Citizen Contribution for Searching for Alternative Antimicrobial Activity Substances in Soil
Abstract
1. Introduction
2. Material and Methods
2.1. MicroMundo: Service-Learning Methodology
2.2. Second Screening of Antimicrobial Activity by the Spot-on-Lawn Method
2.3. Bacterial Identification
2.4. Antibiotic Susceptibility Testing of Antimicrobial-Producing Strains
2.5. Diversity of Antimicrobial-Producing Bacteria and Statistics
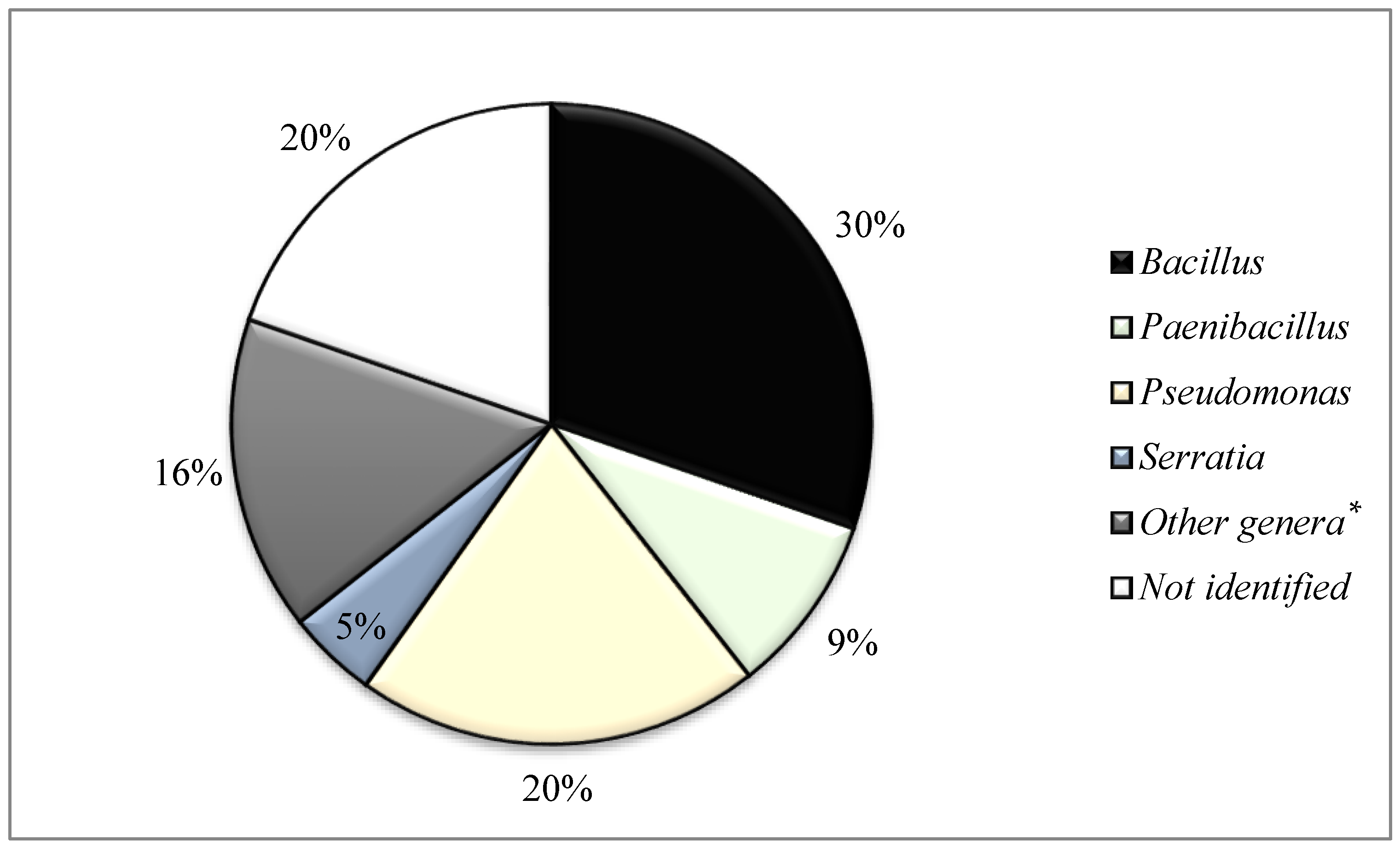
3. Results
3.1. Verification of Antimicrobial Activity of Antimicrobial-Producing Bacteria in a Second Screening Process against 15 Indicator Bacteria
3.2. Antibiotic Resistance Phenotype of the Antimicrobial-Producing Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Yount, N.Y.; Weaver, D.C.; De Anda, J.; Lee, E.Y.; Lee, M.W.; Wong, G.C.L.; Yeaman, M.R. Discovery of Novel Type II Bacteriocins Using a New High-Dimensional Bioinformatic Algorithm. Front. Immunol. 2020, 11, 01873. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Saini, K.C.; Bast, F.; Varjani, S.; Mehariya, S.; Bhatia, S.K.; Sharma, N.; Funk, C. A Review on Microbial Products and Their Perspective Application as Antimicrobial Agents. Biomolecules 2021, 11, 1860. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef] [PubMed]
- Twomey, E.; Hill, C.; Field, D.; Begley, M. Recipe for Success: Suggestions and Recommendations for the Isolation and Characterisation of Bacteriocins. Int. J. Microbiol. 2021, 2021, 1–19. [Google Scholar] [CrossRef]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, Ecology, and Application. Annu. Rev. Microbiol. 2002, 56, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Telhig, S.; Ben Said, L.; Torres, C.; Rebuffat, S.; Zirah, S.; Fliss, I. Evaluating the Potential and Synergetic Effects of Microcins against Multidrug-Resistant Enterobacteriaceae. Microbiol. Spectr. 2022, 10, e02752-21. [Google Scholar] [CrossRef]
- Curtis, T.P.; Sloan, W.T.; Scannell, J.W. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 2002, 99, 10494–10499. [Google Scholar] [CrossRef]
- Schloss, P.; Handelsman, J. Toward a Census of Bacteria in Soil. PLOS Comput. Biol. 2006, 2, e92. [Google Scholar] [CrossRef]
- Hover, B.M.; Kim, S.-H.; Katz, M.; Charlop-Powers, Z.; Owen, J.G.; Ternei, M.A.; Maniko, J.; Estrela, A.B.; Molina, H.; Park, S.; et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018, 3, 415–422. [Google Scholar] [CrossRef]
- Handelsman, J. Small World Initiative: Research Protocols and Research Guide to Microbial and Chemical Diversity; XanEdu Publishing Inc.: Ann Arbor, MI, USA, 2015. [Google Scholar]
- Valderrama, M.J.; González, E.R.; De Pablo, P.C.; Díez-Orejas, R.; Fernández-Acero, T.; Gil-Serna, J.; De Juan, L.; Martín, H.; Molina, M.; Navarro-García, F.; et al. Educating in antimicrobial resistance awareness: Adaptation of the Small World Initiative program to service-learning. FEMS Microbiol. Lett. 2018, 365, fny161. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.; Sloan, T.; Aurelius, K.; Barbour, A.; Bodey, E.; Clark, B.; Dennis, C.; Drown, R.; Fleming, M.; Humbert, A.; et al. Antibiotic discovery throughout the Small World Initiative: A molecular strategy to identify biosynthetic gene clusters involved in antagonistic activity. MicrobiologyOpen 2017, 6, e00435. [Google Scholar] [CrossRef] [PubMed]
- Robredo, B.; Fernández-Fernández, R.; Torres, C. Antimicrobial resistance as a nexus between teaching and research. J. Biol. Educ. 2021, 1–17. [Google Scholar] [CrossRef]
- Ghosh, A.; Dey, N.; Bera, A.; Tiwari, A.; Sathyaniranjan, K.; Chakrabarti, K.; Chattopadhyay, D. Culture independent molecular analysis of bacterial communities in the mangrove sediment of Sundarban, India. Saline Syst. 2010, 6, 1. [Google Scholar] [CrossRef]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 26 November 2022).
- Reddy, B.V.B.; Kallifidas, D.; Kim, J.H.; Charlop-Powers, Z.; Feng, Z.; Brady, S.F. Natural Product Biosynthetic Gene Diversity in Geographically Distinct Soil Microbiomes. Appl. Environ. Microbiol. 2012, 78, 3744–3752. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Liu, B.; Yin, Y.; Liang, F.; Xie, D.; Han, T.; Liu, Y.; Yan, B.; Li, Q.; Huang, Y.; et al. Impact of biocontrol microbes on soil microbial diversity in ginger (Zingiber officinale Roscoe). Pest Manag. Sci. 2021, 77, 5537–5546. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef]
- Balthazar, C.; Novinscak, A.; Cantin, G.; Joly, D.L.; Filion, M. Biocontrol Activity of Bacillus spp. and Pseudomonas spp. Against Botrytis cinerea and Other Cannabis Fungal Pathogens. Phytopathology 2022, 112, 549–560. [Google Scholar] [CrossRef]
- Dimkić, I.; Janakiev, T.; Petrović, M.; Degrassi, G.; Fira, D. Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms—A review. Physiol. Mol. Plant Pathol. 2021, 117, 101754. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, Z.; Bai, X.; Zhang, D.; Zhang, L.; Wang, J.; Wu, B.; Zhu, J.; Yang, Z. A novel of new class II bacteriocin from Bacillus velezensis HN-Q-8 and its antibacterial activity on Streptomyces scabies. Front. Microbiol. 2022, 13, 943232. [Google Scholar] [CrossRef] [PubMed]
- Ghequire, M.G.; De Mot, R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol. Rev. 2014, 38, 523–568. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, E.J.; Rabinovitch, L.; Monnerat, R.G.; Passos, L.K.J.; Zahner, V. Molecular Characterization of Brevibacillus laterosporus and Its Potential Use in Biological Control. Appl. Environ. Microbiol. 2004, 70, 6657–6664. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, G.; Ferrari, C.; Mamberti, S.; Gabrieli, P.; Castelli, M.; Sassera, D.; Ursino, E.; Scoffone, V.C.; Radaelli, G.; Clementi, E.; et al. Identification of a Novel Brevibacillus laterosporus Strain with Insecticidal Activity Against Aedes albopictus Larvae. Front. Microbiol. 2021, 12, 624014. [Google Scholar] [CrossRef] [PubMed]
- Javed, K.; Qiu, D. Protein Elicitor PeBL1 of Brevibacillus laterosporus Enhances Resistance Against Myzus persicae in Tomato. Pathogens 2020, 9, 57. [Google Scholar] [CrossRef]
- Khaled, J.M.; Al-Mekhlafi, F.A.; Mothana, R.A.; Alharbi, N.S.; Alzaharni, K.E.; Sharafaddin, A.H.; Kadaikunnan, S.; Alobaidi, A.S.; Bayaqoob, N.I.; Govindarajan, M.; et al. Brevibacillus laterosporus isolated from the digestive tract of honeybees has high antimicrobial activity and promotes growth and productivity of honeybee’s colonies. Environ. Sci. Pollut. Res. 2017, 25, 10447–10455. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kwon, J.H.; Ahn, S.H.; Lee, S.I.; Han, Y.S.; Choi, Y.O.; Lee, S.Y.; Ahn, K.M.; Ji, G.E. Effect of probiotic mix (Bifidobacterium bifidum, Bifidobacterium lactis, Lactobacillus acidophilus) in the primary prevention of eczema: A double-blind, randomized, placebo-controlled trial. Pediatr. Allergy Immunol. 2010, 21, e386–e393. [Google Scholar] [CrossRef]
- Wilaipun, P.; Zendo, T.; Okuda, K.-I.; Nakayama, J.; Sonomoto, K. Identification of the Nukacin KQU-131, a New Type-A(II) Lantibiotic Produced by Staphylococcus hominis KQU-131 Isolated from Thai Fermented Fish Product (Pla-ra). Biosci. Biotechnol. Biochem. 2008, 72, 2232–2235. [Google Scholar] [CrossRef]
- Fernández-Fernández, R.; Lozano, C.; Eguizábal, P.; Ruiz-Ripa, L.; Martínez-Álvarez, S.; Abdullahi, I.N.; Zarazaga, M.; Torres, C. Bacteriocin-Like Inhibitory Substances in Staphylococci of Different Origins and Species With Activity Against Relevant Pathogens. Front. Microbiol. 2022, 13, 870510. [Google Scholar] [CrossRef]
- Sung, C.; Kim, B.; Kim, S.; Joo, H.; Kim, P. Probiotic potential of Staphylococcus hominis MBBL 2–9 as anti- Staphylococcus aureus agent isolated from the vaginal microbiota of a healthy woman. J. Appl. Microbiol. 2010, 108, 908–916. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef] [PubMed]
- Sumi, C.D.; Yang, B.W.; Yeo, I.-C.; Hahm, Y.T. Antimicrobial peptides of the genus Bacillus: A new era for antibiotics. Can. J. Microbiol. 2015, 61, 93–103. [Google Scholar] [CrossRef] [PubMed]
| Genus | Species | Number of Isolates First Screening | Number of Isolates Second Screening |
|---|---|---|---|
| Acinetobacter | Acinetobacter radioresistens | 1 | |
| Arthrobacter | 2 | ||
| Arthrobacter citreus | 1 | 1 | |
| Arthrobacter ilicis | 1 | ||
| Bacillus | 40 | ||
| Bacillus marisflavi | 1 | ||
| Bacillus atrophaeus | 2 | 2 | |
| Bacillus cereus | 6 | 1 | |
| Bacillus cibi | 1 | ||
| Bacillus megaterium | 4 | 1 | |
| Bacillus mycoides | 4 | 1 | |
| Bacillus pumilus | 7 | 5 | |
| Bacillus safensis | 2 | 2 | |
| Bacillus simplex | 3 | ||
| Bacillus thuringiensis | 2 | ||
| Bacillus weihenstefanensis | 3 | ||
| Bacillus spp. | 5 | 2 | |
| Bradybacterium | Bradybacterium spp. | 1 | 1 |
| Brevibacillus | Brevibacillus laterosoporus | 1 | 1 |
| Enterobacter | Enterobacter cloacae | 3 | |
| Escherichia | Escherichia coli | 2 | |
| Klebsiella | Klebsiella aerogenes | 1 | 1 |
| Microbacterium | Microbacterium arborescensens | 2 | 1 |
| Micrococcus | Micrococcus luteus | 1 | |
| Paenibacillus | 12 | ||
| Paenibacillus amylolyticus | 6 | ||
| Paenibacillus apiarus | 2 | 2 | |
| Paenibacillus gluconolyticus | 1 | ||
| Paenibacillus lautus | 1 | ||
| Paenibacillus polymyxa | 1 | 1 | |
| Paenibacillus xylanilyticus | 1 | ||
| Pseudomonas | 27 | ||
| Pseudomonas brasicacearum | 2 | ||
| Pseudomonas brenneri | 1 | ||
| Pseudomonas caricapapayae | 1 | 1 | |
| Pseudomonas chlororaphis | 3 | 1 | |
| Pseudomonas kilonensis | 5 | 3 | |
| Pseudomonas koreensis | 2 | ||
| Pseudomonas mandelii | 1 | ||
| Pseudomonas mosselii | 1 | ||
| Pseudomonas putida | 1 | ||
| Pseudomonas savastanoi | 1 | ||
| Pseudomonas thivervalensis | 2 | ||
| Pseudomonas umsongensis | 1 | ||
| Pseudomonas spp. | 6 | 2 | |
| Staphylococcus | Staphylococcus hominis | 2 | 1 |
| Serratia | Serratia plymuthica | 6 | |
| Stenotrophomonas | Stenotrophomonas rhizophila | 1 | |
| Streptomyces | Streptomyces avidinii | 1 | 1 |
| Olivibacter | Olivibacter soli | 1 | 1 |
| Variovorax | Variovorax paradoxus | 1 | |
| Viridibacillus | Viridibacillus arenosi | 1 | |
| Not identified | 26 | ||
| Total | 132 | 32 | |
| Antimicrobial Activity a on the Indicator Bacteria b | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Producing Isolate | E. coli | P. aeruginosa | MRSA | MSSA | MRSP | MSSP | S. delphini | S. sciuri | S. epidermidis | E. faecalis | E. faecium c | E. cecorum | E. gallinarum | L. monocytogenes | M. luteus | No (%) |
| A. citreus X7246 | + | + | + | 3 (20) | ||||||||||||
| B. cereus X7247 | + | + | + | + | + | 5 (33) | ||||||||||
| B. safensis X7248 | + | + | + | + | + | 5 (33) | ||||||||||
| B. safensis X7249 | + | + | + | ++ | + | + | 6 (40) | |||||||||
| B. pumilus X7250 | + | + | + | + | + | + | + | + | 8 (53) | |||||||
| B. atrophaeus X7251 | + | + | + | + | + | 5 (33) | ||||||||||
| B. atrophaeus X7252 | + | + | + | + | + | + | + | 7 (47) | ||||||||
| Bacillus spp. X7253 | + | + | + | 3 (20) | ||||||||||||
| Bacillus spp. X7256. | + | + | + | + | + | + | + | + | 8 (53) | |||||||
| B. pumilus X7254 | + | + | + | + | + | + | 6 (40) | |||||||||
| B. megaterium X7255 | + | + | + | 3 (20) | ||||||||||||
| B. pumilus X7257 | + | + | + | 3 (20) | ||||||||||||
| B. mycoides X7258 | + | + | + | + | + | + | + | + | 8 (53) | |||||||
| B. pumilus X7259 | + | + | + | + | + | + | + | + | 8 (53) | |||||||
| B. pumilus X7260 | + | + | + | + | + | + | + | 7 (47) | ||||||||
| Bradybacterium spp. X7261 | + | + | 2 (13) | |||||||||||||
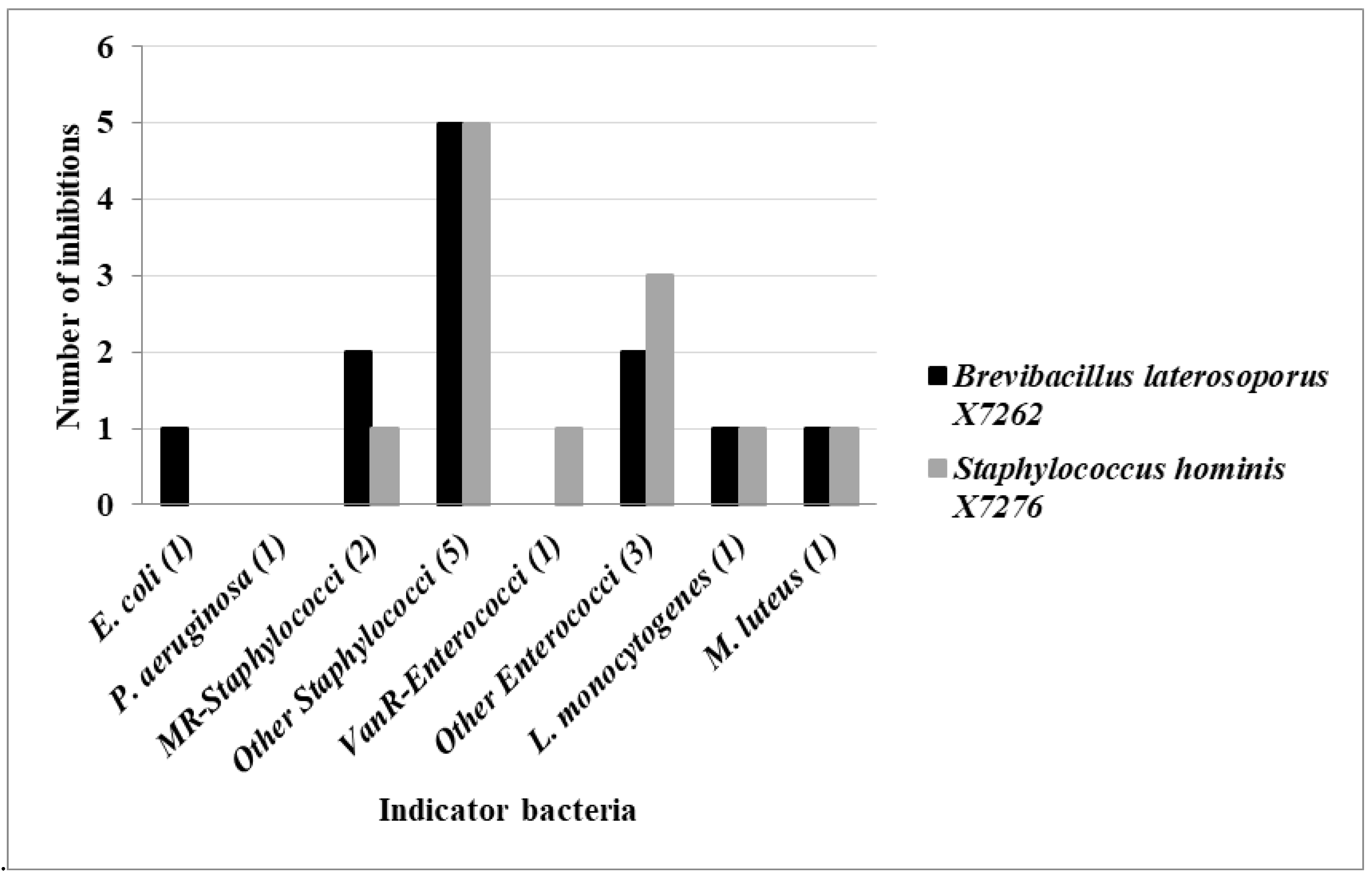
| B. laterosoporus X7262 | + | + | + | + | + | + | + | + | + | + | + | + | 12 (80) | |||
| M. arborescensens X7263 | + | + | + | + | + | + | + | 7 (47) | ||||||||
| P. apiarus X7264 | + | + | + | + | + | 5 (33) | ||||||||||
| P. apiarus X7267 | + | + | + | 3 (20) | ||||||||||||
| P. polymyxa X7268 | + | + | + | + | + | + | + | + | 8 (53) | |||||||
| S. hominis X7276 | + | + | + | + | + | + | + | + | + | + | + | + | 12 (80) | |||
| S. avidinii X7277 | + | + | + | + | 4 (27) | |||||||||||
| Number of inhibitions (%) | 6 (26) | 1 (4) | 18 (78) | 15 (65) | 4 (17) | 15 (65) | 11 (48) | 4 (17) | 23 (100) | 2 (9) | 1 (4) | 12 (52) | 1 (4) | 3 (13) | 22 (92) | |
| O. soli X7265 | + | + | + | 3 (20) | ||||||||||||
| K. aerogenes X7266 | + | + | + | 3 (20) | ||||||||||||
| P. kilonensis X7269 | + | + | + | + | + | + | +++ | + | 8 (53) | |||||||
| P. kilonensis X7270 | + | + | +++ | + | 4 (27) | |||||||||||
| P. kilonensis X7271 | + | + | + | + | 4 (27) | |||||||||||
| Pseudomonas spp. X7272 | + | + | + | 3 (20) | ||||||||||||
| Pseudomonas spp. X7273 | + | + | + | 3 (20) | ||||||||||||
| P. chlororaphis X7274 | + | + | + | + | + | + | + | + | 8 (53) | |||||||
| P. caricapapayae X7275 | + | + | + | + | + | 5 (33) | ||||||||||
| Number of inhibitions (%) | 3 (33) | 2 (22) | 6 (67) | 8 (89) | 2 (22) | 2 (22) | 3 (33) | 1 (11) | 8 (89) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (67) | |
| Number of Positive Results of the Antimicrobial-Producing Isolates against the Following Indicator Bacteria: | |||||||
|---|---|---|---|---|---|---|---|
| Producing Isolates | E. coli (1) | P. aeruginosa (1) | MR-Staphylococcus a (2) | MS-Staphylococcus a (5) | Enterococcus (4) | L. monocytogenes (1) | M. luteus (1) |
| A. citreus (1) | 2 | 1 | |||||
| Bacillus spp. (14) | 3 | 1 | 13 | 41 | 9 | 1 | 14 |
| Bradybacterium spp. (1) | 2 | ||||||
| B. laterosoporus (1) | 1 | 2 | 5 | 2 | 1 | 1 | |
| M. arborescensens (1) | 2 | 4 | 1 | ||||
| Paenibacillus spp. (3) | 2 | 3 | 7 | 1 | 3 | ||
| S. hominis (1) | 1 | 5 | 4 | 1 | 1 | ||
| S. avidinii (1) | 1 | 2 | 1 | ||||
| O. soli (1) | 2 | 1 | |||||
| K. aerogenes (1) | 1 | 2 | |||||
| Pseudomonas spp. (7) | 2 | 2 | 8 | 18 | 5 | ||
| Type of Bacteria | Number of Isolates | Genus | Species a | Antimicrobial Resistance Phenotype b |
|---|---|---|---|---|
| Gram-positive | 1 | Arthrobacter | A. citreus | Susceptible |
| 7 | Bacillus spp. | B. pumilus 2, B. safensis, B. megaterium, B. mycoides, Bacillus spp. 2 | PEN 3-FOX 4-MER 3-IMI 2-S 2-TOB 3-CLI -GEN-SXT-CIP 3 | |
| 7 | Bacillus spp. | B. pumilus 3, B. cereus, B. artrophaeus 2, B. safensis | Susceptible 7 | |
| 1 | Bradybacterium | Bradybacterium spp. | Susceptible | |
| 1 | Brevibacillus | B. laterosporus | Susceptible | |
| 1 | Microbacterium | M. arborescensis | Susceptible | |
| 2 | Paenibacillus | P. apiarus 2 | PEN-FOX-TOB | |
| 1 | Paenibacillus | P. polymyxa | Susceptible | |
| 1 | Staphylococcus | S. hominis | Susceptible | |
| 1 | Streptomyces | S. avidinii | Susceptible | |
| Gram-negative | 1 | Klebsiella | K. aerogenes | AMP-AMC-FOX |
| 1 | Olivibacter | O. soli | AMP-FOX-CTX-CAZ-C-TOB | |
| 4 | Pseudomonas | P. chlororaphis, P. caricapapayae, P. kilonensis 2 | TIC 4-ATM 2 | |
| 3 | Pseudomonas | Pseudomonas spp. 2, P. kilonensis | Susceptible |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Fernández, R.; Robredo, B.; Navajas, E.; Torres, C. Citizen Contribution for Searching for Alternative Antimicrobial Activity Substances in Soil. Antibiotics 2023, 12, 57. https://doi.org/10.3390/antibiotics12010057
Fernández-Fernández R, Robredo B, Navajas E, Torres C. Citizen Contribution for Searching for Alternative Antimicrobial Activity Substances in Soil. Antibiotics. 2023; 12(1):57. https://doi.org/10.3390/antibiotics12010057
Chicago/Turabian StyleFernández-Fernández, Rosa, Beatriz Robredo, Enrique Navajas, and Carmen Torres. 2023. "Citizen Contribution for Searching for Alternative Antimicrobial Activity Substances in Soil" Antibiotics 12, no. 1: 57. https://doi.org/10.3390/antibiotics12010057
APA StyleFernández-Fernández, R., Robredo, B., Navajas, E., & Torres, C. (2023). Citizen Contribution for Searching for Alternative Antimicrobial Activity Substances in Soil. Antibiotics, 12(1), 57. https://doi.org/10.3390/antibiotics12010057







