Abstract
The emergence of multidrug-resistant (MDR) superbugs underlines the urgent need for innovative treatment options to tackle resistant bacterial infections. The clinical efficacy of natural products directed our efforts towards developing new antibacterial leads from naturally abundant known chemical structures. The present study aimed to explore an unusual class of phenylacylphenols (malabaricones) from Myristicamalabarica as antibacterial agents. In vitro antibacterial activity was determined via broth microdilution, cell viability, time–kill kinetics, biofilm eradication, intracellular killing, and checkerboard assays. The efficacy was evaluated in vivo in murine neutropenic thigh and skin infection models. Confocal and SEM analyses were used for mechanistic studies. Among the tested isolates, malabaricone B (NS-7) demonstrated the best activity against S. aureus with a favorable selectivity index and concentration-dependent, rapid bactericidal killing kinetics. It displayed equal efficacy against MDR clinical isolates of S. aureus and Enterococci, efficiently clearing S. aureus in intracellular and biofilm tests, with no detectable resistance. In addition, NS-7 synergized with daptomycin and gentamicin. In vivo, NS-7 exhibited significant efficacy against S. aureus infection. Mechanistically, NS-7 damaged S. aureus membrane integrity, resulting in the release of extracellular ATP. The results indicated that NS-7 can act as a naturally derived bactericidal drug lead for anti-staphylococcal therapy.
1. Introduction
Multidrug-resistant (MDR) Staphylococcus aureus is now widely recognized as a severe burden to the healthcare system owing to its widespread occurrence and drug resistance profile. Nosocomial and community-acquired multidrug-resistant (MDR) S. aureus infections are a significant clinical concern globally, characterized by high morbidity and mortality rates. This is mostly attributed to their intrinsic resistance to commonly used antibiotics for treatment [1,2]. The development of various virulence factors and the rapid acquisition of the MDR phenotype over time make them outwit existing antibiotics, thereby creating life-threatening complications in infection control [3,4]. In light of the emergence of genetically variable epidemic strains of S. aureus, including methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Staphylococcus aureus (VRSA), and their increased incidence, the World Health Organization (WHO) declared them as “high priority” pathogens needing urgent attention [5]. This unceasing emergence and global spread of MDR pathogens necessitates the consistent discovery and development of new drug leads with novel target mechanisms to tackle MDR S. aureus infections.
Natural products and their analogs are pioneers as antibacterial agents and have been approved by the FDA for utilization in clinics [6].The characteristics of naturally derived compounds, such as unique chemo-diversity, bioavailability, target specificity, and microbial metabolism, have proved their ability to address the developing mechanisms of multidrug resistance in order to augment the drug discovery pipeline for antibiotics [7,8,9,10,11]. Therefore, the identification of novel chemical scaffolds from nature and their detailed characterization against predominant MDR strains is of great significance. In addition, their utilization in a synergistic combination with conventional antibiotics has emerged as a highly effective approach for infection control [12,13]. The highly enriched floristic diversity and unremitting legacy of utilizing naturally derived compounds for therapeutic advances [14,15,16] encouraged us to systematically explore the ability of known chemical scaffolds as promising candidates to fight MDR bacterial infections.
The present study investigated the antibacterial properties of a specific group of compounds belonging to the class of phenylacylphenols, derived from the Myristicaceae family, often known as nutmeg. Malabaricones, also known as phenylacylphenols or diarylnonanoids, are a distinctive category of secondary metabolites with bioactive properties. These compounds are synthesized by several plant species that are part of the well-known “Myristicaceae” family. The term “Malabaricones” was assigned to these compounds due to their initial discovery in the fruit rind of the Myristica malabarica, a wild nutmeg species [17]. Subsequently, it was determined that these bioactive elements are also present in other species of the Myristica and Kenema (limited)genera. Myristicamalabarica is a species that is recognized as red-listed and is endemic to specific regions. It is typically found in sub-tropical evergreen forests at altitudes of about 1000 m and in the southern and western ghats within Myristica swamps. Various components of this particular plant, such as the rind, mace, and seed, were discovered to have malabaricones as their primary phytoconstituents [18]. The primary components of malabaricones are acyl phenols of the 2,6-dihydroxyphenyl type and their derivatives. These compounds are characterized by a structure that includes a 2,6-dihydroxyacetophenone group connected to a benzene ring by a C8 alkyl chain [18].
In the present study, our attempts were directed towards developing the naturally derived chemical structure malabaricone B (NS-7) as a promising drug lead to target MDR-S. aureus. Herein, we demonstrated a comprehensive in vitro and in vivo evaluation of NS-7 including the mechanistic studies. Also, for the first time, synergy studies of the compound with clinically used antibiotics were well established.
2. Results
2.1. Isolation of Malabaricones from the Fruit Rinds of M. malabarica Lam
The isolates from M. malabarica (Figure 1) were identified as the known chemical constituents, namely 1-(2,6-dihydroxyphenyl) tetradecan-1-one (NS-1), malabaricone A (NS-3), promalabaricone B (NS-5), malabaricone B (NS-7), malabaricone C (NS-9), and malabaricone D (NS-11), by interpreting their1H NMR, 13C NMR, and HRESI-MS spectroscopic data (available in the Supplementary Materials) and by comparing the results with relevant data from the literature.
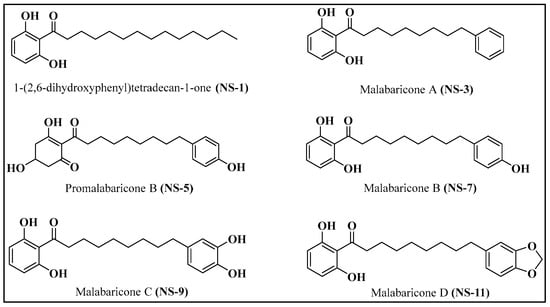
Figure 1.
Structures of compounds isolated from M. malabarica.
2.2. Antibacterial Activity against Clinically Relevant Bacteria
To begin with, the antibacterial efficacy of the isolates was evaluated against ESKAPE pathogens. Amongst the isolates, NS-7 displayed potent and specific activity with an MIC of 0.5 μg/mL, followed by NS-11 and NS-9,against S. aureus. However, a significantly higher MIC (>128 μg/mL, Table 1) value of all tested compounds was a clear indication of their inactivity against Gram-negative bacteria (GNB). To confirm that the presence of an outer membrane (OM) is responsible for the lack of activity of NS-7 against GNBs, its antibacterial activity was assessed with the addition of an OM permeabilizer, polymyxin B nonapeptide (PMBN), at non-lethal concentrations. As seen in Table 2, NS-7 alone is inactive against GNBs with intact an OM but, in the presence of PMBN, its entry is facilitated due to increased membrane permeability, reflected in its lower MICs. The reduction in MIC of NS-7 with the addition of PMBN against GNBs A. baumannii and E. coli (Table 2) clearly indicates that an intact OM is acting as a permeability barrier and that the target for NS-7 is present across both bacterial species.

Table 1.
MIC (µg/mL) table for M. malabarica isolates against ESKAPE pathogen panel.

Table 2.
MIC (µg/mL) table for NS-7 with PMBN against Gram-negative pathogens.
2.3. NS-7 Imparted No Toxicity to Eukaryotic Cells
As we know, the cytotoxic effects of hit compounds have a key role in their further development as antibacterial agents. Intriguingly, the MTT assay results revealed that NS-7 does not impart any toxicity to the host cells (Vero cells, ATCC CCL-81) and possessed an extremely favorable selectivity index (≥80, Table 3) which is highly promising for further characterization. On the other hand, NS-9 and NS-11 were cytotoxic and, thus, did not exhibit a favorable selectivity index, which precluded these compounds from further evaluation.

Table 3.
Selectivity index of NS-7 against Vero cells, ATCC CCL-81.
2.4. NS-7 Is Active against MDR Clinical Strains of Staphylococci and Enterococci
With the selected, active, non-toxic compound NS-7, we further extended the study to explore its spectrum of activity against clinical MDR strains of Staphylococci and Enterococci. NS-7 (MIC 1–2 μg/mL, Table 4) demonstrated equipotent activity against MRSA, VRSA, and vancomycin-resistant Enterococcus (VRE) strains. The admirable activity of NS-7 against various MDR strains proved its ability to escape the prevalent drug resistant mechanisms, thereby acting as a promising candidate for treating MDR S. aureus and Enterococcus infections.

Table 4.
MIC (µg/mL) table for NS-7 and comparators against a panel of S. aureus and Enterococcus MDR clinical strains.
2.5. NS-7 Possesses Rapid Bactericidal Activity
To determine the mode of action of NS-7, a bacterial time–kill kinetics assay with the compound was performed against S. aureus. Levofloxacin and vancomycin served as the reference standards. It was evidenced from the killing kinetics data (Figure 2) that NS-7 demonstrates concentration-dependent, bactericidal activity against S. aureus ATCC 29213. At various MICs, NS-7 showed rapid, bactericidal activity confirmed by complete eradication of viable cultures (~6 log10 cfu/mL, Figure 2a) within 15 min of incubation, and no regrowth in cultures could be seen up to 24 h (Figure 2b). At similar concentrations, levofloxacin caused only ~2 log10 cfu/mL reduction after 1 h of its exposure to S. aureus. The rapid killing kinetics of NS-7 were also better than vancomycin at its 10X MIC (Figure 2b). The extremely fast bactericidal potency of NS-7 with no regrowth was promising enough to carry forward for comprehensive evaluation.
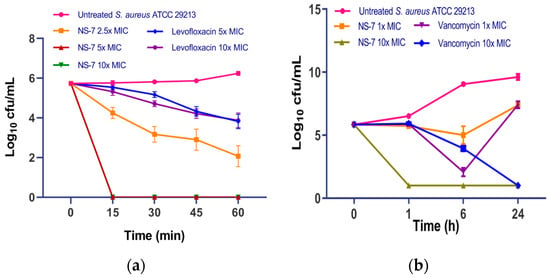
Figure 2.
Bacterial time–kill kinetics of NS-7 and comparators against S. aureus ATCC 29213 for (a) 1 h; (b) 24 h. The bacteria were incubated with various MICs for 24 h, then samples were removed at specified time intervals and the CFU was determined. The error bars indicate the standard deviations (SDs) derived from triplicate samples of each tested antibiotic or compound.
2.6. Efficient Eradication of Preformed S. aureus Biofilm by NS-7
Persistency and recurrence of S. aureus infections are due to its significant virulence mechanism through the formation of microbial biofilms. The presence of inherent tolerance in bacteria within the biofilm, coupled with the restricted diffusion of antibiotics across the surface, is the primary cause of biofilm-mediated AMR. This formidable resistance to conventional antibiotics and therapeutic failure demands an imperative need for new bactericidal agents capable of eradicating the biofilms [19,20]. In this regard, we evaluated the effect of NS-7 in disrupting the S. aureus preformed biofilm. At 1X MIC, NS-7 reduced ~10% of biofilm mass (Figure 3) which was better than vancomycin (~5%) at a similar concentration. Thus, NS-7 exhibits rapid, bactericidal kinetics against planktonic S. aureus ATCC 29213 and is also effective against S. aureus in preformed biofilms.
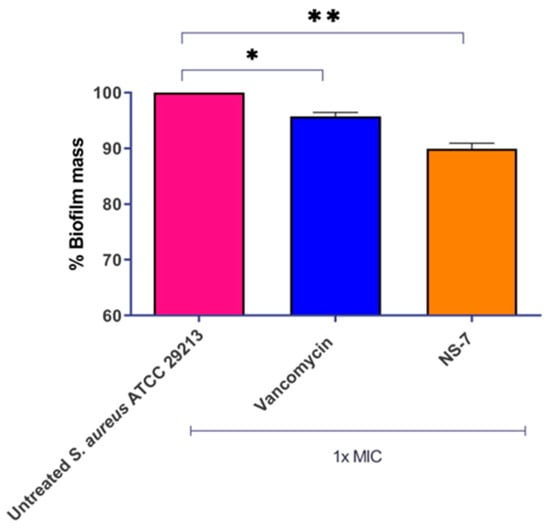
Figure 3.
NS-7 and vancomycin activity against S. aureus ATCC 29213 biofilm. Biofilm mass was stained with crystal violet post-treatment to quantify the % reduction, which is plotted. The error bars indicate the standard deviations (SDs) derived from triplicate samples of each tested antibiotic or compound. * (p < 0.5) and ** (p < 0.05) denotes significance.
2.7. NS-7 Efficiently Cleared Intracellular S. aureus
Bacterial survival within phagocytic cells and inadequate cellular penetration of antibiotics creates serious complications in severe S. aureus infections. There is an urgent need for the development of effective antibacterial drugs that have significant penetrating power to specifically target and eliminate intracellular bacteria. This is crucial in order to avoid infections and combat the growing problem of antibiotic tolerance. Numerous natural and naturally derived compounds, synthetic compounds, and nanoparticles have demonstrated efficacy in the intracellular eradication of bacterial pathogens. [10]. In this context, we assessed the effect of NS-7 against intracellular S. aureus in the murine macrophage cell line J774.A1.Vancomycin was used as the control antibiotic. NS-7 exhibited bactericidal activity that was evidenced by a significant reduction in intracellular bacterial load at its 10X MIC treatment (~0.8 log10 cfu/mL reduction) as compared to untreated, which is better than vancomycin (~0.15 log10 cfu/mL reduction) at 5X MIC (Figure 4). Thus, NS-7 is efficient in clearing intracellular bacteria much more potently than vancomycin. In addition, it also indicates that NS-7 is able to achieve good cellular penetration and accumulation inside the macrophages, and that it is better than vancomycin.
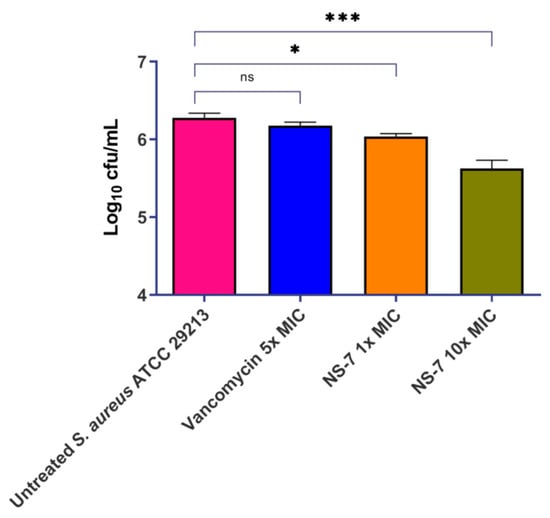
Figure 4.
Activity of NS-7 and comparators against intracellular S. aureus ATCC29213 in J774.A1 mouse macrophages. The error bars indicate the standard deviations (SDs) derived from triplicate samples of each tested antibiotic or compound. * (p < 0.5) and *** (p < 0.05) denotes significance.
2.8. NS-7 Shows Synergism with Daptomycin and Gentamicin
Recent advances in combination therapy to defeat MDR S. aureus infections emphasize the need to evaluate the synergistic interactions of a newly developed antibacterial agent with conventional antibiotics [21]. As such, the potential synergistic interactions between NS-7 and clinically used antibiotics against S. aureus ATCC29213 were evaluated by determining the fractional inhibitory concentration (ΣFIC) using the checkerboard assay [22]. Among the tested antibiotics, NS-7 showed synergistic interactions with gentamicin and daptomycin (FIC 0.265, Table 5). Furthermore, the bacterial killing kinetics of the active combinations were assessed in order to confirm the synergistic interactions. At 1X MIC against S. aureus ATCC 29213, NS-7 + gentamicin caused a significant reduction in cfu/mL, better than either drug alone (Figure 5a), with the complete eradication of all culture in 24 h. When tested against gentamicin-resistant MRSA NRS 119 (Figure 5b), the combination of NS-7 + gentamicin fared better than the individual drugs, with maximum activity achieved after 6 h of treatment (~8.2 log10 cfu/mL reduction as compared to untreated) with slight regrowth observed at 24 h. The combination of NS-7 + daptomycin also performed better than either drug alone at 1X MIC and resulted in a ~9 log10 cfu/mL reduction in S. aureus ATCC 29213 (Figure 5c) in comparison to untreated control growth after 24 h. The results revealed that the antibacterial potential of NS-7 can also be successfully employed with gentamicin and daptomycin to attain the augmented activity, even against gentamicin-resistant MRSA.

Table 5.
Synergy studies of NS-7 with FDA-approved antibiotics against S. aureus ATCC 29213.
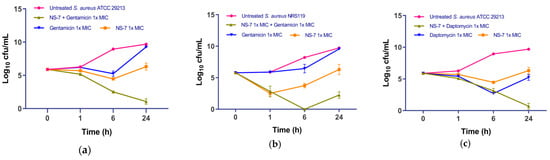
Figure 5.
Bacterial killing kinetics of NS-7 against S. aureus ATCC29213 with (a) gentamicin and (b) daptomycin; (c) bacterial killing kinetics of NS-7 with gentamicin against gentamicinR S. aureus NRS 119.The error bars indicate the standard deviations (SDs) derived from triplicate samples of each tested antibiotic or compound.
2.9. S. aureus Does Not Develop Resistance to NS-7
In view of the escalating AMR globally, it is imperative to evaluate a newly developed antibacterial agent’s propensity for resistance development in bacteria. Therefore, the propensity of S. aureus to develop resistance to NS-7 was evaluated via serial exposure (up to 40 days) to NS-7 at sub-inhibitory concentrations along with levofloxacin as a control. Interestingly, the MIC of NS-7 was not substantially altered after 40 passages, indicating that S. aureus lacks the ability to generate stable resistance to NS-7 (Figure 6). In contrast, the MIC of the clinically used antibiotic levofloxacin increased 256-fold within the same time period, suggesting that NS-7 can be an ideal antibacterial lead candidate and can act as an effective alternative to overcome drug resistance.
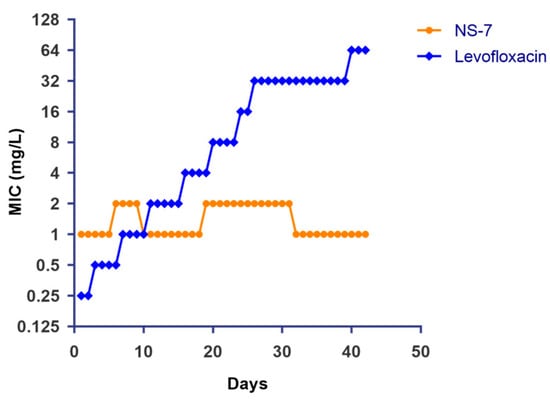
Figure 6.
Induction of resistance in S. aureus ATCC29213 after continuous exposure to NS-7 and levofloxacin at sub-MIC levels. The MIC of NS-7 and levofloxacin against S. aureus during 40 serial passages was determined regularly and plotted.
2.10. NS-7 Displayed Prolonged PAE against S. aureus
A prolonged post-antibiotic effect (PAE) of an active antibacterial agent is an added advantage in reducing the dosage, which in turn guides the selection of antimicrobials for the desired clinical outcome. Herein, we assessed the persistent suppression of S. aureus growth (PAE) in the absence of NS-7 after its 1 h treatment. Promisingly, NS-7 exhibited an extended PAE of 1 h and >22 h when subjected to 1X and 10X MIC treatments, respectively, better than the control antibiotics vancomycin and levofloxacin (Table 6). Taken together, NS-7 exhibits rapid bactericidal killing kinetics with potent activity against bacteria in various growth phases and a prolonged PAE.

Table 6.
Post-antibiotic effect (PAE) of NS-7 against S. aureus ATCC 29213.
2.11. NS-7 Exerts Bactericidal Activity via Cell Membrane Lysis
To obtain an insight into the fast bactericidal activity of NS-7 against S. aureus, confocal microscopic studies were performed using the LIVE/DEAD BacLight bacterial viability kit. It is evident from Figure 7 that dead cells (stained red) were predominant over viable cells upon treatment with 5X MIC of NS-7 for 30 min, indicating a destructive effect on bacterial membrane integrity resulting in rapid bactericidal activity. Consistent with these results, scanning electron microscopy (SEM) images (Figure 7) exhibited the severely damaged morphology of S. aureus ATCC29213 exposed to NS-7 (numerous lysed cells accompanied with cellular debris), with irregular cell wall structures and leakage of intracellular components in contrast to untreated S. aureus (intact morphology).To further ascertain the effect of NS-7 on membrane integrity, the extra-and intracellular ATP levels in S. aureus upon treatment with NS-7 were determined using an ATP bioluminescence assay kit. S. aureus ATCC 29213, when treated with 2.5X and 5X MIC of NS-7 and with melittin as a control, demonstrated a significant dose-dependent decrease in intracellular ATP concomitant with an increase in extracellular ATP release(Figure 8a,b). The observed proportional increase and decrease in extracellular and intracellular ATP, respectively, implied that NS-7 enhanced the permeability of the S. aureus cell membrane, resulting in the escape of ATP molecules. The results were comparable to treatment with melittin (a membrane-puncturing antimicrobial peptide), thus, indicating that the mode of action of the compound is by lysing membranes, leading to cell death. Taken together, the correlation between extracellular and intracellular ATP concentration along with the SEM data indicated that NS-7 targeted the membrane integrity by disruption of the bacterial cell membrane, thus, triggering cell death.
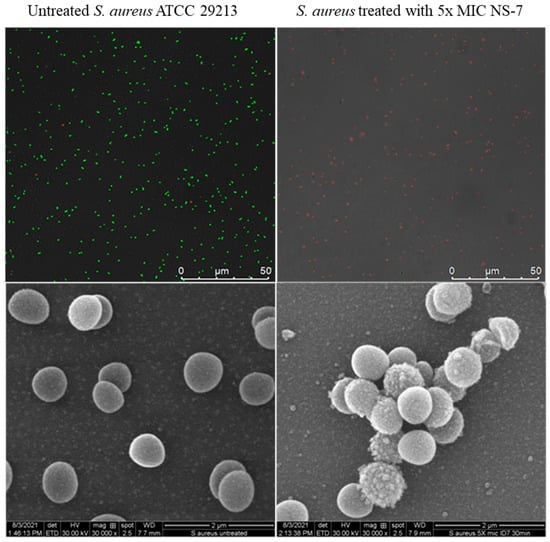
Figure 7.
Confocal (upper panel) and SEM images (lower panel) of untreated and NS-7-treated S. aureus ATCC 29213at 5X MIC for 30 min. The cells were treated with NS-7, exposed to bacterial viability stains for confocal microscopy, and prepared for SEM imaging using the standard protocol. Viable bacterial cells retained SYTO9dye (green) whereas dead cells were stained red by PI, as seen under a confocal microscope. SEM images demonstrated membrane damage.
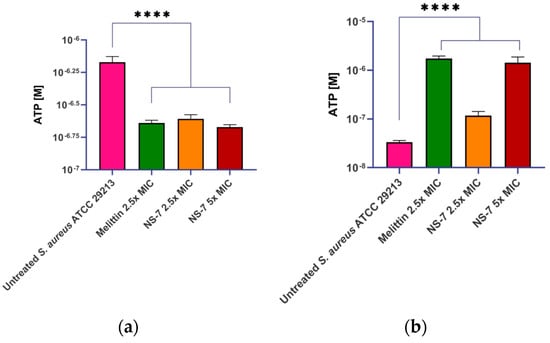
Figure 8.
Treatment with NS-7 leads to (a) intracellular ATP decrease; (b) extracellular ATP increase in NS-7 treated S. aureus ATCC 29213. Melittin was used as the comparator. The error bars indicate the standard deviations (SDs) derived from triplicate samples of each tested antibiotic or compound. **** (p < 0.005) denotes significance.
2.12. NS-7 Demonstrates Significant Efficacy in Murine Infection Models
In view of NS-7’s potent activity against S. aureus in vitro, we tried to further establish its in vivo efficacy in murine infection models. The maximum tolerable dose (MTD) of NS-7 was determined to be ≥250 mg/kg before testing in infection models. NS-7 at 50 mg/kg demonstrated significant in vivo efficacy (Figure 9a) in a murine neutropenic thigh infection model as indicated by reduction of ~0.5 log10cfu/g bacterial load comparable to vancomycin at a b.i.d dose of 25 mg/kg.
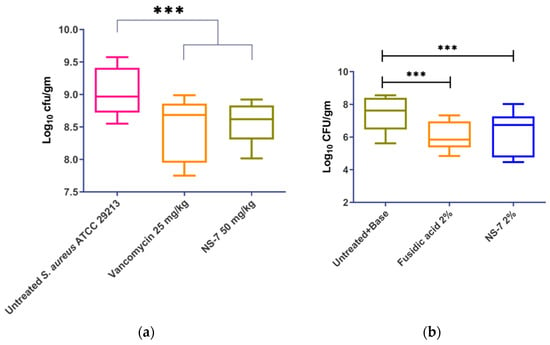
Figure 9.
In vivo efficacy of NS-7 and vancomycin in a murine model: (a) neutropenic thigh infection; (b) skin infection model. The error bars indicate the standard deviations (SDs) derived from triplicate samples of each antibiotic or compound treatment group tested in three independent experiments. *** (p < 0.05) denotes significance.
In the skin infection model, the infected Swiss mice that received treatment twice daily with 2% NS-7 (Figure 9b) exhibited a significant reduction in the bacterial burden (~1 log10cfu/g) of S. aureus ATCC29213 present in skin wounds. The compound exhibited comparable activity with the positive control of 2% fusidic acid that also caused a ~1.2 log10cfu/g reduction in bacterial load. The skin infection model revealed that NS-7 can act as an excellent topical antibacterial agent to trigger wound healing in S. aureus-infected skin wounds, which is an added advantage. Altogether, NS-7 proved its efficacy in both of the in vivo models, which in turn highlights its application as an effective antibiotic lead against severe infections caused by S. aureus. Additionally, NS-7 is naturally derived and does not require any structural modifications, implying the possibility of a cost-effective drug development scheme to treat MDR S. aureus infections.
3. Discussion
The identification and development of new antibacterial agents with novel target mechanisms against the highly virulent MDR S. aureus strains are imperative to avoid the risk associated with resistant infections [23]. The unremitting legacy of utilizing naturally derived compounds for therapeutic advance emphasized exploring the ability of known chemical scaffolds as promising candidates to combat MDR S. aureus infections [14].
Malabaricones have gained adequate attention owing to their structural characteristics and broad array of pharmacological activities, including antioxidant [24], anti-cancer [25,26,27,28], anti-inflammatory [29,30], anti-hypertensive [31], anti-fungal [32], etc. The first report on the antibacterial activity of malabaricones (B and C) isolated from the mace of Myristica fragrans against microorganisms (S. aureus, B. subtilis, S. durans, and C. albicans) appeared many years ago [33], indicating the potent activity of malabaricone B against S. aureus compared to malabaricone C. It also revealed some structure–activity relationships responsible for antibacterial activity. Surprisingly, apart from this preliminary screening, malabaricone B has not been extensively studied to establish its antibacterial properties since then, while few studies have been published regarding the antimicrobial actions of malabaricone C, including Arg-gingipain inhibition [34], anti-quorum sensing activity [35], and larvicidal activity [36].
In this perspective, the present study focused on the detailed biological characterization of the rare phenylacylphenol class compound malabaricone B (NS-7) from the Myristicaceae (the nutmeg) family. To begin with, naturally known abundant malabaricones were isolated from the fruit rind of M. malabarica, which is identified as the reservoir of these secondary metabolites. The hit compound NS-7, with a favorable SI, exhibited excellent activity against multiple MDR strains of S. aureus and Enterococcus, demonstrating its ability to circumvent existing resistance mechanisms and its lack of cross-resistance. In the presence of PMBN, an OM permeabilizer, NS-7 could act against the GNBs E. coli and MDR A. baumannii, indicating its broad-spectrum activity and the probable presence of a common target. Additionally, as NS-7 contains a phenolic part, it might be reduced by the thioredoxin system, which makes Gram-positive bacteria more susceptible to it as compared to Gram-negative bacterial pathogens. NS-7 exhibited rapid bactericidal killing kinetics that were reflected in its low propensity for resistance induction in S. aureus.
NS-7 also caused efficient disruption of established S. aureus biofilm at low concentrations, along with good cellular penetration in acting against intracellular S. aureus. The ability of NS-7 to act against S. aureus’s indifferent metabolic states makes it a good candidate for further development as an antibacterial lead. Its prolonged PAE, which can be attributed to its rapid bactericidal activity, is an added advantage for further developing it as a potent antibacterial drug. A longer PAE favors a short exposure time to attain superior activity with a lower dosage administration [37]. Mechanistic studies via confocal microscopy and SEM indicated membrane damage at bactericidal concentrations of NS-7, corroborated by the extracellular release of large macromolecules, such as ATP, ultimately leading to bacterial cell death.
Recent advances in combination therapy to defeat life-threatening diseases indicate the importance of synergism between drugs for pronounced effect. NS-7 promisingly synergized with gentamicin and daptomycin, outcompeting the drug-alone groups against both sensitive and gentamicin-resistant S. aureus. Its impressive in vitro antibacterial profile prompted us to determine its in vivo efficacy in murine neutropenic thigh and skin infection models. With a maximum tolerable dose of ≥250 mg/kg in mice, NS-7 exhibited good efficacy in both the infection models at much lower doses, indicating its potential as an antibacterial lead candidate.
Thus, we have developed malabaricone B as a new antibacterial drug candidate targeting MDR and persistent S. aureus infections.
4. Materials and Methods
4.1. Plant Material
Myristica malabarica fruits were collected from the Chemunji Hills of the Agasthyamalai Biosphere Reserve in Thiruvananthapuram District, Kerala, India. Dr. Mathew Dan (plant taxonomist, JNTBGRI, Palode, Trivandrum, India) authenticated the collected material, and a voucher specimen [Myristicamalabarica, Chemunji Hills, Trivandrum, Kerala, April, 2017, Govind, 83442 (TBGT)] was deposited in the institute’s herbarium.
4.2. Extraction and Isolation
Initially, rinds were separated from the fruits and air-dried. The grounded material (670 g) was extracted with dichloromethane (2 L × 3 days). The crude extract was obtained by decanting, filtering, and evaporating the supernatant under reduced pressure in a Heidolph rotary evaporator. Further fractionation of the crude extract (25 g) over 100–200 mesh silica gel by gradient elution with hexane/ethylacetate (100:0 to 0:100, v/v) yielded 60 fractions. Based on the similarities in TLC, these fractions were combined together, and repeated column chromatic separation followed by crystallization techniques resulted in the isolation of pure compounds.
4.3. Reagents and Growth Media
Bacterial culture media (MHB, MHBII, and TSB) were purchased from Becton-Dickinson (Franklin Lakes, NJ, USA). All antibiotics and chemicals used in the studies were obtained from Sigma-Aldrich (St. Louis, MO, USA). Eukaryotic cell culture growth media RPMI and FBS were purchased from Gibco (Waltham, MA, USA).
4.4. Bacterial Strains
The ESKAPE panel comprised Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 29213, Klebsiella pneumoniae BAA-1705, Acinetobacter baumannii BAA-1605, Pseudomonas aeruginosa ATCC 27853, and Enterococcus NR 31903. The clinical MDR S. aureus and Enterococcus strain details are provided in Table 4. All strains were procured from the American Type Culture Collection (ATCC, Manassas, VA, USA) and Biodefense and Emerging Infectious Disease/Network on Antimicrobial Resistance in Staphylococcus aureus (BEI/NARSA), and were routinely cultivated on appropriate culture media, namely MHB II, MHA, TSB, or TSA.
4.5. Antibacterial Susceptibility Testing against ESKAPE Panel of Bacteria
The antibacterial activity of test compounds was assessed against an ESKAPE pathogen panel via broth microdilution assay using standard CLSI guidelines [38], as described before. The MIC of each test compound was determined through three replicate measurements.
4.6. Outer Membrane Susceptibility Assay
As previously described, polymyxin B nonapeptide (PMBN) was used at 10 µg/mL in the culture medium to determine the susceptibility of Gram-negative bacteria-E. coli and A. baumannii towards the tested compounds via broth microdilution assay [39]. Levofloxacin, vancomycin, and rifampicin served as controls.
4.7. Cell Viability Assay in Vero Cells
To test the effect of compounds on the growth of mammalian cells, a cell viability assay was performed using MTT, as per published protocols [40]. The SI (selectivity index) was calculated as CC50/MIC, where CC50 is defined as the compound concentration resulting in 50% reduction in cell viability post-treatment. The experiments were performed thrice, and the average mean was used to calculate CC50 and SI.
4.8. Activity against MDR Strains of S. aureus and Enterococcus Panel
NS-7 was tested against clinical MDR S. aureus and Enterococcus strains comprising of MSSA, MRSA, VRSA, VSE, and VRE via broth microdilution assay. The resistance profile of each strain is described in Table 4.
4.9. Time–Kill Kinetics Study
The killing kinetics of compound were determined via time–kill kinetics assay according to CLSI guidelines, following a previously described protocol [40]. Treated and untreated S. aureus samples were withdrawn at 0 min, 15 min, 30 min, 45 min, 60 min, 6 h and 24 h, and the cfu/mL at each time point was determined and plotted with respect to time. Experiments were performed in triplicate and the mean data with SD were plotted.
4.10. Biofilm Eradication Assay
In vitro S. aureus biofilms were grown and treated with NS-7 and vancomycin at 1X MIC using the method described before [41]. A total of 0.06% crystal violet was used to stain the remaining biofilm mass post-treatment, while 30% acetic acid (0.2 mL/well) was utilized to perform the elution, and quantification was performed by measuring the absorbance at 600 nm. Experiments were repeated thrice, and the average mean was plotted with SD.
4.11. Intracellular Activity Assay
The murine macrophage cell line J774.A1 was used to establish intracellular S. aureus infection, and the ability of NS-7 to kill S. aureus inside host eukaryotic cells was determined according to the previously described protocol [42]. Macrophages were treated with 1X and 10X MIC of NS-7, while vancomycin at 5X MIC and untreated macrophages served as controls. Experiments were repeated thrice, and the average mean along with the SD were plotted for each group.
4.12. Synergy Screening of NS-7 with Frontline Antibiotics
A checkerboard assay was used to determine fractional inhibitory concentrations (ΣFICs) as a measure of synergy between the test compound and clinical antibiotics in combination [40]. The antibiotics that were examined in this study encompassed daptomycin, gentamicin, levofloxacin, linezolid, meropenem, minocycline, rifampicin, and vancomycin. The summation of fractional inhibitory concentrations (ΣFIC) was determined using the following formula: ΣFIC = FIC A + FIC B. Here, FIC A represents the ratio of the MIC of compound “A” in combination with antibiotic “B” to the MIC of compound “A” alone, while FIC B represents the ratio of the MIC of antibiotic “B” in combination with compound “A” to the MIC of antibiotic “B” alone. The combination is deemed synergistic when the ΣFIC is equal to or less than 0.5. It is considered indifferent when the ΣFIC is greater than 0.5 but less than or equal to 4. Lastly, it is classified as antagonistic when the ΣFIC exceeds 4 [22].
4.13. Induced Resistant Mutant Generation Studies
S. aureus ATCC29213 was subjected to 40 days of serial passaging in the presence of subinhibitory concentrations of NS-7 and levofloxacin to induce resistance. The changes in MIC were monitored every third passage and plotted against the number of passages, and the fold increase in MIC was calculated as previously described [43].
4.14. Determination of Post-Antibiotic Effect (PAE)
S. aureus ATCC29213 grown overnight and subsequently diluted to ~105 cfu/mL was subjected to 1X and 5X MIC of NS-7 and the control antibiotics vancomycin and levofloxacin for 1 h. PAE was assessed in accordance with the previously outlined protocol [37,41].
4.15. Bacterial LIVE/DEAD BacLight Assay
The LIVE/DEAD BacLight bacterial viability kit (Invitrogen, Waltham, MA, USA, catalogue no. L7007) was used to assess the viability of S. aureus ATCC29213 upon treatment with NS-7, as described in the manufacturer guidelines. The method consists of two fluorescent probes SYT09 (the green intercalating stain permeates the membranes of both live and dead cells) and propidium iodide (PI; the red intercalating stain enters only the cells with damaged membranes) with excitation/emission wavelengths corresponding to 488/530 nm and 488/620 nm, respectively. Initially, S. aureus cells were adjusted to OD600 = 0.2 in PBS buffer and were exposed to NS-7 at 5X MIC for 10−30 min. After the compound exposure the bacterial suspensions were washed in PBS buffer and incubated with dye mixture of SYTO9 and PI (mixing equal volume of each) for 20 min in the dark at 37 °C. An aliquot of the stained sample was taken, and the fluorescent images were examined by using a Leica SP8 laser scanning confocal microscope (Leica, Wetzlar, Germany).
4.16. Scanning Electron Microscopy (SEM)
Scanning electron microscopy was used to examine the surface morphological changes in S. aureus ATCC29213 cells. Briefly, the mid-logarithmic-phase S. aureus cells (~108 cfu/mL) were treated with 5X MIC of NS-7 at 37 °C for 30 min with untreated bacterial cells as controls. After the incubation period, the bacterial cells were washed two times by using PBS (pH 7.4) and fixed overnight with 2.5% glutaraldehyde in PBS at 4 °C. The next day, the cells were dehydrated in a series of graded ethanol (20–95%) and finally dissolved in pure ethanol and vacuum-dried. The dried cells were gold-coated with an automatic sputter coater and, finally, the images of the samples were recorded by using Quanta 3D 250 field-emission scanning electron microscopy at 30 kV for S. aureus.
4.17. Determination of Extracellular and Intracellular ATP
An ATP Bioluminescence Assay Kit CLS II (Roche, Basel, Switzerland, Cat. No. 11699695001) was used to measure the extracellular and intracellular ATP levels of S. aureus in accordance with the manufacturer’s instructions and the previously described protocol [40]. Briefly, S. aureus ATCC29213 culture in log phase was treated with 2.5X and 5X MIC of NS-7 and 2.5X melittin to determine extracellular and intracellular ATP levels. The luciferase agent was added to the extracellular and intracellular samples in equal volumes. This addition was carried out using automated injection into 96-well white plates. The signals obtained were then integrated for a duration of 1 to 10 s using the Promega GloMax®® 96 Microplate Luminometer.
4.18. Murine Neutropenic Thigh Infection Model
Here, 4–5 weeks old BALB/c mice weighing~22–24 g were grouped together in a set of 5/per cage. Mice were rendered neutropenic, infected with (107–109 cfu/mL) S. aureus intramuscularly, treated, and sacrificed according to the previously described protocol [44]. Experimental groups were NS-7 (50 mg/kg), vancomycin (25 mg/Kg),and untreated controls that were administered saline. Post-sacrifice, each removed thigh was homogenized in 5 mL of PBS, serial dilutions were performed, and the homogenized samples were plated on Mueller–Hinton agar plates to quantify the colony-forming units (cfu). Following incubation at a temperature of 37 °C for a duration of 18–24 h, the cfu were quantified. The experiments were conducted in triplicate, and the resulting mean data were graphed along with standard deviation (SD) from all three experiments.
4.19. Murine Skin Infection Model
This in vivo study utilized 6-week-old male BALB/c mice in accordance with the previously established protocol [40]. The experimental groups consisted of untreated control mice, mice treated with 2% fusidic acid (serving as the positive control), mice treated with 2% NS-7 (the test drug), and mice treated with the vehicle (base). The experiments were conducted three times, and the mean log10cfu/g was determined for each experiment. The average mean and SD were then graphed based on the results obtained from all three experiments.
5. Conclusions
To conclude, the present study illustrates the development of a known natural product, malabaricone B (NS-7), a phenylacylphenol from the fruit rinds of Myristic malabarica, as a membrane-active, rapid, bactericidal agent equipotently active against MDR S. aureus without any inducible resistance. S-7 demonstrates significant activity against clinical MDR strains of S. aureus and Enterococcus, lacks cytotoxicity towards eukaryotic cells, efficiently eradicates preformed biofilms, and is highly efficient in killing intracellular S. aureus. In addition, NS-7 exhibits excellent synergistic interactions with daptomycin and gentamicin against S. aureus ATCC 29213, including gentamicin-resistant MRSA NRS119, and demonstrates its in vivo potential by reducing the bacterial load in both S. aureus-infected neutropenic thigh infection and skin infection models. The rapid bactericidal potential of NS-7 proceeds via lysing the bacterial cell membrane suggesting it to be a membrane-active antibacterial agent. Taken together, NS-7 exhibits all the desired properties to be efficiently translated as a potent anti-staphylococcal therapeutic.
6. Patents
The work reported here has been duly filed for an Indian patent with application number 202111044825.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12101483/s1, Figure S1: Schematic representation of extraction and isolation procedure from rind; Figures S2, S4, S6, S8, S10, and S12: 1H NMR spectrum of NS-1, NS-3, NS-5, NS-7, NS-9, and NS-11, respectively; Figures S3, S5, S7, S9, S11, and S13: 13C NMR spectrum of NS-1, NS-3, NS-5, NS-7, NS-9, and NS-11, respectively.
Author Contributions
Conceptualization, N.S., G.K., M.G.G., M.D., K.V.R. and S.C.; methodology, N.S., G.K., M.D., A.A. and M.S.; software, N.S. and G.K.; validation, K.V.R. and S.C.; formal analysis, N.S. and G.K.; investigation, N.S., G.K., M.G.G., M.D., M.S. and A.A.; resources, K.V.R. and S.C.; data curation, N.S. and G.K.; writing—original draft preparation, N.S. and G.K.; writing—review and editing, K.V.R. and S.C.; visualization, N.S. and G.K.; supervision, K.V.R. and S.C.; project administration, K.V.R. and S.C.; funding acquisition, K.V.R. and S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CSIR Intramural funds to K.V.R. and S.C.
Institutional Review Board Statement
The animal study protocol was approved by the institutional review board (or ethics committee) of the CSIR-CENTRAL DRUG RESEARCH INSTITUTE, LUCKNOW, UP, INDIA (IAEC/2019/2/Renew-1/Dated-22 June 2020) for studies involving animals.
Data Availability Statement
The data presented in this study are available in the manuscript and supplementary material.
Acknowledgments
N.S., M.S. and A.A. thank UGC for the funding, G.K. thanks DST INSPIRE, and M.G.G thanks the University of Kerala for their fellowship. The following reagents used in the study were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, and NIH: NR100, NR119, NR129, NR186, NR191, NR192, NR193, NR194, NR198, VRS1, VRS4, VRS12, NR31884, NR31885, NR31886, NR31887, NR31888, NR31903, NR31909, and NR31912. This manuscript bears the CSIR-CDRI communication number 10601.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-Resistant Staphylococcus aureus: An Overview of Basic and Clinical Research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Katayama, Y.; Matsuo, M.; Sasaki, T.; Morimoto, Y.; Sekiguchi, A.; Baba, T. Multi-Drug-Resistant Staphylococcus aureus and Future Chemotherapy. J. Infect. Chemother. 2014, 20, 593–601. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Wright, G.D. Molecular Mechanisms of Antibiotic Resistance. Chem. Commun. 2011, 47, 4055–4061. [Google Scholar] [CrossRef]
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Chopra, B.; Dhingra, A.K. Natural Products: A Lead for Drug Discovery and Development. Phytother. Res. 2021, 35, 4660–4702. [Google Scholar] [CrossRef] [PubMed]
- Rossiter, S.E.; Fletcher, M.H.; Wuest, W.M. Natural Products as Platforms to Overcome Antibiotic Resistance. Chem. Rev. 2017, 117, 12415–12474. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-C.; Liu, F.; Zhu, K.; Shen, J.-Z. Natural Products That Target Virulence Factors in Antibiotic-Resistant Staphylococcus aureus. J. Agric. Food Chem. 2019, 67, 13195–13211. [Google Scholar] [CrossRef]
- Porras, G.; Chassagne, F.; Lyles, J.T.; Marquez, L.; Dettweiler, M.; Salam, A.M.; Samarakoon, T.; Shabih, S.; Farrokhi, D.R.; Quave, C.L. Ethnobotany and the Role of Plant Natural Products in Antibiotic Drug Discovery. Chem. Rev. 2020, 121, 3495–3560. [Google Scholar] [CrossRef]
- Liu, K.; Huigens, R.W., III. Instructive Advances in Chemical Microbiology Inspired by Nature’s Diverse Inventory of Molecules. ACS Infect. Dis. 2019, 6, 541–562. [Google Scholar] [CrossRef]
- Abouelhassan, Y.; Garrison, A.T.; Yang, H.; Chávez-Riveros, A.; Burch, G.M.; Huigens, R.W., III. Recent Progress in Natural-Product-Inspired Programs Aimed to Address Antibiotic Resistance and Tolerance. J. Med. Chem. 2019, 62, 7618–7642. [Google Scholar] [CrossRef]
- Tyers, M.; Wright, G.D. Drug Combinations: A Strategy to Extend the Life of Antibiotics in the 21st Century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Kaul, G.; Shukla, M.; Dasgupta, A.; Chopra, S. Update on Drug-Repurposing: Is It Useful for Tackling Antimicrobial Resistance? Future Microbiol. 2019, 14, 829–831. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Prabhakaran, V.-S.; Kim, K. The Multi-Faceted Potential of Plant-Derived Metabolites as Antimicrobial Agents against Multidrug-Resistant Pathogens. MicrobPathog 2018, 116, 209–214. [Google Scholar] [CrossRef]
- Silva, L.N.; Zimmer, K.R.; Macedo, A.J.; Trentin, D.S. Plant Natural Products Targeting Bacterial Virulence Factors. Chem. Rev. 2016, 116, 9162–9236. [Google Scholar] [CrossRef] [PubMed]
- Davison, E.K.; Brimble, M.A. Natural Product Derived Privileged Scaffolds in Drug Discovery. Curr. Opin. Chem. Biol. 2019, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, K.K.; Sarada, A.; Connolly, J.D. Malabaricones A–D, Novel Diarylnonanoids from Myristica malabarica Lam (Myristicaceae). J. Chem. Soc. Perkin Trans. 1 1977, 587–588. [Google Scholar] [CrossRef]
- Chelladurai, P.K.; Ramalingam, R. Myristica malabarica: A Comprehensive Review. J. Pharmacogn. Phytochem. 2017, 6, 255–258. [Google Scholar]
- Yin, W.; Xu, S.; Wang, Y.; Zhang, Y.; Chou, S.H.; Galperin, M.Y.; He, J. Ways to Control Harmful Biofilms: Prevention, Inhibition, and Eradication. Crit. Rev. Microbiol. 2021, 47, 57. [Google Scholar] [CrossRef]
- Craft, K.M.; Nguyen, J.M.; Berg, L.J.; Townsend, S.D. Methicillin-Resistant Staphylococcus aureus (MRSA): Antibiotic-Resistance and the Biofilm Phenotype. Medchemcomm 2019, 10, 1231–1241. [Google Scholar] [CrossRef]
- Ayaz, M.; Ullah, F.; Sadiq, A.; Ullah, F.; Ovais, M.; Ahmed, J.; Devkota, H.P. Synergistic Interactions of Phytochemicals with Antimicrobial Agents: Potential Strategy to Counteract Drug Resistance. Chem. Biol. Interact. 2019, 308, 294–303. [Google Scholar] [CrossRef]
- Odds, F.C. Synergy, Antagonism, and What the Chequerboard Puts between Them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Beyer, P.; Paulin, S. The Antibacterial Research and Development Pipeline Needs Urgent Solutions. ACS Infect. Dis. 2020, 6, 1289–1291. [Google Scholar] [CrossRef]
- Patro, B.S.; Bauri, A.K.; Mishra, S.; Chattopadhyay, S. Antioxidant Activity of Myristica malabarica Extracts and Their Constituents. J. Agric. Food Chem. 2005, 53, 6912–6918. [Google Scholar] [CrossRef]
- Tyagi, M.; Patro, B.S.; Chattopadhyay, S. Mechanism of the Malabaricone C-Induced Toxicity to the MCF-7 Cell Line. Free Radic. Res. 2014, 48, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Patro, B.S.; Tyagi, M.; Saha, J.; Chattopadhyay, S. Comparative Nuclease and Anti-Cancer Properties of the Naturally Occurring Malabaricones. Bioorg. Med. Chem. 2010, 18, 7043–7051. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Bhattacharyya, R.; Bauri, A.K.; Patro, B.S.; Chattopadhyay, S. DNA Damage Dependent Activation of Checkpoint Kinase-1 and Mitogen-Activated Protein Kinase-P38 Are Required in Malabaricone C-Induced Mitochondrial Cell Death. Biochim. Et. Biophys. Acta BBA-Gen. Subj. 2014, 1840, 1014–1027. [Google Scholar] [CrossRef]
- Manna, A.; De Sarkar, S.; De, S.; Bauri, A.K.; Chattopadhyay, S.; Chatterjee, M. Impact of MAPK and PI3K/AKT Signaling Pathways on Malabaricone-A Induced Cytotoxicity in U937, a Histiocytic Lymphoma Cell Line. Int. Immunopharmacol. 2016, 39, 34–40. [Google Scholar] [CrossRef]
- Kang, J.; Tae, N.; Min, B.S.; Choe, J.; Lee, J.-H. Malabaricone C Suppresses Lipopolysaccharide-Induced Inflammatory Responses via Inhibiting ROS-Mediated Akt/IKK/NF-ΚB Signaling in Murine Macrophages. Int. Immunopharmacol. 2012, 14, 302–310. [Google Scholar] [CrossRef]
- Maity, B.; Yadav, S.K.; Patro, B.S.; Tyagi, M.; Bandyopadhyay, S.K.; Chattopadhyay, S. Molecular Mechanism of the Anti-Inflammatory Activity of a Natural Diarylnonanoid, Malabaricone C. Free Radic. Biol. Med. 2012, 52, 1680–1691. [Google Scholar] [CrossRef]
- Rathee, J.S.; Patro, B.S.; Brown, L.; Chattopadhyay, S. Mechanism of the Anti-Hypertensive Property of the Naturally Occurring Phenolic, Malabaricone C in DOCA-Salt Rats. Free Radic. Res. 2016, 50, 111–121. [Google Scholar] [CrossRef]
- Choi, N.H.; Choi, G.J.; Jang, K.S.; Choi, Y.H.; Lee, S.O.; Choi, J.E.; Kim, J.C. Antifungal Activity of the Methanol Extract of Myristica malabarica Fruit Rinds and the Active Ingredients Malabaricones against Phytopathogenic Fungi. Plant Pathol. J. 2008, 24, 317–321. [Google Scholar] [CrossRef]
- Orabi, K.Y.; Mossa, J.S.; El-Feraly, F.S. Isolation and Characterization of Two Antimicrobial Agents from Mace (Myristica fragrans). J. Nat. Prod. 1991, 54, 856–859. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, C.; Mori, S.; Ando, T.; Tsuji, T. Arg-Gingipain Inhibition and Anti-Bacterial Activity Selective for Porphyromonas Gingivalis by Malabaricone C. Biosci. Biotechnol. Biochem. 1999, 63, 1475–1477. [Google Scholar]
- Chong, Y.M.; Yin, W.F.; Ho, C.Y.; Mustafa, M.R.; Hadi, A.H.A.; Awang, K.; Narrima, P.; Koh, C.-L.; Appleton, D.R.; Chan, K.-G. Malabaricone C from Myristica Cinnamomea Exhibits Anti-Quorum Sensing Activity. J. Nat. Prod. 2011, 74, 2261–2264. [Google Scholar] [CrossRef]
- Nakamura, N.; Kiuchi, F.; Tsuda, Y.; Kondo, K. Studies on Crude Drugs Effective on Visceral Larva Migrans. V.: The Larvicidal Principle in Mace (Aril of Myristica fragrans). Chem. Pharm. Bull. 1988, 36, 2685–2688. [Google Scholar] [CrossRef]
- Craig, W.A. The Postantibiotic Effect. Clin. Microbiol. Newsl. 1991, 13, 121–124. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing—30th Edition: M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2000. [Google Scholar]
- Thangamani, S.; Mohammad, H.; Abushahba, M.F.N.; Sobreira, T.J.P.; Hedrick, V.E.; Paul, L.N.; Seleem, M.N. Antibacterial Activity and Mechanism of Action of Auranofin against Multi-Drug Resistant Bacterial Pathogens. Sci. Rep. 2016, 6, 22571. [Google Scholar] [CrossRef]
- Meenu, M.T.; Kaul, G.; Akhir, A.; Shukla, M.; Radhakrishnan, K.V.; Chopra, S. Developing the Natural Prenylflavone Artocarpin from Artocarpus Hirsutus as a Potential Lead Targeting Pathogenic, Multidrug-Resistant Staphylococcus aureus, Persisters and Biofilms with No Detectable Resistance. J. Nat. Prod. 2022, 85, 2413–2423. [Google Scholar] [CrossRef]
- Grace, K.; Abdul, A.; Manjulika, S.; Hasham, S.; Ravikumar, A.; Gaurav, P.; Mahammad, G.; Nanduri, S.; Sidharth, C. Oxiconazole Potentiates Gentamicin against Gentamicin-Resistant Staphylococcus aureus In Vitro and In Vivo. Microbiol. Spectr. 2023, 11, e05031-22. [Google Scholar] [CrossRef]
- Seral, C.; Van Bambeke, F.; Tulkens, P.M. Quantitative Analysis of Gentamicin, Azithromycin, Telithromycin, Ciprofloxacin, Moxifloxacin, and Oritavancin (LY333328) Activities against Intracellular Staphylococcus aureus in Mouse J774 Macrophages. Antimicrob. Agents Chemother. 2003, 47, 2283–2292. [Google Scholar] [CrossRef]
- Suller, M.T.E.; Lloyd, D. The Antibacterial Activity of Vancomycin towards Staphylococcus aureus under Aerobic and Anaerobic Conditions. J. Appl. Microbiol. 2002, 92, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Kaul, G.; Akhir, A.; Shukla, M.; Rawat, K.S.; Sharma, C.P.; Sangu, K.G.; Rode, H.B.; Goel, A.; Chopra, S. Nitazoxanide Potentiates Linezolid against Linezolid-Resistant Staphylococcus aureus In Vitro and In Vivo. J. Antimicrob. Chemother. 2022, 77, 2456–2460. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).