. The cell wall polymer lipoteichoic acid becomes nonessential in Staphylococcus aureus cells lacking the ClpX chaperone. MBio 2016, 7, e01228-16. [Google Scholar] [CrossRef] [PubMed]
- Frees, D.; Qazi, S.N.A.; Hill, P.J.; Ingmer, H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 2003, 48, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Bæk, K.T.; Gründling, A.; Mogensen, R.G.; Thøgersen, L.; Petersen, A.; Paulander, W.; Frees, D. β-lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrob. Agents Chemother. 2014, 58, 4593–4603. [Google Scholar] [CrossRef]
- Jelsbak, L.; Ingmer, H.; Valihrach, L.; Cohn, M.T.; Christiansen, M.H.G.; Kallipolitis, B.H.; Frees, D. The Chaperone ClpX Stimulates Expression of Staphylococcus aureus Protein A by Rot Dependent and Independent Pathways. PLoS ONE 2010, 5, e12752. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Abbott, J.C.; Burhenne, H.; Kaever, V.; Gründling, A. c-di-AMP Is a New Second Messenger in Staphylococcus aureus with a Role in Controlling Cell Size and Envelope Stress. PLoS Pathog. 2011, 7, e1002217. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Gründling, A. Cyclic di-AMP: Another second messenger enters the fray. Nat. Rev. Microbiol. 2013, 11, 513–524. [Google Scholar] [CrossRef]
- Griffiths, J.M.; O’Neill, A.J. Loss of Function of the GdpP Protein Leads to Joint β-Lactam/Glycopeptide Tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 579–581. [Google Scholar] [CrossRef]
- Argudín, M.A.; Roisin, S.; Nienhaus, L.; Dodémont, M.; de Mendonça, R.; Nonhoff, C.; Deplano, A.; Denis, O. Genetic Diversity among Staphylococcus aureus Isolates Showing Oxacillin and/or Cefoxitin Resistance Not Linked to the Presence of mec Genes. Antimicrob. Agents Chemother. 2018, 62, e00091-18. [Google Scholar] [CrossRef]
- Banerjee, R.; Gretes, M.; Harlem, C.; Basuino, L.; Chambers, H.F. A mec A-Negative Strain of Methicillin-Resistant S taphylococcus aureus with High-Level β-Lactam Resistance Contains Mutations in Three Genes. Antimicrob. Agents Chemother. 2010, 54, 4900–4902. [Google Scholar] [CrossRef]
- Maalej, S.M.; Rhimi, F.M.; Fines, M.; Mnif, B.; Leclercq, R.; Hammami, A. Analysis of Borderline Oxacillin-Resistant Staphylococcus aureus (BORSA) Strains Isolated in Tunisia. J. Clin. Microbiol. 2012, 50, 3345–3348. [Google Scholar] [CrossRef] [PubMed]
- Poon, R.; Basuino, L.; Satishkumar, N.; Chatterjee, A.; Mukkayyan, N.; Buggeln, E.; Huang, L.; Nair, V.; Argudín, M.A.; Datta, S.K.; et al. Loss of GdpP Function in Staphylococcus aureus Leads to β-Lactam Tolerance and Enhanced Evolution of β-Lactam Resistance. Antimicrob. Agents Chemother. 2022, 66, e01431-21. [Google Scholar] [CrossRef]
- Panchal, V.V.; Griffiths, C.; Mosaei, H.; Bilyk, B.; Sutton, J.A.F.; Carnell, O.T.; Hornby, D.P.; Green, J.; Hobbs, J.K.; Kelley, W.L.; et al. Evolving MRSA: High-level β-lactam resistance in Staphylococcus aureus is associated with RNA Polymerase alterations and fine tuning of gene expression. PLoS Pathog. 2020, 16, e1008672. [Google Scholar] [CrossRef] [PubMed]
- Aiba, Y.; Katayama, Y.; Hishinuma, T.; Murakami-Kuroda, H.; Cui, L.; Hiramatsu, K. Mutation of RNA Polymerase β-Subunit Gene Promotes Heterogeneous-to-Homogeneous Conversion of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 4861–4871. [Google Scholar] [CrossRef] [PubMed]
- Le, K.Y.; Otto, M. Quorum-sensing regulation in staphylococci—An overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef]
- Duran, S.P.; Kayser, F.H.; Berger-Bachi, B. Impact of sar and agr on methicillin resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 1996, 141, 255–260. [Google Scholar] [CrossRef][Green Version]
- Song, H.-S.; Choi, T.-R.; Han, Y.-H.; Park, Y.-L.; Park, J.Y.; Yang, S.-Y.; Bhatia, S.K.; Gurav, R.; Kim, Y.-G.; Kim, J.-S.; et al. Increased resistance of a methicillin-resistant Staphylococcus aureus Δagr mutant with modified control in fatty acid metabolism. AMB Express 2020, 10, 64. [Google Scholar] [CrossRef]
- Plata, K.B.; Rosato, R.R.; Rosato, A.E. Fate of Mutation Rate Depends on agr Locus Expression during Oxacillin-Mediated Heterogeneous-Homogeneous Selection in Methicillin-Resistant Staphylococcus aureus Clinical Strains. Antimicrob. Agents Chemother. 2011, 55, 3176–3186. [Google Scholar] [CrossRef]
- Cheung, A.L.; Nishina, K.; Manna, A.C. SarA of Staphylococcus aureus Binds to the sarA Promoter To Regulate Gene Expression. J. Bacteriol. 2008, 190, 2239–2243. [Google Scholar] [CrossRef]
- Liu, Y.; Manna, A.C.; Pan, C.-H.; Kriksunov, I.A.; Thiel, D.J.; Cheung, A.L.; Zhang, G. Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2006, 103, 2392–2397. [Google Scholar] [CrossRef]
- Cheung, A.L.; Koomey, J.M.; Butler, C.A.; Projan, S.J.; Fischetti, V.A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 1992, 89, 6462–6466. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.L.; Eberhardt, K.; Heinrichs, J.H. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 1997, 65, 2243–2249. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cheung, A.; Bayer, A.S.; Chen, L.; Abdelhady, W.; Kreiswirth, B.N.; Yeaman, M.R.; Xiong, Y.Q. The Global Regulon sarA Regulates β-Lactam Antibiotic Resistance in Methicillin-Resistant Staphylococcus aureus In Vitro and in Endovascular Infections. J. Infect. Dis. 2016, 214, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Lee, H.-J.; Jo, S.-H.; Kim, M.-G.; Lee, Y.; Lee, W.; Kim, W.; Joo, H.-S.; Kim, Y.-G.; Kim, J.-S.; et al. Study of SarA by DNA Affinity Capture Assay (DACA) Employing Three Promoters of Key Virulence and Resistance Genes in Methicillin-Resistant Staphylococcus aureus. Antibiotics 2022, 11, 1714. [Google Scholar] [CrossRef]
- Wang, G.; Li, L.; Wang, X.; Li, X.; Zhang, Y.; Yu, J.; Jiang, J.; You, X.; Xiong, Y.Q. Hypericin enhances β-lactam antibiotics activity by inhibiting sarA expression in methicillin-resistant Staphylococcus aureus. Acta Pharm. Sin. B 2019, 9, 1174–1182. [Google Scholar] [CrossRef]
- Jarick, M.; Bertsche, U.; Stahl, M.; Schultz, D.; Methling, K.; Lalk, M.; Stigloher, C.; Steger, M.; Schlosser, A.; Ohlsen, K. The serine/threonine kinase Stk and the phosphatase Stp regulate cell wall synthesis in Staphylococcus aureus. Sci. Rep. 2018, 8, 13693. [Google Scholar] [CrossRef]
- Beltramini, A.M.; Mukhopadhyay, C.D.; Pancholi, V. Modulation of Cell Wall Structure and Antimicrobial Susceptibility by a Staphylococcus aureus Eukaryote-Like Serine/Threonine Kinase and Phosphatase. Infect. Immun. 2009, 77, 1406–1416. [Google Scholar] [CrossRef]
- Tamber, S.; Schwartzman, J.; Cheung, A.L. Role of PknB Kinase in Antibiotic Resistance and Virulence in Community-Acquired Methicillin-Resistant Staphylococcus aureus Strain USA300. Infect. Immun. 2010, 78, 3637–3646. [Google Scholar] [CrossRef]
- Ohlsen, K.; Donat, S. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int. J. Med. Microbiol. 2010, 300, 137–141. [Google Scholar] [CrossRef]
- Débarbouillé, M.; Dramsi, S.; Dussurget, O.; Nahori, M.-A.; Vaganay, E.; Jouvion, G.; Cozzone, A.; Msadek, T.; Duclos, B. Characterization of a Serine/Threonine Kinase Involved in Virulence of Staphylococcus aureus. J. Bacteriol. 2009, 191, 4070–4081. [Google Scholar] [CrossRef]
- Sun, F.; Ding, Y.; Ji, Q.; Liang, Z.; Deng, X.; Wong, C.C.L.; Yi, C.; Zhang, L.; Xie, S.; Alvarez, S.; et al. Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 15461–15466. [Google Scholar] [CrossRef]
- Chatterjee, A.; Poon, R.; Chatterjee, S.S. Stp1 Loss of Function Promotes β-Lactam Resistance in Staphylococcus aureus That Is Independent of Classical Genes. Antimicrob. Agents Chemother. 2020, 64, e02222-19. [Google Scholar] [CrossRef]
- Lithgow, J.K.; Ingham, E.; Foster, S.J. Role of the hprT–ftsH locus in Staphylococcus aureus. Microbiology 2004, 150, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Yeo, W.-S.; Jeong, B.; Ullah, N.; Shah, M.; Ali, A.; Kim, K.; Bae, T. FtsH Sensitizes Methicillin-Resistant Staphylococcus aureus to β-Lactam Antibiotics by Degrading YpfP, a Lipoteichoic Acid Synthesis Enzyme. Antibiotics 2021, 10, 1198. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, J.; Strässle, A.; Hächler, H.; Kayser, F.H.; Berger-Bächi, B. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J. Bacteriol. 1994, 176, 1460–1467. [Google Scholar] [CrossRef] [PubMed]
- Kondo, N.; Kuwahara-Arai, K.; Kuroda-Murakami, H.; Tateda-Suzuki, E.; Hiramatsu, K. Eagle-Type Methicillin Resistance: New Phenotype of High Methicillin Resistance under mec Regulator Gene Control. Antimicrob. Agents Chemother. 2001, 45, 815–824. [Google Scholar] [CrossRef]
- Komatsuzawa, H.; Ohta, K.; Fujiwara, T.; Choi, G.H.; Labischinski, H.; Sugai, M. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 2001, 203, 49–54. [Google Scholar] [CrossRef]
- Komatsuzawa, H.; Ohta, K.; Sugai, M.; Fujiwara, T.; Glanzmann, P.; Berger-Bachi, B.; Suginaka, H. Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 2000, 45, 421–431. [Google Scholar] [CrossRef]
- Mikkelsen, K.; Sirisarn, W.; Alharbi, O.; Alharbi, M.; Liu, H.; Nøhr-Meldgaard, K.; Mayer, K.; Vestergaard, M.; Gallagher, L.A.; Derrick, J.P.; et al. The Novel Membrane-Associated Auxiliary Factors AuxA and AuxB Modulate β-lactam Resistance in MRSA by stabilizing Lipoteichoic Acids. Int. J. Antimicrob. Agents 2021, 57, 106283. [Google Scholar] [CrossRef]
- Jousselin, A.; Manzano, C.; Biette, A.; Reed, P.; Pinho, M.G.; Rosato, A.E.; Kelley, W.L.; Renzoni, A. The Staphylococcus aureus Chaperone PrsA Is a New Auxiliary Factor of Oxacillin Resistance Affecting Penicillin-Binding Protein 2A. Antimicrob. Agents Chemother. 2016, 60, 1656–1666. [Google Scholar] [CrossRef]
- Roch, M.; Lelong, E.; Panasenko, O.O.; Sierra, R.; Renzoni, A.; Kelley, W.L. Thermosensitive PBP2a requires extracellular folding factors PrsA and HtrA1 for Staphylococcus aureus MRSA β-lactam resistance. Commun. Biol. 2019, 2, 417. [Google Scholar] [CrossRef] [PubMed]
- Jousselin, A.; Renzoni, A.; Andrey, D.O.; Monod, A.; Lew, D.P.; Kelley, W.L. The Posttranslocational Chaperone Lipoprotein PrsA Is Involved in both Glycopeptide and Oxacillin Resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 3629–3640. [Google Scholar] [CrossRef] [PubMed]
- Navratna, V.; Nadig, S.; Sood, V.; Prasad, K.; Arakere, G.; Gopal, B. Molecular basis for the role of Staphylococcus aureus penicillin binding protein 4 in antimicrobial resistance. J. Bacteriol. 2010, 192, 134–144. [Google Scholar] [CrossRef]
- Memmi, G.; Filipe, S.R.; Pinho, M.G.; Fu, Z.; Cheung, A. Staphylococcus aureus PBP4 Is Essential for β-Lactam Resistance in Community-Acquired Methicillin-Resistant Strains. Antimicrob. Agents Chemother. 2008, 52, 3955–3966. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.S.; Chen, L.; Gilbert, A.; da Costa, T.M.; Nair, V.; Datta, S.K.; Kreiswirth, B.N.; Chambers, H.F. PBP4 Mediates β-Lactam Resistance by Altered Function. Antimicrob. Agents Chemother. 2017, 61, e00932-17. [Google Scholar] [CrossRef]
- Silber, N.; Matos de Opitz, C.L.; Mayer, C.; Sass, P. Cell division protein FtsZ: From structure and mechanism to antibiotic target. Future Microbiol. 2020, 15, 801–831. [Google Scholar] [CrossRef]
- Lee, S.H.; Jarantow, L.W.; Wang, H.; Sillaots, S.; Cheng, H.; Meredith, T.C.; Thompson, J.; Roemer, T. Antagonism of Chemical Genetic Interaction Networks Resensitize MRSA to β-Lactam Antibiotics. Chem. Biol. 2011, 18, 1379–1389. [Google Scholar] [CrossRef]
- Ferrer-González, E.; Huh, H.; Al-Tameemi, H.M.; Boyd, J.M.; Lee, S.-H.; Pilch, D.S. Impact of FtsZ Inhibition on the Localization of the Penicillin Binding Proteins in Methicillin-Resistant Staphylococcus aureus. J. Bacteriol. 2021, 203, 204–225. [Google Scholar] [CrossRef]
- Neuhaus, F.C.; Baddiley, J. A Continuum of Anionic Charge: Structures and Functions of D-Alanyl Teichoic Acids in Gram-Positive Bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef]
- Coupri, D.; Verneuil, N.; Hartke, A.; Liebaut, A.; Lequeux, T.; Pfund, E.; Budin-Verneuil, A. Inhibition of D-alanylation of teichoic acids overcomes resistance of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2021, 76, 2778–2786. [Google Scholar] [CrossRef]
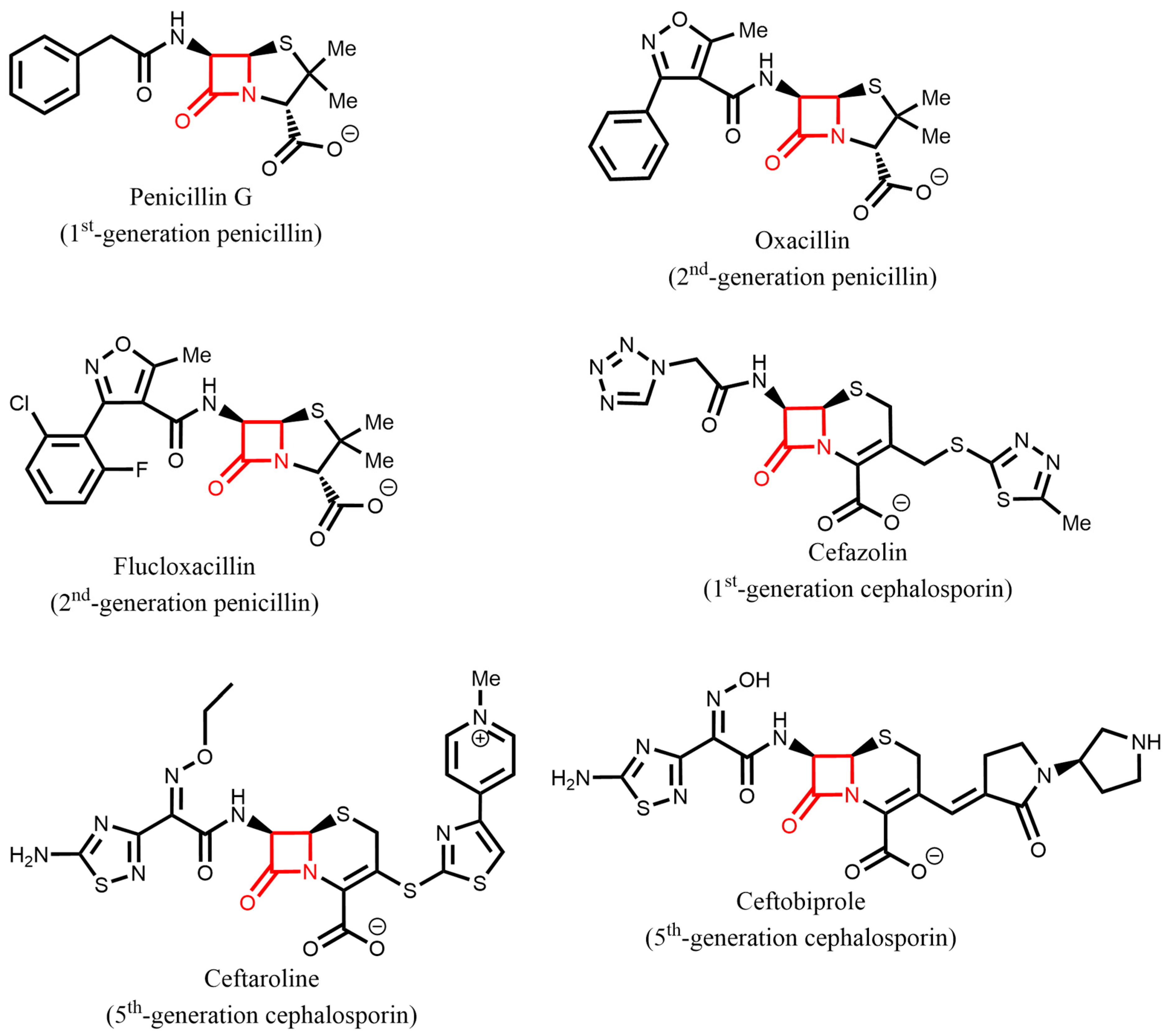
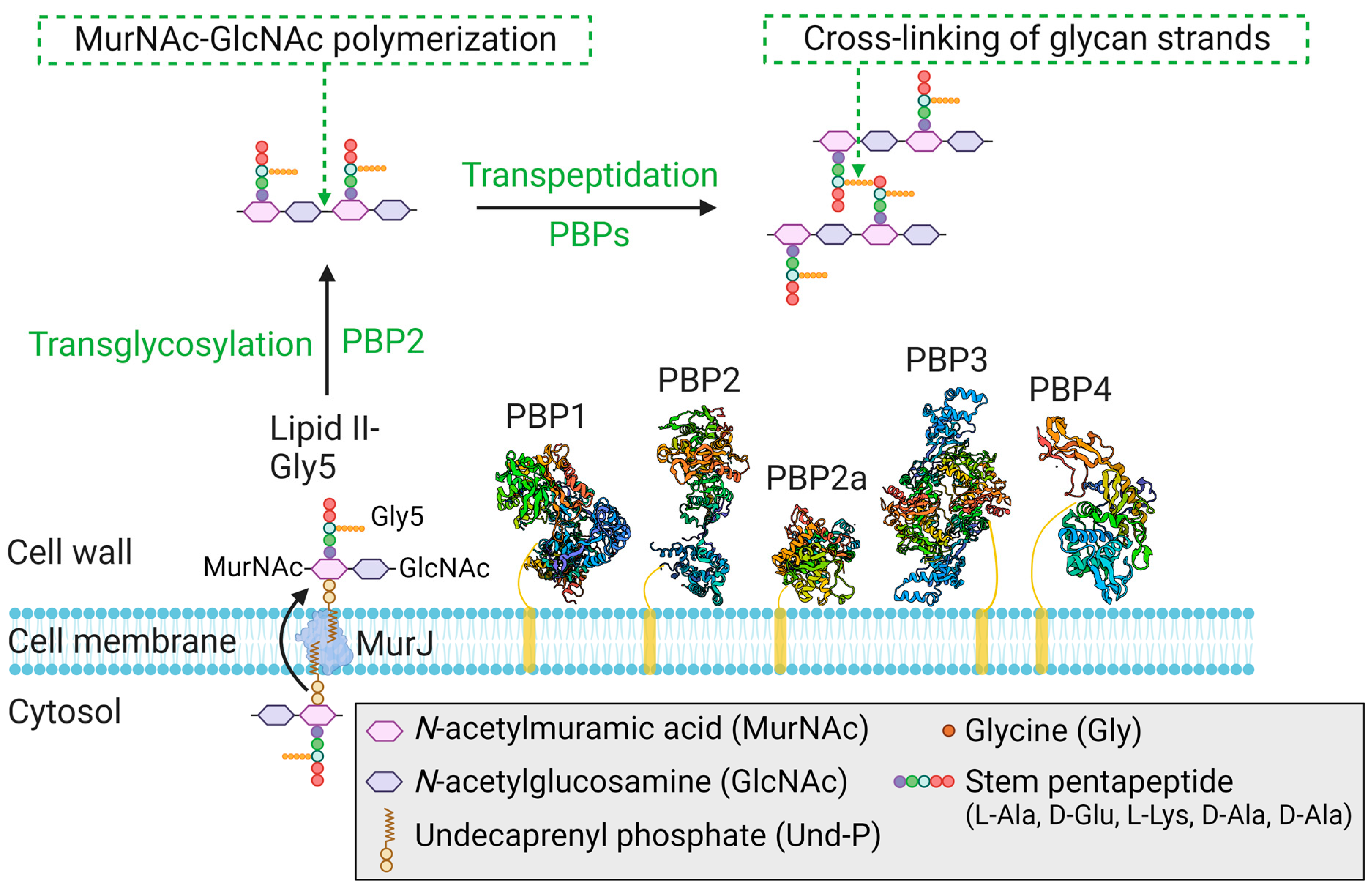
| Protein(s) | Gene(s) | Class | Activity | Function/Relevant Features |
|---|---|---|---|---|
| PBP1 | pbp1 | HMM class B | TPase | PBP1 is an essential protein that plays a crucial role in cell division and cell wall integrity [69,74]. |
| PBP2 | pbp2 | HMM class A | TPase and TGase | PBP2 is an essential protein that acts as the major peptidoglycan synthase responsible for cell wall synthesis [75,76]. |
| PBP2a | mecA | HMM class B | TPase | PBP2a is an acquired protein [12] that compensates for the loss of endogenous PBP2 TPase activity in the presence of β-lactams [77]. |
| PBP3 | pbp3 | HMM class B | TPase | PBP3 is a nonessential protein for growth [78] and is associated with septal localization of RodA [79]; how PBP3 modulates RodA activity (cell elongation and maintenance) remains unclear. |
| PBP4 | pbp4 | LMM class C | TPase | PBP4 is a nonessential protein for growth and is involved in the regulation of cross-linking within peptidoglycan [70,80]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lade, H.; Kim, J.-S. Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review. Antibiotics 2023, 12, 1362. https://doi.org/10.3390/antibiotics12091362
Lade H, Kim J-S. Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review. Antibiotics. 2023; 12(9):1362. https://doi.org/10.3390/antibiotics12091362
Chicago/Turabian StyleLade, Harshad, and Jae-Seok Kim. 2023. "Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review" Antibiotics 12, no. 9: 1362. https://doi.org/10.3390/antibiotics12091362
APA StyleLade, H., & Kim, J.-S. (2023). Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review. Antibiotics, 12(9), 1362. https://doi.org/10.3390/antibiotics12091362







