Integrating Network Pharmacology Approaches to Decipher the Multi-Target Pharmacological Mechanism of Microbial Biosurfactants as Novel Green Antimicrobials against Listeriosis
Abstract
1. Introduction
2. Results
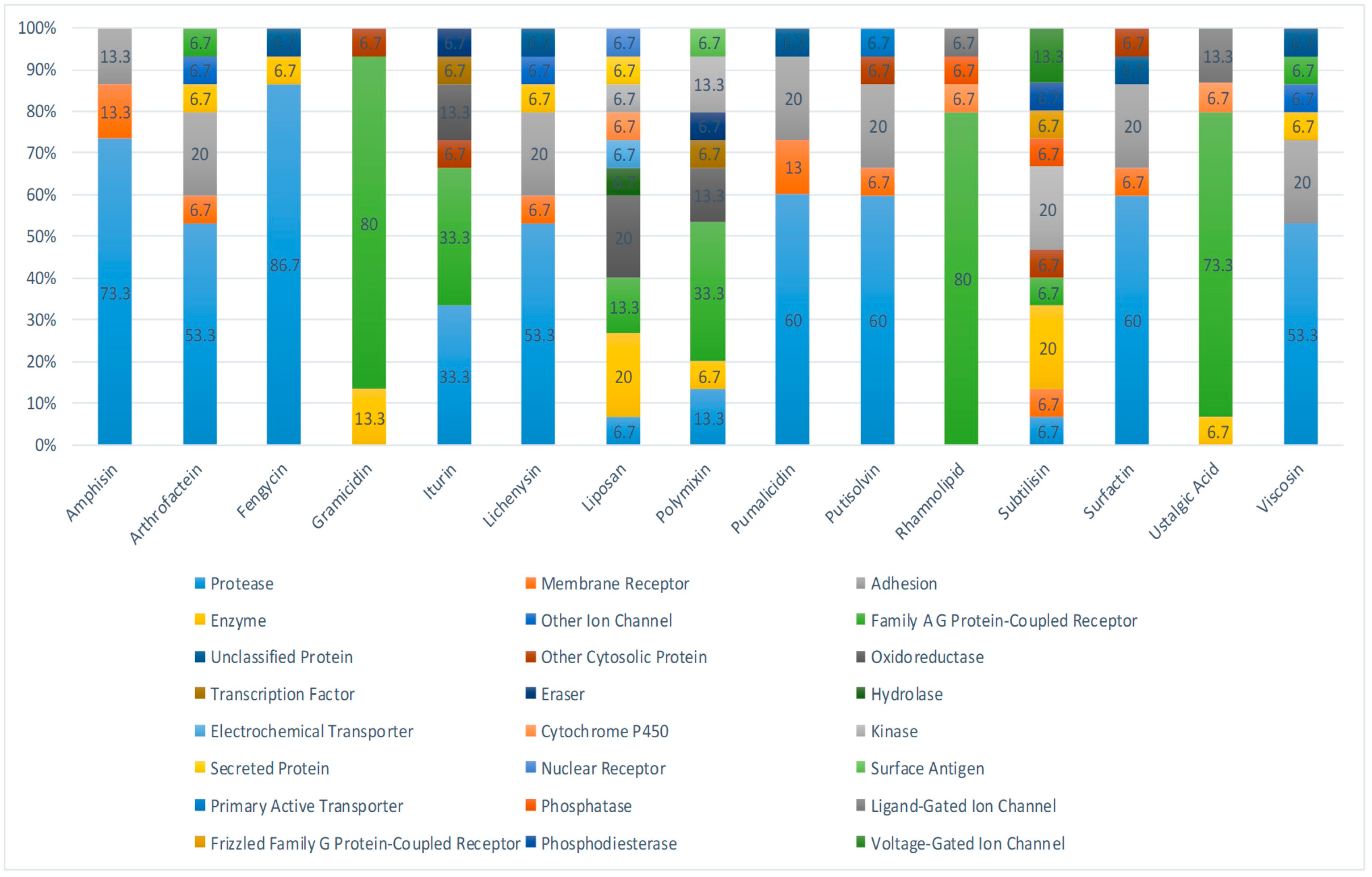
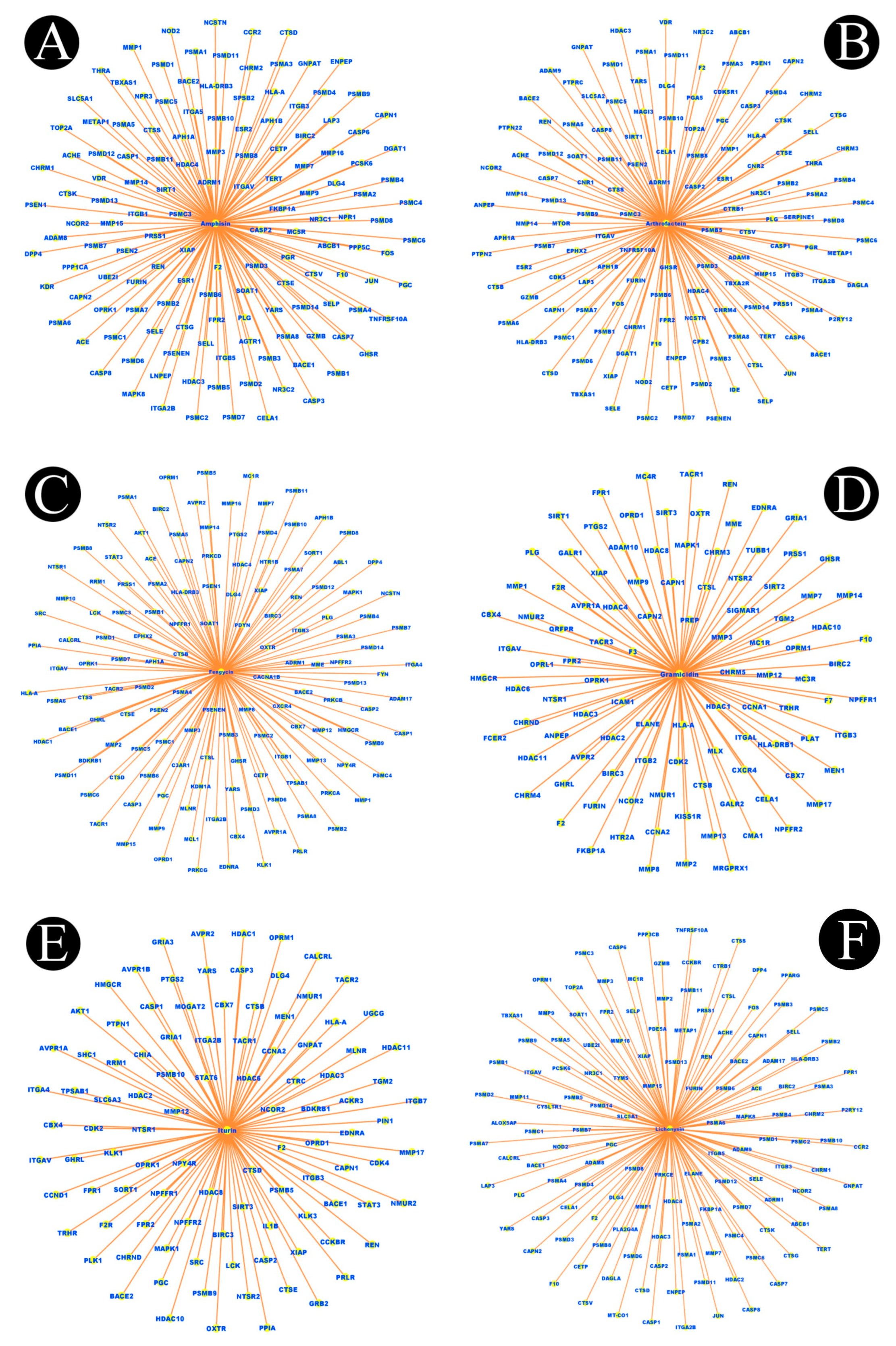
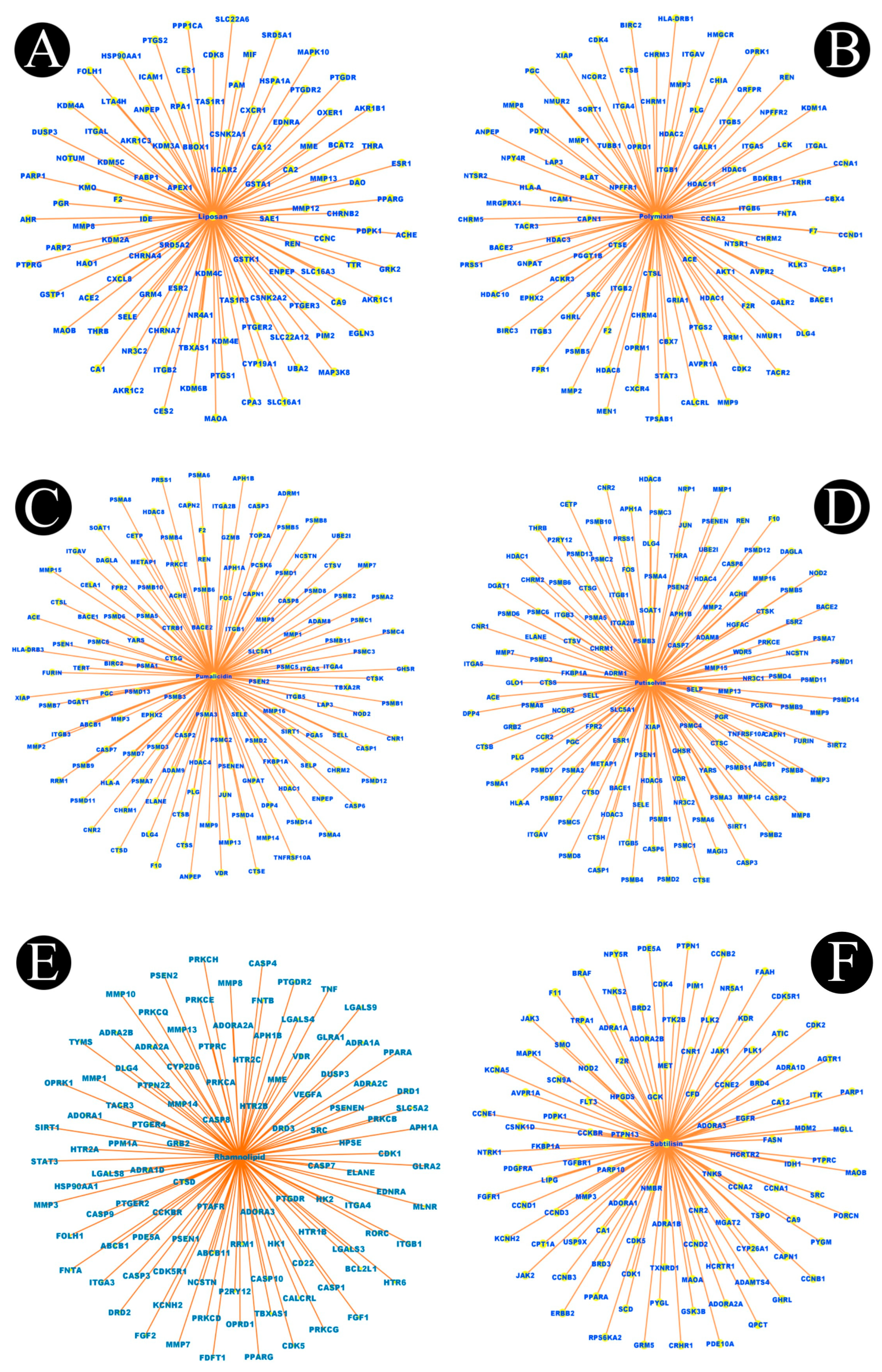
2.1. Identification of Active Components of Biosurfactants
2.2. Listeriosis and Intersection Target
2.3. Construction of Protein–Protein Interaction Network (PPI) and Key Targets
2.4. Functional GO and KEGG Pathways
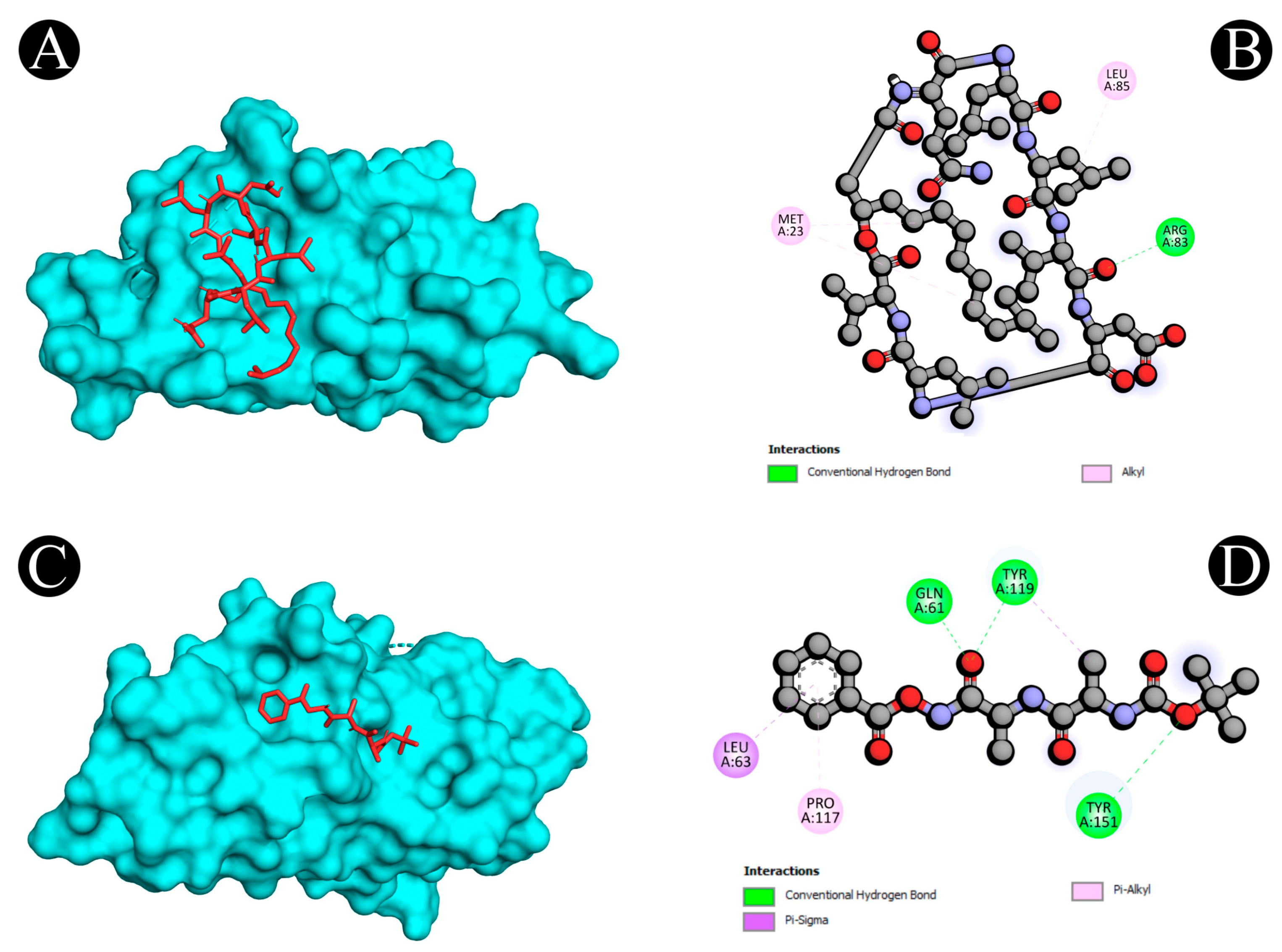
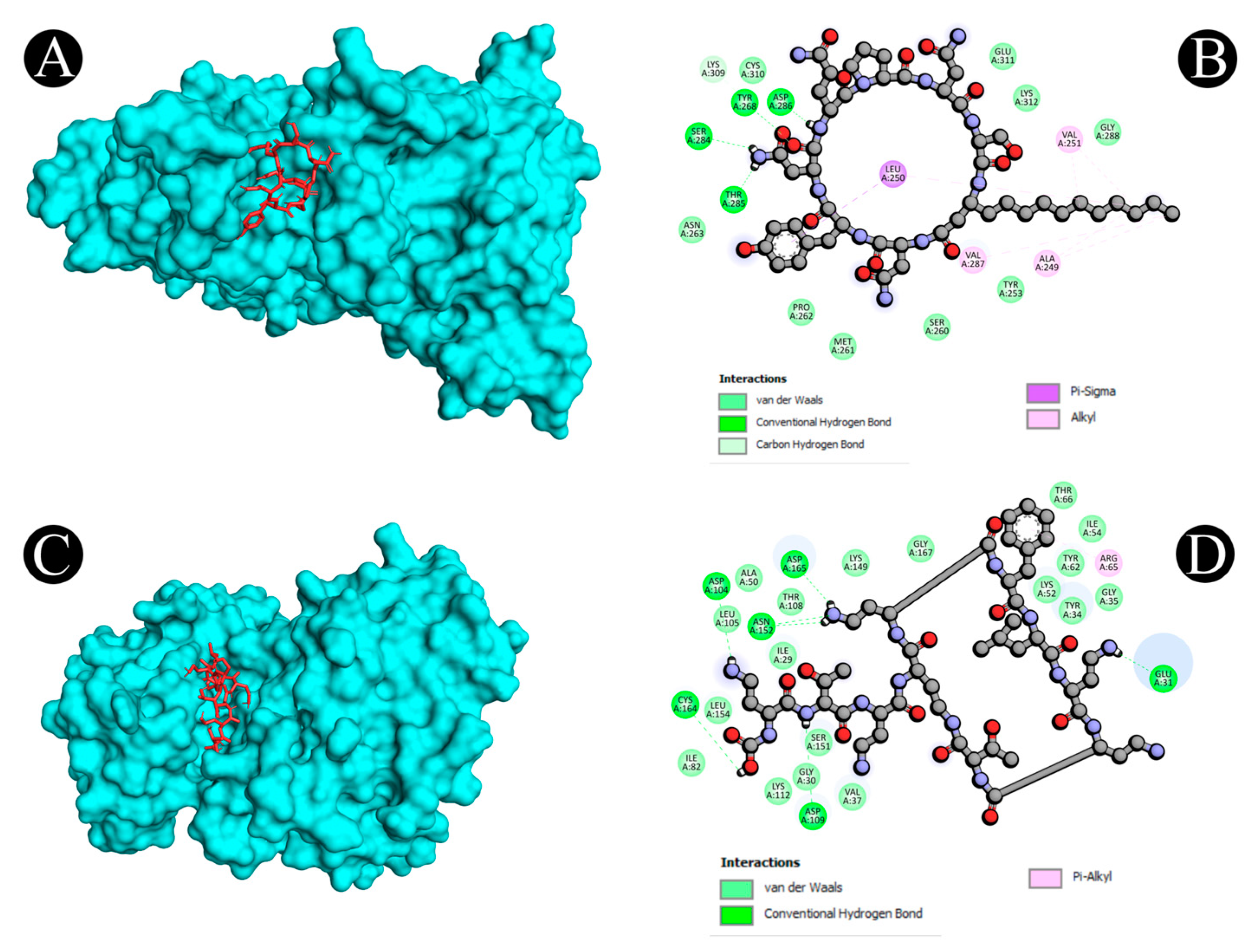
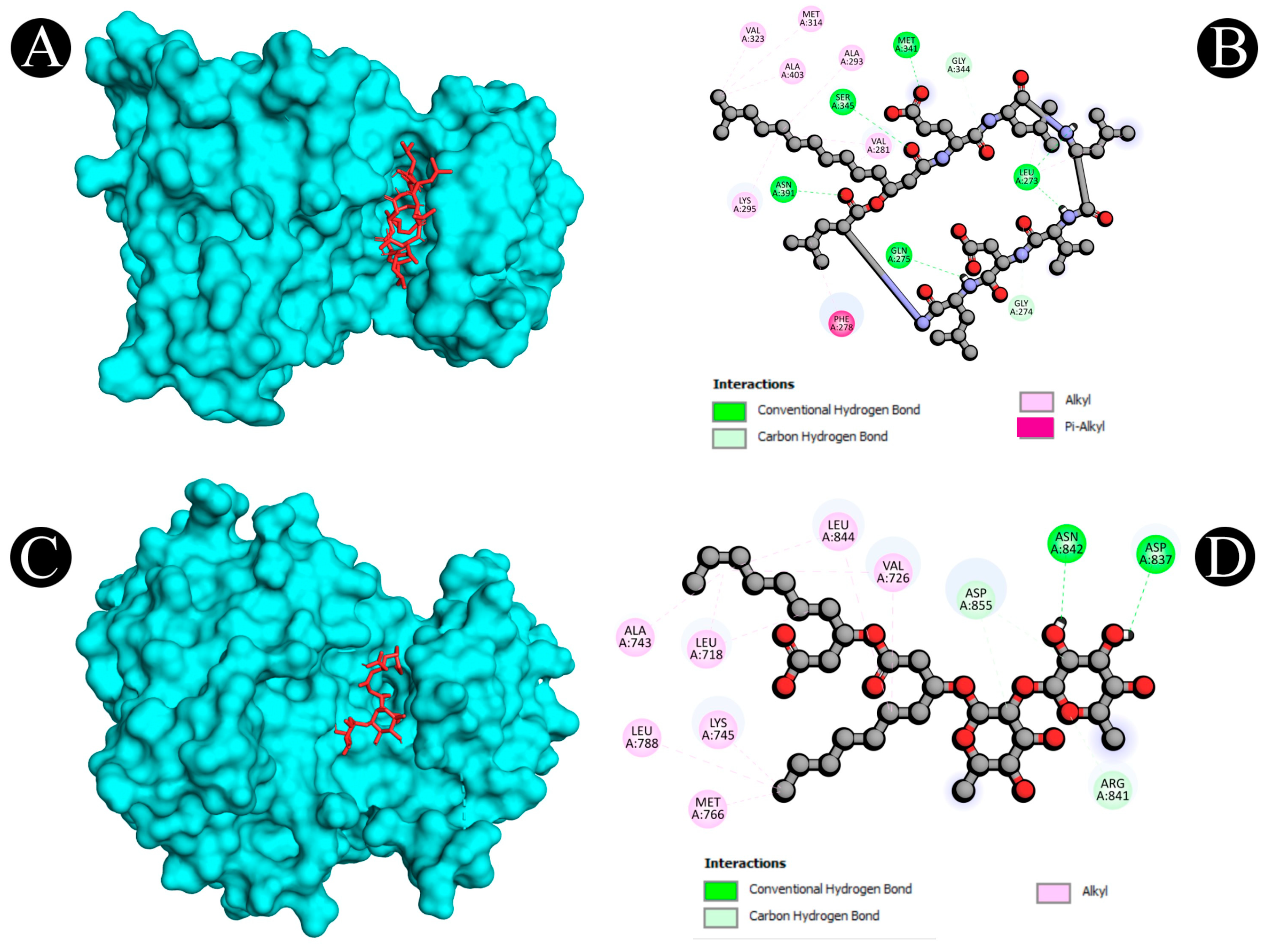
2.5. Molecular Docking
3. Discussion
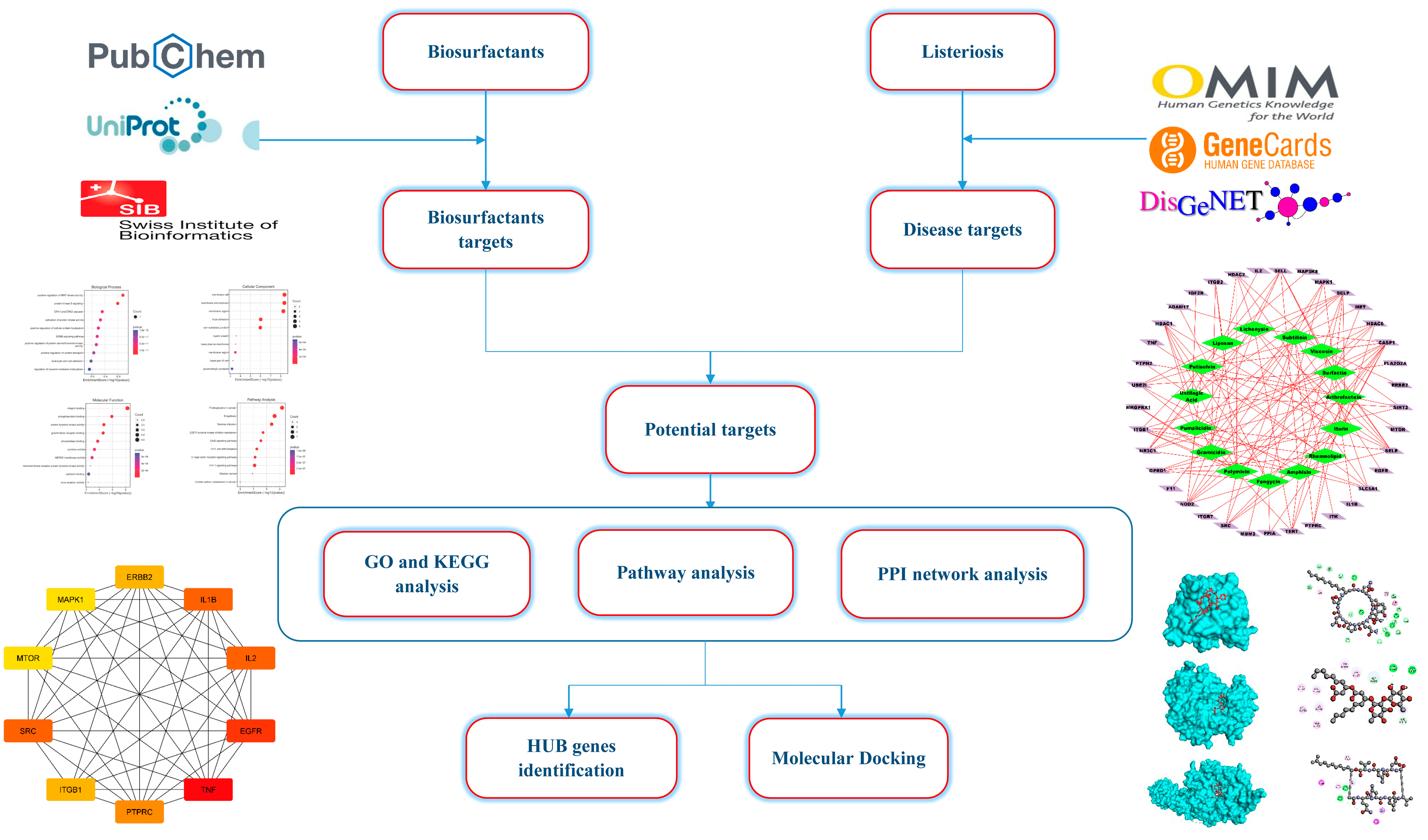
4. Materials and Methods
4.1. Biosurfactants Target Prediction
4.2. Network Construction for Compound–Targets
4.3. Protein Targets Associated with Listeriosis
4.4. Target Screening and Network Construction for Biosurfactants and Listeriosis
4.5. Protein–Protein Interaction Network (PPI) Construction and Target Identification
4.6. Analysis of Gene Ontology (GO) Function and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment
4.7. Construction of Target-Path/Functional Networks
4.8. Findings of Hub Genes
4.9. Molecular Docking Analysis
4.9.1. Protein and Ligand Structures
4.9.2. Ligand Preparation
4.9.3. Prediction of Binding Site
4.9.4. Molecular Docking
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ge, H.; Wang, Y.; Zhao, X. Research on the Drug Resistance Mechanism of Foodborne Pathogens. Microb. Pathog. 2022, 162, 105306. [Google Scholar] [CrossRef] [PubMed]
- Nyachuba, D.G. Foodborne Illness: Is It on the Rise? Nutr. Rev. 2010, 68, 257–269. [Google Scholar] [CrossRef] [PubMed]
- White, D.G.; Zhao, S.; Simjee, S.; Wagner, D.D.; McDermott, P.F. Antimicrobial Resistance of Foodborne Pathogens. Microbes Infect. 2002, 4, 405–412. [Google Scholar] [CrossRef]
- Jones, G.S.; Bussell, K.M.; Myers-Morales, T.; Fieldhouse, A.M.; Bou Ghanem, E.N.; D’Orazio, S.E. Intracellular Listeria monocytogenes comprises a minimal but vital fraction of the intestinal burden following foodborne infection. Infect. Immun. 2015, 83, 3146–3156. [Google Scholar] [PubMed]
- Rouquette, C.; Berche, P. The pathogenesis of infection by Listeria monocytogenes. Microbiologia 1996, 12, 245–258. [Google Scholar]
- Kazmierczak, M.J.; Wiedmann, M.; Boor, K.J. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 2005, 69, 527–543. [Google Scholar] [CrossRef]
- Mansfield, B.E.; Freitag, N.E. Listeria monocytogenes pathogenesis: Exploration of alternative hosts. In Abstracts of the General Meeting of the American Society for Microbiology; American Society for Microbiology: Washington, DC, USA, 2003; Volume 103, p. B-186. [Google Scholar]
- Gandhi, M.; Chikindas, M.L. Listeria: A Foodborne Pathogen That Knows How to Survive. Int. J. Food Microbiol. 2007, 113, 1–15. [Google Scholar]
- Voronina, O.L.; Tartakovsky, I.S.; Yuyshuk, N.D.; Ryzhova, N.N.; Aksenova, E.I.; Kunda, M.S.; Kutuzova, A.V.; Melkumyan, A.R.; Karpova, T.I.; Gruzdeva, O.A.; et al. Analysis of Sporadic Cases of Invasive Listeriosis in a Metropolis. J. Microbiol. Epidemiol. Immun. 2020, 97, 546–555. [Google Scholar] [CrossRef]
- Noor, F.; Noor, A.; Ishaq, A.R.; Farzeen, I.; Saleem, M.H.; Ghaffar, K.; Aslam, M.F.; Aslam, S.; Chen, J.-T. Recent Advances in Diagnostic and Therapeutic Approaches for Breast Cancer: A Comprehensive Review. Curr. Pharm. Des. 2021, 27, 2344–2365. [Google Scholar] [CrossRef]
- Noor, F.; Saleem, M.H.; Chen, J.-T.; Javed, M.R.; Al-Megrin, W.A.; Aslam, S. Integrative Bioinformatics Approaches to Map Key Biological Markers and Therapeutic Drugs in Extramammary Paget’s Disease of the Scrotum. PLoS ONE 2021, 16, e0259408. [Google Scholar]
- Kashif, A.; Rehman, R.; Fuwad, A.; Shahid, M.K.; Dayarathne, H.N.P.; Jamal, A.; Aftab, M.N.; Mainali, B.; Choi, Y. Current advances in the classification, production, properties and applications of microbial biosurfactants—A critical review. Adv. Colloid Inter. Sci. 2022, 306, 102718. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Siddiqui, A.J.; Hamadou, W.S.; Surti, M.; Awadelkareem, A.M.; Ashraf, S.A.; Alreshidi, M.; Snoussi, M.; Rizvi, S.M.D.; Bardakci, F. Inhibition of Bacterial Adhesion and Antibiofilm Activities of a Glycolipid Biosurfactant from Lactobacillus Rhamnosus with Its Physicochemical and Functional Properties. Antibiotics 2021, 10, 1546. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Siddiqui, A.J.; Hamadou, W.S.; Ashraf, S.A.; Hassan, M.I.; Snoussi, M.; Badraoui, R.; Jamal, A.; Bardakci, F.; Awadelkareem, A.M. Functional and Structural Characterization of Pediococcus pentosaceus-Derived Biosurfactant and Its Biomedical Potential against Bacterial Adhesion, Quorum Sensing, and Biofilm Formation. Antibiotics 2021, 10, 1371. [Google Scholar] [CrossRef]
- Magalhães, L.; Nitschke, M. Antimicrobial Activity of Rhamnolipids against Listeria monocytogenes and Their Synergistic Interaction with Nisin. Food Control 2013, 29, 138–142. [Google Scholar] [CrossRef]
- Boezio, B.; Audouze, K.; Ducrot, P.; Taboureau, O. Network-based Approaches in Pharmacology. Mol. Inform. 2017, 36, 1700048. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Chen, B.; Chen, S.; Lin, M.; Chen, Y.; Jin, S.; Chen, W.; Zhang, Y. Applications of Network Pharmacology in Traditional Chinese Medicine Research. Evid. Based. Complement. Altern. Med. 2020, 2020, 1646905. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Park, S.-Y.; Lee, H.-J.; Kim, C.E. A Systems-Level Analysis of Mechanisms of Platycodon grandiflorum Based on a Network Pharmacological Approach. Molecules 2018, 23, 2841. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weaponsfor plant disease biocontrol. Trends. Microbiol. 2007, 16, 115–125. [Google Scholar] [CrossRef]
- Pathak, K.V.; Keharia, H. Application of extracellular lipopeptide bio-surfactant produced by endophytic Bacillus subtilis K1 isolated from aerial rootsof banyan (Ficus benghalensis) in microbially enhanced oil recovery (MEOR). 3 Biotech 2014, 4, 41–48. [Google Scholar] [CrossRef]
- Rahman, K.S.M.; Thahira-Rahman, J.; McClean, S.; Marchant, R.; Banat, I.M. Rhamnolipid biosurfactants production by strains of Pseudomonas aeruginosa using low cost materials. Biotechnol. Prog. 2002, 18, 1277–1281. [Google Scholar] [CrossRef]
- Alsohim, A.S.; Taylor, T.B.; Barrett, G.A.; Gallie, J.; Zhang, X.X.; Altamirano-Junqueira, A.E.; Johnson, L.J.; Rainey, P.B.; Jackson, R.W. The biosurfactant viscosin produced by P seudomonas fluorescens SBW 25 aids spreading motility and plant growth promotion. Environ. Microbiol. 2014, 16, 2267–2281. [Google Scholar] [CrossRef] [PubMed]
- Rufino, R.D.; Luna, J.M.; Sarubbo, L.A.; Rodrigues, L.R.M.; Teixeira, J.A.C.; Campos-Takaki, G.M. Antimicrobial and anti-adhesive potential of a biosurfactant Rufisan produced by Candida lipolytica UCP 0988. Colloids Surf. B Biointerfaces 2011, 84, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Yeak, K.Y.C.; Perko, M.; Staring, G.; Fernandez-Ciruelos, B.M.; Wells, J.M.; Abee, T.; Wells-Bennik, M.H. Lichenysin Production by Bacillus licheniformis Food Isolates and Toxicity to Human Cells. Front. Microbiol. 2022, 13, 831033. [Google Scholar] [CrossRef] [PubMed]
- Arrebola, E.; Jacobs, R.; Korsten, L. Iturin A is the principal inhibitor inthe biocontrol activity of Bacillus amyloliquefaciens PPCB004 against posthar-vest fungal pathogens. J. Appl. Microbiol. 2010, 108, 386–395. [Google Scholar] [CrossRef]
- Morikawa, M.; Daido, H.; Takao, T.; Murata, S.; Shimonishi, Y.; Imanaka, T. A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. J. Bacteriol. 1993, 175, 6459–6466. [Google Scholar] [CrossRef]
- Groboillot, A.; Portet-Koltalo, F.; Le Derf, F.; Feuilloley, M.J.; Orange, N.; Poc, C.D. Novel application of cyclolipopeptide amphisin: Feasibility study as additive to remediate polycyclic aromatic hydrocarbon (PAH) contaminated sediments. Int. J. Mol. Sci. 2011, 12, 1787–1806. [Google Scholar] [CrossRef]
- Kuiper, I.; Lagendijk, E.L.; Pickford, R.; Derrick, J.P.; Lamers, G.E.; Thomas-Oates, J.E.; Lugtenberg, B.J.; Bloemberg, G.V. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 2004, 51, 97–113. [Google Scholar] [CrossRef]
- Hewald, S.; Josephs, K.; Bölker, M. Genetic analysis of biosurfactant production in Ustilago maydis. Appl. Environ. Microbiol. 2005, 71, 3033–3040. [Google Scholar] [CrossRef]
- Saggese, A.; Culurciello, R.; Casillo, A.; Corsaro, M.M.; Ricca, E.; Baccigalupi, L. A marine isolate of Bacillus pumilus secretes a pumilacidin active against Staphylococcus aureus. Mar. Drugs 2018, 16, 180. [Google Scholar] [CrossRef]
- Gudiña, E.J.; Fernandes, E.C.; Rodrigues, A.I.; Teixeira, J.A.; Rodrigues, L.R. Biosurfactant production by Bacillus subtilis using corn steep liquor as culture medium. Front. Microbiol. 2015, 6, 59. [Google Scholar]
- Abdelhamid, H.N.; Khan, M.S.; Wu, H.F. Graphene oxide as a nanocarrier for gramicidin (GOGD) for high antibacterial performance. RSC Adv. 2014, 4, 50035–50046. [Google Scholar] [CrossRef]
- Eras-Muñoz, E.; Farré, A.; Sánchez, A.; Font, X.; Gea, T. Microbial biosurfactants: A review of recent environmental applications. Bioengineered 2022, 13, 12365–12391. [Google Scholar] [CrossRef] [PubMed]
- Bodro, M.; Paterson, D.L. Listeriosis in Patients Receiving Biologic Therapies. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1225–1230. [Google Scholar] [CrossRef] [PubMed]
- Allerberger, F.; Wagner, M. Listeriosis: A Resurgent Foodborne Infection. Clin. Microbiol. Infect. 2010, 16, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Melo, J.; Andrew, P.W.; Faleiro, M.L. Listeria monocytogenes in Cheese and the Dairy Environment Remains a Food Safety Challenge: The Role of Stress Responses. Food. Res. Int. 2015, 67, 75–90. [Google Scholar] [CrossRef]
- Benincasa, M.; Abalos, A.; Oliveira, I.; Manresa, A. Chemical Structure, Surface Properties and Biological Activities of the Biosurfactant Produced by Pseudomonas aeruginosa LBI from Soapstock. Antonie Van Leeuwenhoek 2004, 85, 1–8. [Google Scholar] [CrossRef]
- Haba, E.; Pinazo, A.; Jauregui, O.; Espuny, M.J.; Infante, M.R.; Manresa, A. Physicochemical Characterization and Antimicrobial Properties of Rhamnolipids Produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 2003, 81, 316–322. [Google Scholar] [CrossRef]
- Ortiz, S.; López-Alonso, V.; Rodríguez, P.; Martínez-Suárez, J.V. The Connection between Persistent, Disinfectant-Resistant Listeria Monocytogenes Strains from Two Geographically Separate Iberian Pork Processing Plants: Evidence from Comparative Genome Analysis. Appl. Environ. Microbiol. 2016, 82, 308–317. [Google Scholar] [CrossRef]
- De Araujo, L.V.; Guimarães, C.R.; da Silva Marquita, R.L.; Santiago, V.M.J.; de Souza, M.P.; Nitschke, M.; Freire, D.M.G. Rhamnolipid and Surfactin: Anti-Adhesion/Antibiofilm and Antimicrobial Effects. Food Control 2016, 63, 171–178. [Google Scholar] [CrossRef]
- Chen, W.; Ding, C.; Yu, J.; Wang, C.; Wan, L.; Hu, H.; Ma, J. Wuzi Yanzong Pill—Based on Network Pharmacology and In Vivo Evidence—Protects Against Spermatogenesis Disorder via the Regulation of the Apoptosis Pathway. Front. Pharmacol. 2020, 11, 592827. [Google Scholar] [CrossRef]
- Yap, P.-C.; MatRahim, N.-A.; AbuBakar, S.; Lee, H.Y. Antilisterial Potential of Lactic Acid Bacteria in Eliminating Listeria monocytogenes in Host and Ready-to-Eat Food Application. Microbiol. Res. 2021, 12, 17. [Google Scholar] [CrossRef]
- Almeida, M.T.; Mesquita, F.S.; Cruz, R.; Osório, H.; Custódio, R.; Brito, C.; Vingadassalom, D.; Martins, M.; Leong, J.M.; Holden, D.W. Src-Dependent Tyrosine Phosphorylation of Non-Muscle Myosin Heavy Chain-IIA Restricts Listeria Monocytogenes Cellular Infection. J. Biol. Chem. 2015, 290, 8383–8395. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, M.; Ceballos-Olvera, I.; del Barrio, L.; Re, F. Role of the Inflammasome, IL-1. Sci. World J. 2011, 11, 2037–2050. [Google Scholar] [CrossRef] [PubMed]
- Cossart, P.; Pizarro-Cerdá, J.; Lecuit, M. Invasion of Mammalian Cells by Listeria monocytogenes: Functional Mimicry to Subvert Cellular Functions. Trends. Cell. Biol. 2003, 13, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.J.; Lauwaet, T.; de Bruyne, G.; Mareel, M.; Leroy, A. Listeria monocytogenes Produces a Pro-Invasive Factor That Signals via ErbB2/ErbB3 Heterodimers. J. Cancer Res. Clin. Oncol. 2005, 131, 49–59. [Google Scholar] [CrossRef]
- Isberg, R.R.; Barnes, P. Subversion of Integrins by Enteropathogenic Yersinia. J. Cell. Sci. 2001, 114, 21–28. [Google Scholar] [CrossRef]
- Fowler, T.; Wann, E.R.; Joh, D.; Johansson, S.; Foster, T.J.; Höök, M. Cellular Invasion by Staphylococcus Aureus Involves a Fibronectin Bridge between the Bacterial Fibronectin-Binding MSCRAMMs and Host Cell Β1 Integrins. Eur. J. Cell. Biol. 2000, 79, 672–679. [Google Scholar] [CrossRef]
- Hauck, C.R.; Ohlsen, K. Sticky Connections: Extracellular Matrix Protein Recognition and Integrin-Mediated Cellular Invasion by Staphylococcus Aureus. Curr. Opin. Microbiol. 2006, 9, 5–11. [Google Scholar] [CrossRef]
- Van Putten, J.P.M.; Duensing, T.D.; Cole, R.L. Entry of OpaA+ Gonococci into HEp-2 Cells Requires Concerted Action of Glycosaminoglycans, Fibronectin and Integrin Receptors. Mol. Microbiol. 1998, 29, 369–379. [Google Scholar] [CrossRef]
- Izquierdo, M.; Alvestegui, A.; Nataro, J.P.; Ruiz-Perez, F.; Farfan, M.J. Participation of Integrin A5β1 in the Fibronectin-Mediated Adherence of Enteroaggregative Escherichia coli to Intestinal Cells. Biomed. Res. Int. 2014, 2014, 781246. [Google Scholar] [CrossRef]
- Hashino, M.; Tachibana, M.; Nishida, T.; Hara, H.; Tsuchiya, K.; Mitsuyama, M.; Watanabe, K.; Shimizu, T.; Watarai, M. Inactivation of the MAPK Signaling Pathway by Listeria Monocytogenes Infection Promotes Trophoblast Giant Cell Death. Front. Microbiol. 2015, 6, 1145. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, J.D.; Riminton, D.S.; Cyster, J.G.; Körner, H. Tumor necrosis factor: A master-regulator of leukocyte movement. Immunol. Today 2000, 21, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Tripp, C.S.; Wolf, S.F.; Unanue, E.R. Interleukin 12 and tumor necrosis factor alpha are costimulators of interferon gamma production by natural killer cells in severe combined immunodeficiency mice with listeriosis, and interleukin 10 is a physiologic antagonist. Proc. Natl. Acad. Sci. USA 1993, 90, 3725–3729. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, G.J.; Sheehan, K.C.; Schreiber, R.D.; Unanue, E.R. Tumor necrosis factor is involved in the T cell-independent pathway of macrophage activation in scid mice. J. Immunol. 1989, 143, 127–130. [Google Scholar] [PubMed]
- Li, X.; Körner, H.; Liu, X. Susceptibility to Intracellular Infections: Contributions of TNF to Immune Defense. Front. Microbiol. 2020, 11, 1643. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB Signalling Network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Garrett, T.P.J.; McKern, N.M.; Lou, M.; Elleman, T.C.; Adams, T.E.; Lovrecz, G.O.; Zhu, H.-J.; Walker, F.; Frenkel, M.J.; Hoyne, P.A. Crystal Structure of a Truncated Epidermal Growth Factor Receptor Extracellular Domain Bound to Transforming Growth Factor α. Cell 2002, 110, 763–773. [Google Scholar] [CrossRef]
- Ogiso, H.; Ishitani, R.; Nureki, O.; Fukai, S.; Yamanaka, M.; Kim, J.-H.; Saito, K.; Sakamoto, A.; Inoue, M.; Shirouzu, M. Crystal Structure of the Complex of Human Epidermal Growth Factor and Receptor Extracellular Domains. Cell 2002, 110, 775–787. [Google Scholar] [CrossRef]
- Cho, H.-S.; Mason, K.; Ramyar, K.X.; Stanley, A.M.; Gabelli, S.B.; Denney, D.W.; Leahy, D.J. Structure of the Extracellular Region of HER2 Alone and in Complex with the Herceptin Fab. Nature 2003, 421, 756–760. [Google Scholar] [CrossRef]
- Olayioye, M.A.; Neve, R.M.; Lane, H.A.; Hynes, N.E. The ErbB Signaling Network: Receptor Heterodimerization in Development and Cancer. EMBO J. 2000, 19, 3159–3167. [Google Scholar] [CrossRef]
- Hoffmann, I.; Eugène, E.; Nassif, X.; Couraud, P.-O.; Bourdoulous, S. Activation of ErbB2 Receptor Tyrosine Kinase Supports Invasion of Endothelial Cells by Neisseria meningitidis. J. Cell Biol. 2001, 155, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huong, S.-M.; Chiu, M.L.; Raab-Traub, N.; Huang, E.S. Epidermal Growth Factor Receptor Is a Cellular Receptor for Human Cytomegalovirus. Nature 2003, 424, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Conner, S.D.; Schmid, S.L. Regulated Portals of Entry into the Cell. Nature 2003, 422, 37–44. [Google Scholar] [CrossRef]
- Nabi, I.R.; Le, P.U. Caveolae/Raft-Dependent Endocytosis. J. Cell Biol. 2003, 161, 673–677. [Google Scholar] [CrossRef] [PubMed]
- McMahon, H.T.; Boucrot, E. Molecular Mechanism and Physiological Functions of Clathrin-Mediated Endocytosis. Nat. Rev. Mol. Cell Biol. 2011, 12, 517–533. [Google Scholar] [CrossRef]
- Jubelin, G.; Taieb, F.; Duda, D.M.; Hsu, Y.; Samba-Louaka, A.; Nobe, R.; Penary, M.; Watrin, C.; Nougayrède, J.-P.; Schulman, B.A. Pathogenic Bacteria Target NEDD8-Conjugated Cullins to Hijack Host-Cell Signaling Pathways. PLoS Pathog. 2010, 6, e1001128. [Google Scholar] [CrossRef] [PubMed]
- Hoving, J.C.; Wilson, G.J.; Brown, G.D. Signalling C-type Lectin Receptors, Microbial Recognition and Immunity. Cell. Microbiol. 2014, 16, 185–194. [Google Scholar] [CrossRef]
- Schmidt-Weber, C.B.; Akdis, M.; Akdis, C.A. TH17 Cells in the Big Picture of Immunology. J. Allergy Clin. Immunol. 2007, 120, 247–254. [Google Scholar] [CrossRef]
- Galanis, A.; Pappa, A.; Giannakakis, A.; Lanitis, E.; Dangaj, D.; Sandaltzopoulos, R. Reactive Oxygen Species and HIF-1 Signalling in Cancer. Cancer Lett. 2008, 266, 12–20. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTarget Prediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the Interaction Landscape of Bioactive Molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef]
- Zhang, J.; Li, H.; Zhang, Y.; Zhao, C.; Zhu, Y.; Han, M. Uncovering the Pharmacological Mechanism of Stemazole in the Treatment of Neurodegenerative Diseases Based on a Network Pharmacology Approach. Int. J. Mol. Res. 2020, 21, 427. [Google Scholar]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The Human Gene Integrator. Database 2010, 2010, baq020. [Google Scholar] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A Comprehensive Platform Integrating Information on Human Disease-Associated Genes and Variants. Nucleic Acids Res. 2016, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET Knowledge Platform for Disease Genomics: 2019 Update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar]
- UniProt Consortium. Uniport: A Hub for Protein Information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA Update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Olson, A.J. Using Autodock for Ligand-receptor Docking. Curr. Protoc. Bioinformatics 2008, 24, 8.14.1–8.14.40. [Google Scholar] [CrossRef]
- Accelrys. Discovery Studio, 2.1; Accelrys: San Diego, CA, USA, 2008. [Google Scholar]
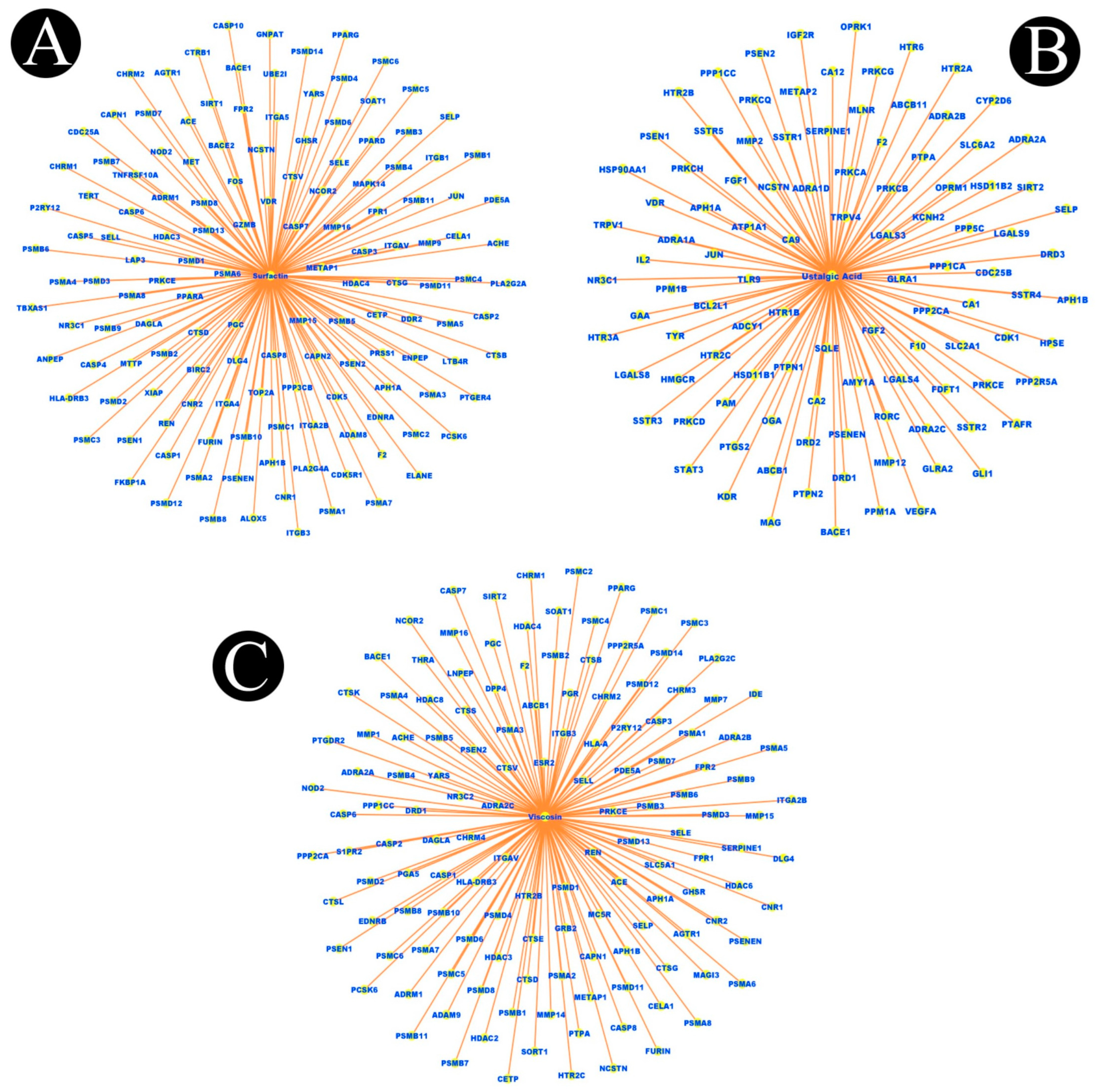

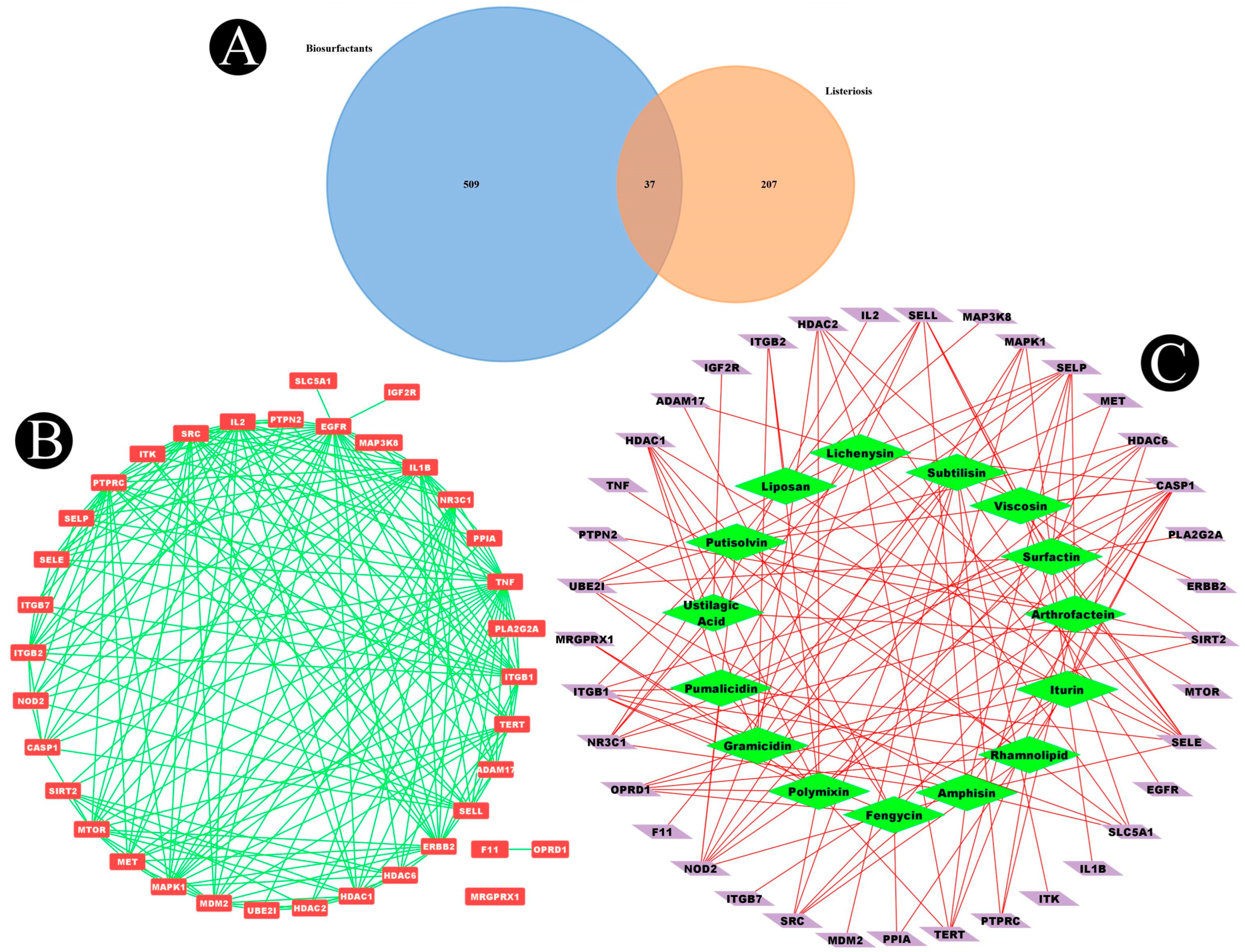
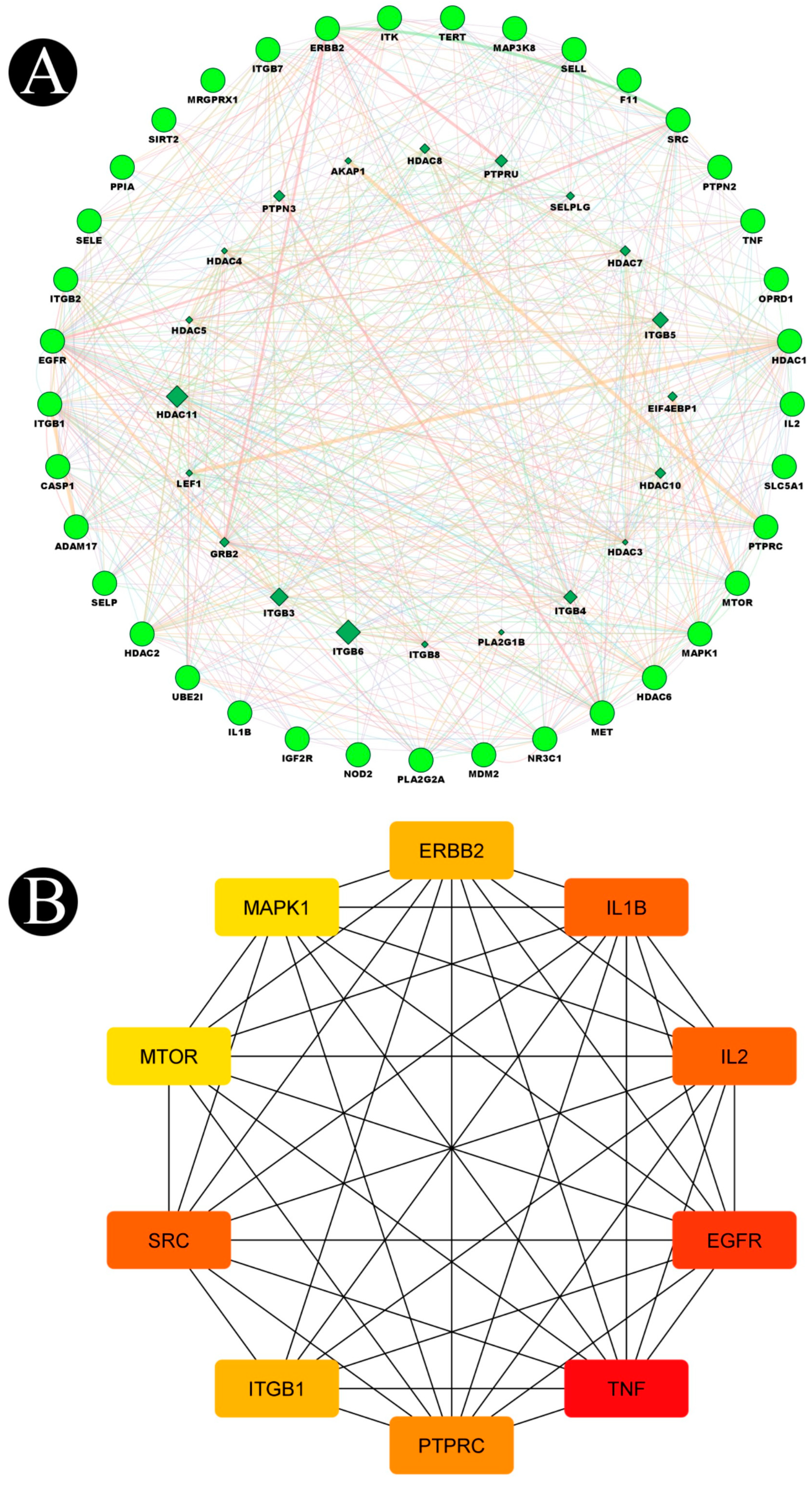
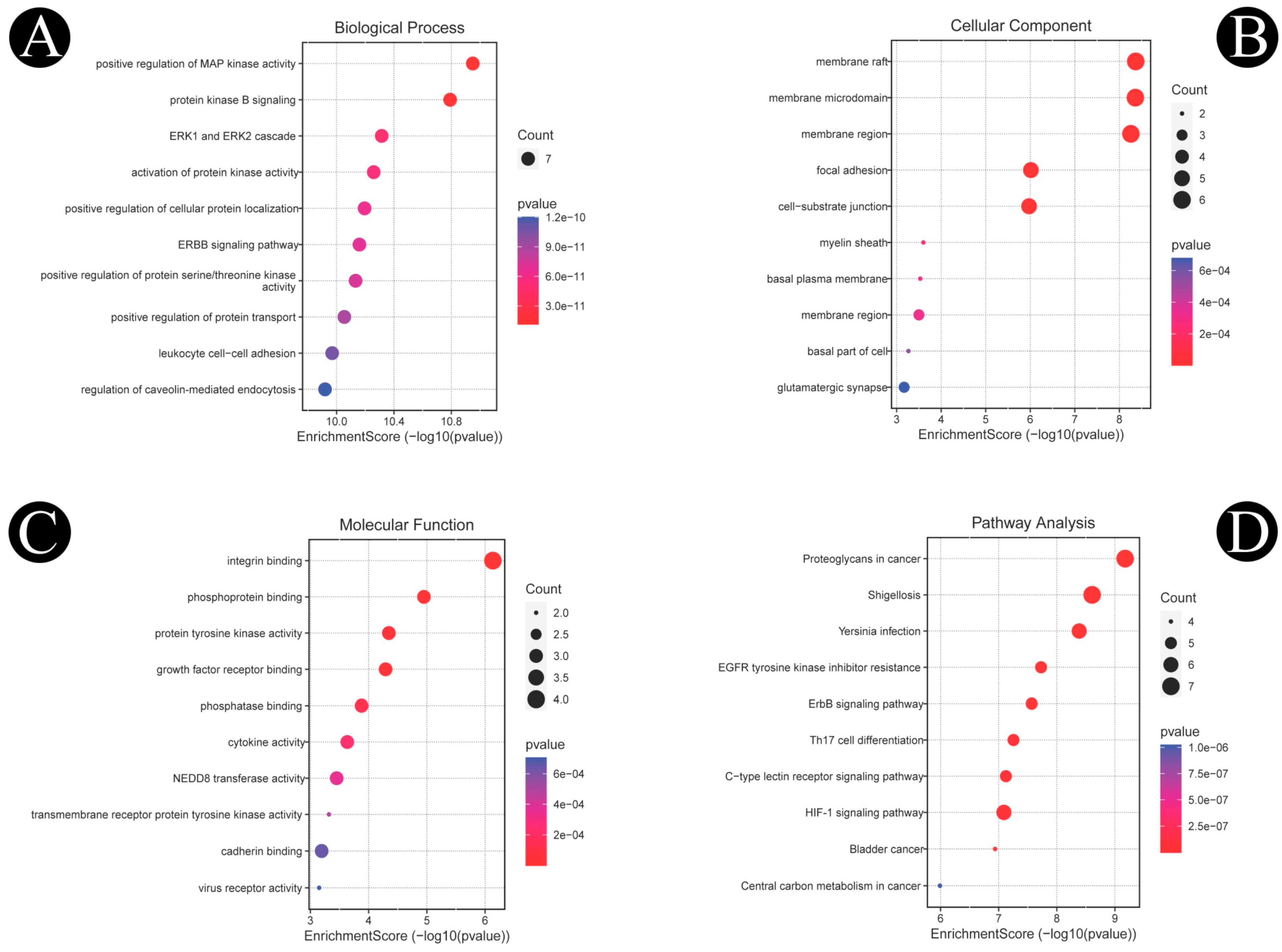
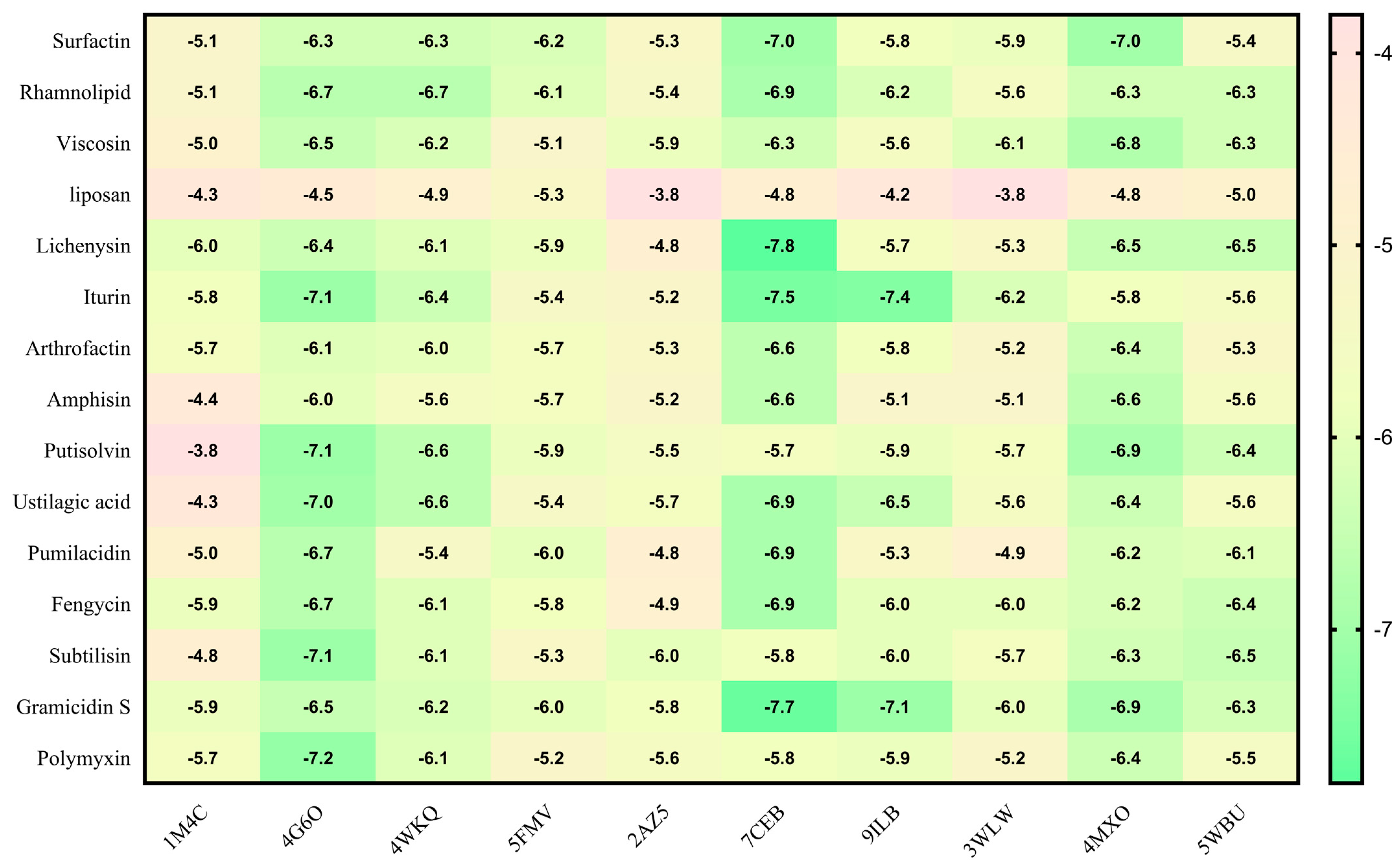


| Sr. No. | Biosurfactant | Microbial Origin | References | Molecular Formula | PubChem | Canonical SMILE |
|---|---|---|---|---|---|---|
| 1 | Surfactin | Bacillus subtilis Bacillus siamensis | [19,20] | C53H93N7O13 | 5066078 | CC(C)CCCCCCCCCC1CC(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)O1)CC(C)C)CC(C)C)CC(=O)O)C(C)C)CC(C)C)CC(C)C)CCC(=O)O |
| 2 | Rhamnolipid | Pseudomonas aeruginosa | [21] | C32H58O13 | 5458394 | CCCCCCCC(CC(=O)O)OC(=O)CC(CCCCCCC)OC1C(C(C(C(O1)C)O)O)OC2C(C(C(C(O2)C)O)O)O |
| 3 | Viscosin | Pseudomonas fluorescens | [22] | C54H95N9O16 | 72937 | CCCCCCCC(CC(=O)NC(CC(C)C)C(=O)NC(CCC(=O)O)C(=O)NC1C(OC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C(NC1=O)C(C)C)CC(C)C)CO)CC(C)C)CO)C(C)CC)C)O |
| 4 | Liposan | Candida lipolytica | [23] | C8H14O2S2 | 864 | C1CSSC1CCCCC(=O)O |
| 5 | Lichenysin | Bacillus licheniformis | [24] | C51H90N8O12 | 11804102 | CC(C)CCCCCCCCC1CC(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)O1)C(C)C)CC(C)C)CC(=O)O)C(C)C)CC(C)C)CC(C)C)CCC(=O)N |
| 6 | Iturin | Bacillus subtilis Bacillus amyloliquefaciens | [25] | C48H74N12O14 | 158570 | CCCCCCCCCCCC1CC(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)N2CCCC2C(=O)NC(C(=O)NC(C(=O)N1)CO)CC(=O)N)CCC(=O)N)CC(=O)N)CC3=CC=C(C=C3)O)CC(=O)N |
| 7 | Arthrofactin | Arthrobacter sp. strain MIS38 | [26] | C64H111N11O20 | 23724538 | CCCCCCCC1CC(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)O1)CC(=O)O)C(C)CC)C(C)CC)CO)CC(C)C)CO)CC(C)C)CC(C)C)C(C)O)CC(=O)O)CC(C)C |
| 8 | Amphisin | Pseudomonasfluorescens | [27] | C66H114N12O20 | 101134740 | CCCCCCCC(CC(=O)NC(CC(C)C)C(=O)NC(CC(=O)O)C(=O)NC1C(OC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C(NC1=O)CC(C)C)CC(C)C)CO)CC(C)C)CCC(=O)N)CC(C)C)C(C)CC)CC(=O)O)C)O |
| 9 | Putisolvin | Pseudomonas putida | [28] | C65H113N13O19 | 139588800 | CCCCCC(=O)NC(CC(C)C)C(=O)NC(CCC(=O)O)C(=O)NC(CC(C)C)C(=O)NC(C(C)CC)C(=O)NC(CCC(=O)N)C(=O)NC(CO)C(=O)NC(C(C)C)C(=O)NC(C(C)CC)C(=O)NC1COC(=O)C(NC(=O)C(NC(=O)C(NC1=O)CC(C)C)C(C)C)CO |
| 10 | Ustilagic Acid | Ustilago maydis | [29] | C36H64O18 | 52922086 | CCCC(CC(=O)OC1C(C(C(OC1OC2C(OC(C(C2O)O)OCC(CCCCCCCCCCCCC(C(=O)O)O)O)COC(=O)C)CO)O)O)O |
| 11 | Pumilacidin | Bacillus pumilus | [30] | C55H99N7O12 | 101174694 | CCC(C)C1C(=O)OC(CC(=O)NC(C(=O)NC(C(=O)NC(CNC(C(=O)NC(C(=O)NC(C(=O)N1)CC(C)C)CC(=O)O)CC(C)C)CC(C)C)CC(C)C)CCC(=O)O)CCCCCCCCCCC(C)C |
| 12 | Fengycin | Bacillus subtilis | [25] | C72H110N12O20 | 443591 | CCCCCCCCCCCCCC(CC(=O)NC(CCC(=O)O)C(=O)NC(CCCN)C(=O)NC1CC2=CC=C(C=C2)OC(=O)C(NC(=O)C(NC(=O)C(NC(=O)C3CCCN3C(=O)C(NC(=O)C(NC(=O)C(NC1=O)C(C)O)CCC(=O)O)C)CCC(=O)N)CC4=CC=C(C=C4)O)C(C)CC)O |
| 13 | Subtilisin | Bacillus subtilis | [31] | C18H25N3O6 | 92174084 | CC(C(=O)NOC(=O)C1=CC=CC=C1)NC(=O)C(C)NC(=O)OC(C)(C)C |
| 14 | Gramicidin S | Brevibacillus brevis | [32] | C60H92N12O10 | 73357 | CC(C)CC1C(=O)NC(C(=O)N2CCCC2C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NC(C(=O)N3CCCC3C(=O)NC(C(=O)NC(C(=O)N1)CCCN)C(C)C)CC4=CC=CC=C4)CC(C)C)CCCN)C(C)C)CC5=CC=CC=C5 |
| 15 | Polymyxin | Paenibacillus polymyxa | [33] | C48H82N16O14 | 3083714 | CC(C)CC1C(=O)NC(C(=O)NC(C(=O)NC(C(=O)NCCC(C(=O)NC(C(=O)NC(C(=O)N1)CC2=CC=CC=C2)CCN)NC(=O)C(CCN)NC(=O)C(C(C)O)NC(=O)C(CCN)NC(=O)O)C(C)O)CCN)CCN |
| Sr. No. | Biosurfactant | Degree | Betweenness | Closeness |
|---|---|---|---|---|
| 1 | Putisolvin | 12 | 218.86357 | 0.44347826 |
| 2 | Surfactin | 11 | 260.51117 | 0.42857143 |
| 3 | Lichenysin | 11 | 180.90701 | 0.43589744 |
| 4 | Arthrofactin | 10 | 295.14645 | 0.4214876 |
| 5 | Amphisin | 10 | 98.94803 | 0.4214876 |
| 6 | Iturin | 10 | 312.37665 | 0.39534885 |
| 7 | Pumalicidin | 10 | 114.981255 | 0.42857143 |
| 8 | Subtilisin | 10 | 543.5165 | 0.39534885 |
| 9 | Polymyxin | 9 | 186.33307 | 0.4015748 |
| 10 | Viscosin | 9 | 126.53945 | 0.4214876 |
| 11 | Fengycin | 8 | 143.53413 | 0.38345864 |
| 12 | Gramicidin | 8 | 158.42682 | 0.38345864 |
| 13 | Ustilagic Acid | 6 | 220.03003 | 0.3167702 |
| 14 | Rhamnolipid | 6 | 151.10707 | 0.37226278 |
| 15 | Liposan | 3 | 112.77881 | 0.3090909 |
| Sr. No. | Genes | Degree | Betweenness | Closeness |
|---|---|---|---|---|
| 1 | TNF | 27 | 157.15123 | 0.24 |
| 2 | EGFR | 26 | 212.85284 | 0.23841059 |
| 3 | SRC | 21 | 59.026463 | 0.23076923 |
| 4 | IL2 | 21 | 49.672054 | 0.23076923 |
| 5 | IL1B | 21 | 72.07377 | 0.23076923 |
| 6 | PTPRC | 19 | 35.360935 | 0.2264151 |
| 7 | ITGB1 | 17 | 31.785282 | 0.22360249 |
| 8 | ERBB2 | 17 | 19.92101 | 0.225 |
| 9 | MAPK1 | 15 | 20.884993 | 0.22222222 |
| 10 | MTOR | 15 | 14.559942 | 0.22222222 |
| 11 | MDM2 | 14 | 31.841478 | 0.2208589 |
| 12 | ITGB2 | 13 | 11.806349 | 0.21818182 |
| 13 | HDAC1 | 13 | 26.021725 | 0.2195122 |
| 14 | NR3C1 | 12 | 25.276262 | 0.21818182 |
| 15 | TERT | 12 | 2.5834055 | 0.21818182 |
| 16 | MET | 11 | 4.9985447 | 0.21686748 |
| 17 | SELL | 11 | 11.046661 | 0.21301775 |
| 18 | CASP1 | 9 | 2.6095238 | 0.21301775 |
| 19 | HDAC6 | 9 | 5.4996777 | 0.21301775 |
| 20 | NOD2 | 8 | 3.3137822 | 0.20930232 |
| 21 | SELE | 8 | 0 | 0.21176471 |
| 22 | SELP | 8 | 0 | 0.21176471 |
| 23 | ADAM17 | 7 | 1.1746032 | 0.21052632 |
| 24 | SIRT2 | 7 | 2.0468254 | 0.20571429 |
| 25 | ITK | 7 | 6.074603 | 0.20809248 |
| 26 | HDAC2 | 7 | 4.533211 | 0.20571429 |
| 27 | PTPN2 | 6 | 2.3246753 | 0.20571429 |
| 28 | ITGB7 | 5 | 0.22222222 | 0.2 |
| 29 | PLA2G2A | 5 | 0.2 | 0.20809248 |
| 30 | UBE2I | 4 | 0 | 0.18947369 |
| 31 | PPIA | 4 | 1.137931 | 0.20224719 |
| 32 | MAP3K8 | 3 | 0 | 0.20454545 |
| 33 | IGF2R | 1 | 0 | 0.19672132 |
| 34 | OPRD1 | 1 | 0 | 0.027777778 |
| 35 | SLC5A1 | 1 | 0 | 0.19672132 |
| 36 | F11 | 1 | 0 | 0.027777778 |
| 37 | MRGPRX1 | 0 | 0 | 0.027027028 |
| Sr. No. | Protein | Receptor–Ligand | Interaction Type | Distance |
|---|---|---|---|---|
| 1 | 1M4C | A:ARG83:HN2 - :UNL1:O | Conventional Hydrogen Bond | 2.07493 |
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 1.65486 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 1.62878 | ||
| A:MET23 - :UNL1 | Alkyl | 5.36375 | ||
| UNL1 - A:MET23 | Alkyl | 5.22848 | ||
| UNL1 - A:LEU85 | Alkyl | 4.12048 | ||
| 2 | 3WLW | A:TYR268:HH - N:UNK1:O | Conventional Hydrogen Bond | 2.03597 |
| N:UNK1:H - A:ASP286:OD1 | Conventional Hydrogen Bond | 2.62308 | ||
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 2.48544 | ||
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 2.1777 | ||
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 2.83049 | ||
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 1.55546 | ||
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 2.51507 | ||
| N:UNK1:H - A:THR285:O | Conventional Hydrogen Bond | 2.22765 | ||
| N:UNK1:H - A:SER284:O | Conventional Hydrogen Bond | 3.005 | ||
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 2.72822 | ||
| A:LYS309:CE - N:UNK1:O | Carbon Hydrogen Bond | 3.69856 | ||
| A:LEU250:CB - N:UNK1 | Pi-Sigma | 3.92346 | ||
| A:ALA249 - N:UNK1 | Alkyl | 4.39901 | ||
| A:ALA249 - N:UNK1:C | Alkyl | 4.10378 | ||
| A:VAL251 - N:UNK1 | Alkyl | 5.23527 | ||
| A:VAL251 - N:UNK1 | Alkyl | 5.01063 | ||
| N:UNK1 - A:LEU250 | Alkyl | 4.96966 | ||
| N:UNK1:C - A:VAL287 | Alkyl | 5.09767 | ||
| 3 | 4MXO | A:MET341:HN - N:UNK1:O | Conventional Hydrogen Bond | 1.97267 |
| A:SER345:HN - N:UNK1:O | Conventional Hydrogen Bond | 2.85586 | ||
| A:ASN391:HD21 - N:UNK1:O | Conventional Hydrogen Bond | 2.986 | ||
| A:ASN391:HD22 - N:UNK1:O | Conventional Hydrogen Bond | 2.8606 | ||
| N:UNK1:H - A:LEU273:O | Conventional Hydrogen Bond | 2.23507 | ||
| N:UNK1:H - A:LEU273:O | Conventional Hydrogen Bond | 2.69853 | ||
| N:UNK1:H - A:GLN275:O | Conventional Hydrogen Bond | 2.83421 | ||
| A:GLY274:CA - N:UNK1:O | Carbon Hydrogen Bond | 3.14193 | ||
| A:GLY344:CA - N:UNK1:O | Carbon Hydrogen Bond | 3.14994 | ||
| N:UNK1:C - N:UNK1:O | Carbon Hydrogen Bond | 3.54737 | ||
| A:VAL281 - N:UNK1 | Alkyl | 4.86428 | ||
| A:VAL281 - N:UNK1 | Alkyl | 5.14814 | ||
| A:ALA293 - N:UNK1 | Alkyl | 4.50016 | ||
| A:LYS295 - N:UNK1 | Alkyl | 5.25134 | ||
| A:ALA403 - N:UNK1:C | Alkyl | 3.80138 | ||
| N:UNK1:C - A:MET314 | Alkyl | 4.94124 | ||
| N:UNK1:C - A:VAL323 | Alkyl | 3.63942 | ||
| N:UNK1 - A:LEU273 | Alkyl | 4.79106 | ||
| N:UNK1:C - A:LEU273 | Alkyl | 4.84683 | ||
| A:PHE278 - N:UNK1 | Pi-Alkyl | 5.36838 | ||
| 4 | 5FMV | N:UNK1:H - A:ASP508:OD2 | Salt Bridge | 2.60084 |
| N:UNK1:H - A:ASP508:OD2 | Conventional Hydrogen Bond | 2.42554 | ||
| N:UNK1:H - A:ASP508:OD1 | Conventional Hydrogen Bond | 2.71193 | ||
| A:LYS448 - N:UNK1 | Alkyl | 4.81167 | ||
| A:PRO449 - N:UNK1 | Alkyl | 4.98618 | ||
| A:HIS404 - N:UNK1 | Pi-Alkyl | 4.73078 | ||
| A:TRP487 - N:UNK1 | Pi-Alkyl | 4.80377 | ||
| A:TRP487 - N:UNK1 | Pi-Alkyl | 4.83494 | ||
| A:TRP487 - N:UNK1 | Pi-Alkyl | 4.37643 | ||
| 5 | 9ILB | N:UNK1:H - A:THR79:OG1 | Conventional Hydrogen Bond | 2.04809 |
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 2.48545 | ||
| N:UNK1:H - A:GLU25:OE2 | Conventional Hydrogen Bond | 3.01317 | ||
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 2.08891 | ||
| N:UNK1:H - N:UNK1:O | Conventional Hydrogen Bond | 1.5553 | ||
| N:UNK1:H - A:LEU134:O | Conventional Hydrogen Bond | 2.12807 | ||
| N:UNK1:H - A:VAL132:O | Conventional Hydrogen Bond | 2.61684 | ||
| N:UNK1:H - A:LEU80:O | Conventional Hydrogen Bond | 2.59836 | ||
| N:UNK1:C - N:UNK1:O | Carbon Hydrogen Bond | 3.5902 | ||
| A:PHE133 - N:UNK1 | Pi-Pi Stacked | 3.75995 | ||
| A:TYR24 - N:UNK1 | Pi-Alkyl | 4.38175 | ||
| A:TYR24 - N:UNK1:C | Pi-Alkyl | 4.05141 | ||
| N:UNK1 - A:PRO131 | Pi-Alkyl | 5.36192 | ||
| 6 | 4G6O | A:ASN152:HD22 - :UNL1:N | Conventional Hydrogen Bond | 2.68599 |
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 1.97392 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 1.72687 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.6197 | ||
| UNL1:H - A:ASP109:OD2 | Conventional Hydrogen Bond | 2.68182 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.16901 | ||
| UNL1:H - A:CYS164:SG | Conventional Hydrogen Bond | 3.02337 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.8299 | ||
| UNL1:H - A:ASP104:O | Conventional Hydrogen Bond | 2.77312 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.25806 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.61859 | ||
| UNL1:H - A:GLU31:OE1 | Conventional Hydrogen Bond | 2.17908 | ||
| UNL1:H - A:ASN152:OD1 | Conventional Hydrogen Bond | 2.75093 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.45343 | ||
| UNL1:H - A:ASP165:OD1 | Conventional Hydrogen Bond | 2.30444 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.29142 | ||
| UNL1 - A:ARG65 | Pi-Alkyl | 5.08193 | ||
| 7 | 4WKQ | UNL1:H - A:ASN842:OD1 | Conventional Hydrogen Bond | 2.34513 |
| UNL1:H - A:ASP837:OD2 | Conventional Hydrogen Bond | 2.64181 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.7793 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 2.61272 | ||
| A:ARG841:CD - :UNL1:O | Carbon Hydrogen Bond | 3.48781 | ||
| UNL1:C - A:ASP855:OD2 | Carbon Hydrogen Bond | 3.36649 | ||
| UNL1:C - A:ASP855:OD2 | Carbon Hydrogen Bond | 2.93591 | ||
| A:LEU718 - :UNL1 | Alkyl | 4.75503 | ||
| A:LEU718 - :UNL1 | Alkyl | 4.63701 | ||
| A:VAL726 - :UNL1 | Alkyl | 4.56822 | ||
| A:VAL726 - :UNL1 | Alkyl | 5.49427 | ||
| A:ALA743 - :UNL1 | Alkyl | 4.47905 | ||
| A:LEU844 - :UNL1 | Alkyl | 5.36797 | ||
| A:LEU844 - :UNL1 | Alkyl | 5.0695 | ||
| UNL1:C - A:LYS745 | Alkyl | 4.04982 | ||
| UNL1:C - A:MET766 | Alkyl | 5.00922 | ||
| UNL1:C - A:LEU788 | Alkyl | 4.51178 | ||
| 8 | 5WBU | UNL1:H - A:MET2345:SD | Conventional Hydrogen Bond | 2.81877 |
| A:ILE2356:CG2 - :UNL1 | Pi-Sigma | 3.70617 | ||
| A:TYR2225 - :UNL1 | Pi-Pi T-shaped | 4.91263 | ||
| UNL1:C - A:PRO2169 | Alkyl | 4.70722 | ||
| 9 | 7CEB | A:TYR295:HH - :UNL1:O | Conventional Hydrogen Bond | 2.30825 |
| A:TYR411:HH - :UNL1:O | Conventional Hydrogen Bond | 2.74757 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 1.65568 | ||
| UNL1:H - :UNL1:O | Conventional Hydrogen Bond | 1.6278 | ||
| UNL1:C - A:TYR234 | Pi-Sigma | 3.72179 | ||
| UNL1:C - A:ILE356 | Alkyl | 4.29736 | ||
| UNL1:C - A:PRO185 | Alkyl | 4.4092 | ||
| A:TRP91 - :UNL1 | Pi-Alkyl | 5.11969 | ||
| A:TRP91 - :UNL1 | Pi-Alkyl | 5.3081 | ||
| A:HIS110 - :UNL1 | Pi-Alkyl | 5.20706 | ||
| A:PHE237 - :UNL1 | Pi-Alkyl | 5.3189 | ||
| A:PHE237 - :UNL1 | Pi-Alkyl | 4.60088 | ||
| A:TYR295 - :UNL1 | Pi-Alkyl | 4.91972 | ||
| A:TYR295 - :UNL1:C | Pi-Alkyl | 5.27292 | ||
| A:TYR411 - :UNL1:C | Pi-Alkyl | 4.32441 | ||
| 10 | 2AZ5 | A:GLN61:HE12 - :UNL1:O | Conventional Hydrogen Bond | 2.61257 |
| A:TYR119:HH - :UNL1:O | Conventional Hydrogen Bond | 2.86757 | ||
| A:TYR151:HH - :UNL1:O | Conventional Hydrogen Bond | 2.59785 | ||
| A:LEU63:CD1 - :UNL1 | Pi-Sigma | 3.80396 | ||
| A:LEU63:CD2 - :UNL1 | Pi-Sigma | 3.73556 | ||
| UNL1:C - A:TYR119 | Pi-Sigma | 3.95427 | ||
| UNL1 - A:PRO117 | Pi-Alkyl | 4.8335 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adnan, M.; Siddiqui, A.J.; Noumi, E.; Hannachi, S.; Ashraf, S.A.; Awadelkareem, A.M.; Snoussi, M.; Badraoui, R.; Bardakci, F.; Sachidanandan, M.; et al. Integrating Network Pharmacology Approaches to Decipher the Multi-Target Pharmacological Mechanism of Microbial Biosurfactants as Novel Green Antimicrobials against Listeriosis. Antibiotics 2023, 12, 5. https://doi.org/10.3390/antibiotics12010005
Adnan M, Siddiqui AJ, Noumi E, Hannachi S, Ashraf SA, Awadelkareem AM, Snoussi M, Badraoui R, Bardakci F, Sachidanandan M, et al. Integrating Network Pharmacology Approaches to Decipher the Multi-Target Pharmacological Mechanism of Microbial Biosurfactants as Novel Green Antimicrobials against Listeriosis. Antibiotics. 2023; 12(1):5. https://doi.org/10.3390/antibiotics12010005
Chicago/Turabian StyleAdnan, Mohd, Arif Jamal Siddiqui, Emira Noumi, Sami Hannachi, Syed Amir Ashraf, Amir Mahgoub Awadelkareem, Mejdi Snoussi, Riadh Badraoui, Fevzi Bardakci, Manojkumar Sachidanandan, and et al. 2023. "Integrating Network Pharmacology Approaches to Decipher the Multi-Target Pharmacological Mechanism of Microbial Biosurfactants as Novel Green Antimicrobials against Listeriosis" Antibiotics 12, no. 1: 5. https://doi.org/10.3390/antibiotics12010005
APA StyleAdnan, M., Siddiqui, A. J., Noumi, E., Hannachi, S., Ashraf, S. A., Awadelkareem, A. M., Snoussi, M., Badraoui, R., Bardakci, F., Sachidanandan, M., Patel, M., & Patel, M. (2023). Integrating Network Pharmacology Approaches to Decipher the Multi-Target Pharmacological Mechanism of Microbial Biosurfactants as Novel Green Antimicrobials against Listeriosis. Antibiotics, 12(1), 5. https://doi.org/10.3390/antibiotics12010005














