Abstract
Resistance to antibiotic treatment developed by bacteria in humans and animals occurs when the microorganisms resist treatment with clinically approved antibiotics. Actions must be implemented to stop the further development of antibiotic resistance and the subsequent emergence of superbugs. Medication repurposing/repositioning is one strategy that can help find new antibiotics, as it speeds up drug development phases. Among them, the Zn2+ ion binders, such as sulfonamides and their bioisosteres, are considered the most promising compounds to obtain novel antibacterials, thus avoiding antibiotic resistance. Sulfonamides and their bioisosteres have drug-like properties well-known for decades and are suitable lead compounds for developing new pharmacological agent families for inhibiting carbonic anhydrases (CAs). CAs are a superfamily of metalloenzymes catalyzing the reversible reaction of CO2 hydration to HCO3− and H+, being present in most bacteria in multiple genetic families (α-, β-, γ- and ι-classes). These enzymes, acting as CO2 transducers, are promising drug targets because their activity influences microbe proliferation, biosynthetic pathways, and pathogen persistence in the host. In their natural or slightly modified scaffolds, sulfonamides/sulfamates/sulamides inhibit CAs in vitro and in vivo, in mouse models infected with antibiotic-resistant strains, confirming thus their role in contrasting bacterial antibiotic resistance.
1. Introduction
Antibiotic resistance is a worldwide emergency that kills more people than HIV/AIDS and malaria combined [1]. It kills around 30,000 people in Europe each year, with Italy accounting for one-third of them [2,3]. In addition, antibiotic-resistant illnesses have a significant effect on public health services [4]. Antibiotic resistance is the ability of bacteria to counteract the action of one or more of the commune FDA-approved antibiotics (Figure 1) [4,5]. The antibiotics reported in Figure 1 act as suppressors of cell wall synthesis, inhibitors of proteins or nucleic acids synthesis, membrane destroyers, antimetabolites, and competitive antagonists of substrates used in biosynthetic reactions [6]. It is essential to understand that humans and other animals do not acquire antibiotic resistance; instead, this phenomenon is developed by bacteria harbored in humans and animals [3]. When placed under selective pressure due to antibiotics, bacteria that have acquired a greater capacity for resistance (by DNA mutation or DNA genetic transfer) will have a greater chance of surviving and instead will occupy the environment vacated by bacteria that have been eliminated by therapy [7].
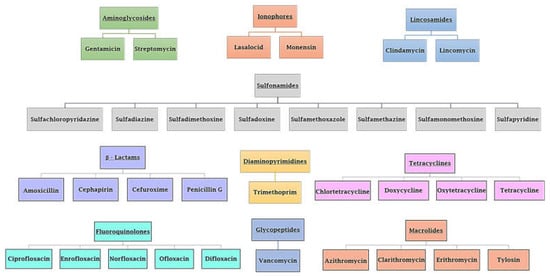
Figure 1.
Various types of FDA-approved antibiotics. Class and antibiotic name are reported.
Thus, to treat infections caused by those resistant bacteria, one strategy is to administer other antibiotics to which they are sensitive. However, they might also acquire resistance to the new class of antibiotics (multi-resistant organisms), and so switching to a new type of antibiotic is required until we arrive at bacteria resistant to all antibiotics (pan-resistant microorganisms) [8]. The antibiotic resistance phenomenon is associated with the misuse and overprescribing of these drugs, as well as the inappropriate administration of antibiotics to companion animals and animals in the agriculture industry [9,10]. In livestock farms or aquacultures all over the world, antibiotics are routinely used not only to treat diseases, as is the case in human medicine but also to prevent diseases and as promoters of animal growth [11]. Furthermore, the rising discharge of antibiotics into waterways and soils poses a risk to all microorganisms in these habitats [11]. Thus, bacteria can become resistant and infect people who came in touch with the polluted environment, animals, or meat. Therefore, policies must be put in place to combat both the spread and future development of antibiotic resistance, as well as the oncoming wave of superbugs [12].
How do we intervene to stop antibiotic resistance? There is no shortage of strategies for dealing with the antibiotic resistance [13,14,15,16]. Among them, one may consider: (i) community- and healthcare-based approaches for infection control and prevention; (ii) vaccine preparation, which may have a good chance of preventing bacterial illnesses (up to date, only for Streptococcus pneumoniae, one of the six most hazardous antibiotic-resistant bacteria, exists such a vaccine); (iii) reduction of the use of antibiotics in non-human infection-treatment contexts, such as livestock farms; (iv) appropriate antibiotic use, as well as stopping their use for the management of viral infections; (v) maintenance of investments to make second-line antibiotics available, and the development of novel antibiotics and especially those with novel modes of action less susceptible to the onset of resistance, which can save lives [13,14,15,16]. This last strategy is fundamental, but new drug development takes a long time; most candidate compounds were in the research and development pipelines for over a decade before they made it to market [17]. Moreover, the most critical points are that the majority of the new antibiotics in development are variants of preexisting antibiotic classes, do not have a novel mechanism of action, and only a small number of them are expected to be effective against the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacter). Finally, the future of antibiotics is shaky because of questions about their effectiveness and safety [18,19].
In this context, the drug repurposing or drug repositioning strategy offers the potential to “fast-track” the identification of novel antibiotics, since it speeds up the drug research process and reduces the time to market [20]. FDA-approved medications are considered the “existing drugs,” and their repurposing reduces clinical development risk, making them attractive candidates [20].
2. A Superfamily of CO2 Transducer Biomolecules: The Carbonic Anhydrase
2.1. Bacterial Carbonic Anhydrase
Carbon dioxide (CO2) is a gas released into the atmosphere due to cellular respiration and oxidative metabolism, and all living creatures are responsible for its production [21]. In most cases, the process of transporting this waste gas out of cells is carried out by passive diffusion [22]. In many cases, the CO2 channels, controlled by CO2 levels inside the cell, make this transfer easier. However, CO2 is not merely a waste product; it can also trigger various cellular signaling pathways to increase bacteria virulence and pathogenicity [23]. Bacteria can adapt to their environment by sensing and responding to CO2, enhancing thus their chances of survival. This may be crucial because bacteria have to adapt to the relatively low CO2 levels of the outer atmosphere for the higher CO2 levels found inside most multicellular host organisms [23]. Bacteria, for example, may upregulate virulence factors at host physiologic CO2 levels rather than ambient CO2 levels to aid colonization or infection. Several such examples are Vibrio cholerae, which causes cholera, and produces enterotoxin as carbon dioxide levels rise, whereas bicarbonate produced by the CO2 hydration is the first positive effector of the primary V. cholerae virulence gene transcription activator (ToxT), responsible for the cholera virulence cascade [24]. Pseudomonas aeruginosa, which can lead to infections in the blood, lungs (pneumonia), or other regions of the body following surgery, lives in vastly varying CO2 settings depending on whether or not it is colonizing a host [25].
Biomolecules in microbes related to CO2-sensitive pathways or acting as a CO2 transducer have been proposed as appealing targets for medicines, since they control cell development and the subsequent synthesis of chemicals, enhancing the pathogen persistence in the host [26,27]. In this context, a crucial role is played by a superfamily of molecules known as carbonic anhydrases (CAs, EC 4.2.1.1). CAs can be thought as molecules that, rather than instantly detecting a change in CO2, serve as CO2 transducers, adjusting its levels [23,28]. With their activity, the CAs encoded by the bacterial genome of pathogenic and non-pathogenic bacteria provide the indispensable CO2 and HCO3−/protons to microbial biosynthetic pathways, catalyzing the reversible reaction of CO2 hydration to HCO3− and H+ (CO2 + H2O ⇋ HCO3−+ H+) [28]. Here, we stress the fact that the non-catalytic CO2 hydration/dehydration reaction is too slow at physiological pH values to fulfill the organism’s metabolic demands (kcat (hydration) = 0.15 s−1; kcat (dehydration) = 50.0 s−1) [29].
The classification system for CAs uses the Greek letters to represent the eight distinct families (or classes): α, β, γ, δ, ζ, η, θ, and ι [29,30,31,32,33]. The eight distinct CA-classes descend from the same ancestor, yet exhibit significant evolutionary diversity. The representative amino acid sequences of each CA-class show low sequence similarity, characteristic folds, and structures compared to the polypeptide chain of other CAs belonging to a different class [34,35,36,37,38]. In contrast, the mechanism involved in the reversible hydration of CO2 is strictly conserved across all CA-classes, illustrating the CA superfamily’s convergent evolution [34,35,36,37,38]. CAs are generally metalloenzymes with catalytic sites that contain a metal ion cofactor required for catalysis (Figure 2). The ion cofactor in many CAs is Zn2+, which is coordinated by three amino acid residues from the protein backbone [29,30,39]. The fourth metal ion ligand is a water molecule/hydroxide ion that acts as the nucleophile in the enzyme’s catalytic cycle. Metal ions other than Zn2+, such as Co2+, Cd2+, Fe2+, and Mn2+, can be coordinated by several CA-classes [40,41,42,43,44,45,46,47]. Recently, it has been demonstrated that the newly discovered CA-class, the ι-CA, shows catalytic activity without the need for metal ions, as shown from the X-ray crystal structure of Anabaena sp. [48]. The CA-classes differ in the amino acid residues involved in metal coordination [49,50,51,52]. For example, the ion metal is coordinated by three His residues in the α, β, and γ-CAs and presumably θ-classes [42,45,53,54]; one His and two Cys residues in the β- and ζ-CAs [41]; and two His and one Gln residue in the η-class. α-CAs usually act as monomers or dimers; β-CAs only behave as dimers, tetramers, or octamers. To perform their catalytic activity, the γ-CAs must be trimers [55]. A tandemly repeated hexapeptide characterizes γ-CA monomers and is required for the left-hand fold of trimeric-helix structures. The X-ray structure of the θ-CAs was remarkably similar to that of some β-CAs [42,45,53,54]. The crystal structure of ζ-CA showed three slightly different active sites on the same polypeptide chain. Regarding the structural organization of δ- and η-CAs, no data are currently available, and a homology modelling of the η-CA was built [56]. Interestingly, only the α, η, θ, and ι -CAs have been shown to catalyze the esters/thioesters hydrolysis, while the other CA families lacked any detectable esterase activity [29,30,39,57,58,59]. Presently, four CA-classes (α, β, γ, and ι) have been demonstrated to exist in bacteria, and their distribution is noteworthy [34,35,36,37,38]. In many cases, the bacterial genome encodes for the three CA-classes (α, β, and γ) and rarely for the ι- class. However, it is common to find bacteria whose genomes encodes just one or two CAs, and very rarely none [27,29,33,49]. Moreover, the structural variations between bacterial and human α-CAs allow for the synthesis of inhibitors that target the bacterial enzyme but not the mammalian ones. Again, in some cases, the pathogenic bacterial genome encodes for CA-classes, which are absent in mammals, whose genome encodes only for α-CA, increasing the likelihood of success in treating the bacterial illness with compounds that act only on the bacterial CAs. Interestingly, for each CA-class has been obtained the X-ray crystallographic structures (Figure 3). It has been observed that α-CAs reside in the periplasmic region of bacteria cells and prevent CO2 loss from bacteria by converting CO2 into bicarbonate, which is then transported inside the cytoplasm by bicarbonate transporters [29,30,31,33]. On the other hand, cytoplasmic β- and γ -CA classes are responsible for carrying out intracellular functions such as maintaining CO2 and HCO3− equilibrium and regulating pH. Recently, β, γ, and ι-CAs with signal peptide at N-terminus have been found, attributing them a putative periplasmic localization and a physiological role similar to those mentioned above for the α-CAs [31,32,39,57,58,59].
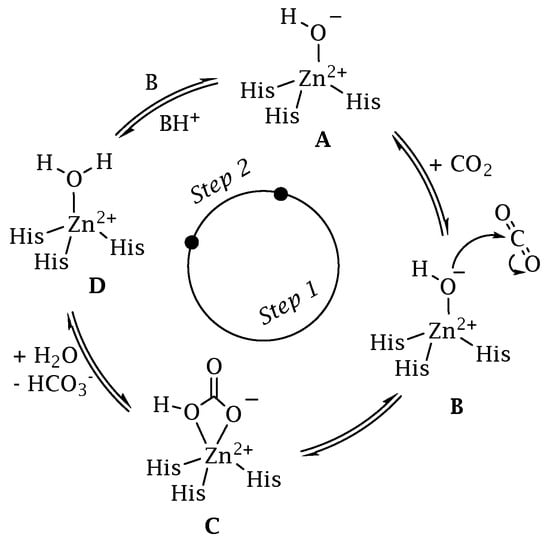
Figure 2.
CA catalytic mechanism schematically represented for a α-class isoform.
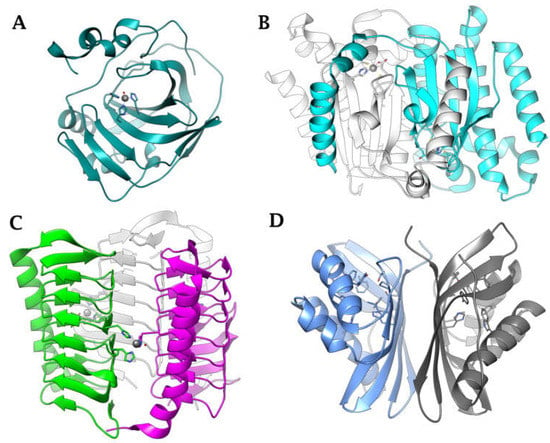
Figure 3.
Ribbon view of (A) α-CA from Neisseria gonorrhoeae (PDB 1KOQ), (B) β-CA from Escherichia coli (PDB 1I6O), (C) γ-CA from Burkholderia pseudomallei (PDB 7ZW9), (D) ι-CA from B. territorii built by homology using PDB 3H51 as a template.
2.2. CAs Help Bacteria to Survive
In the literature, many shreds of evidence support the opinion that the activity of CAs is connected to the survival of microbes because these enzymes are essential for supporting numerous physiological functions involving dissolved inorganic carbon, such as transport and supply of CO2 or HCO3−, pH homeostasis, secretion of electrolytes/toxins, and biosynthetic processes [29,33,60]. For example, it has been proven in vivo that bacterial growth at ambient CO2 concentrations is dependent on CA activity in bacteria such as Ralstonia eutropha (Gram-negative bacterium found in soil and water) and Escherichia coli (Gram-negative bacterium) [61,62,63]. In E. coli, the two β-CAs (CynT and CynT2) generate HCO3− to prevent bicarbonate depletion from cyanate breakdown and bacterial expansion at atmospheric CO2 concentrations, respectively [62,63]. More intriguing is the in vivo evidence that CAs have a role in the proliferation of harmful bacteria such as Mycobacterium tuberculosis [64,65,66,67,68], Helicobacter pylori [69,70,71], Vibrio cholerae [72], Brucella suis [65,66,67,68], Salmonella enterica [73,74,75], and Pseudomonas aeruginosa [76]. CAs encoded by the genome of H. pylori, a Gram-negative, microaerophilic bacteria that colonizes the human stomach, are essential for the pathogen’s acid acclimation and, consequently, survival in the severe environment typical of this organ, with pH values as low as 1.5–2.0 [69,70,71]. In addition to CAs, urease is the other enzymatic system used by the microbe for growing in this extreme environment. Under acidic conditions, urea goes into the cytoplasm through the urea channel. In the bacterial cytoplasm, 2NH3 and CO2 are produced by the hydrolysis of urea [69,70,71]. The resulting CO2 is then hydrated by β-CA, while the periplasmic α-CA hydrates the CO2 diffused in the periplasm. The produced ions (H+) by the CA-catalyzed reaction are used to form NH4+ by reacting with NH3+ in the periplasm and cytoplasm, which neutralizes the entering acid in the above environments [69,70,71]. In the case of the pathogenic bacterium Vibrio cholerae, a Gram-negative bacterium already mentioned above, CAs are involved in the production of sodium bicarbonate, which stimulates the development of cholera toxin [24]. It has been proposed that V. cholerae employs CAs to colonize the host [72]. Again, the brucellosis causal agent, Brucella suis, a non-motile Gram-negative coccobacillus, and Mycobacterium tuberculosis, a pathogenic bacterium that causes tuberculosis, were demonstrated to require functional CAs to proliferate [64,65,66,67,68]. Furthermore, through in vivo gene expression investigations on the bacterium Salmonella enterica, the MIG5 gene, which encodes for a CA that is significantly expressed during bacterial infection, has been found [73,74,75]. The deletion of the gene encoding this CA (psCA1) in the Pseudomonas aeruginosa reduced pathogenicity by decreasing calcium salt depositions [77].
2.3. Carbonic Anhydrase Sulfonamide Inhibitors
Because the many biochemical processes mentioned above involve the activity of bacterial CAs, their inhibition may reduce the pathogen’s survival and fitness. The good news is that CA inhibition suppresses bacterial growth differently from those demonstrated by traditional antibiotics, toward which the bacteria have developed or are developing antibiotic resistance. As reported in the scientific literature, many unique chemical classes of CA inhibitors (CAIs) exist [60]. The CAIs are classified into four distinct types based on how the inhibitors bind and inhibit the CA metalloenzymes. Four types of inhibitor-enzyme binding are currently known, based on whether the binding involves the catalytic metal ion or the metal coordinated-water molecule or how the active site is obstructed [78]. Therefore, there are metal ion binders (anion, sulfonamides and their bioisosteres, dithiocarbamates, xanthates, and so on); chemicals that bind to the zinc-coordinated water molecule/hydroxide ion (phenols, polyamines, thioxocoumarins, sulfocumarins); compounds that obstruct the active site entrance (coumarins and related isosteres); and compounds that bind out of the active site (carboxylate) [78].
Among these types, the Zn2+ ion binders, in particular, the sulfonamides and their bioisosteres, are considered the most promising compounds for the realization of novel antibacterials, avoiding antibiotic resistance [78,79,80,81]. Here, we emphasize that the sulfonamides/sulfamates/sulfamides able to inhibit the CA specifically are nonantibiotic inhibitors characterized by a primary sulfonamide moiety, having the following chemical formula: R-X-SO2-NH2, where R can be an aromatic, heterocyclic, aliphatic, or sugar scaffold, X = nothing, O or NH. Thus, sulfanilamide led to the discovery of the sulfa drugs and benzenesulfonamide CAIs of the type. They constitute an important class of drugs since they have drug-like properties well-known for decades and are suitable lead compounds for developing new pharmacological agent families for inhibiting CAs. Among them are the commercial derivatives 1–24 (Figure 4) and the clinically used agent AAZ-EPA (Figure 5). The series AAZ-EPA include acetazolamide (AAZ), methazolamide (MZA), ethoxzolamide (EZA), and dichlorophenamide (DCP) are classical, systemically working antiglaucoma CAIs. Dorzolamide (DZA) and brinzolamide (BRZ) are topically acting antiglaucoma agents. Benzolamide (BZA) is an orphan drug belonging to this class of pharmacological agents. Zonisamide (ZNS), sulthiame (SLT), and sulfamic acid ester topiramate (TPM) are widely used antiepileptic drugs. Sulpiride (SLP) and indisulam (IND) were also shown by our group to belong to this class of pharmacological agents, together with the COX2 selective inhibitors celecoxib (CLX) and valdecoxib (VLX). Saccharin (SAC) and the diuretic hydrochlorothiazide (HCT) are also known to act as CAIs. Famotidine (FAM) and epacadostat (EPA) are CAI sulfamide drugs clinically used respectively as a histamine H2 receptor antagonist and a selective indoleamine-2,3-dioxygenase 1 inhibitor. These inhibitors bind Zn (II) in a tetrahedral geometry, forming an extended network of hydrogen bonds with the enzyme amino acid residues, whereas the aromatic/heterocyclic portions of the inhibitor interact with the hydrophilic and hydrophobic residues found in the enzyme catalytic cavity, according to enzyme-inhibitor X-ray crystallographic data (Figure 6) [60].
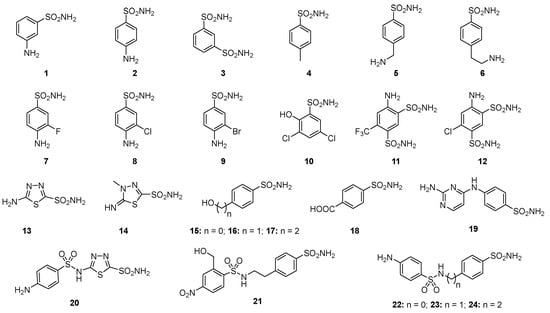
Figure 4.
Structure of commercially available sulfonamide derivatives 1–24.
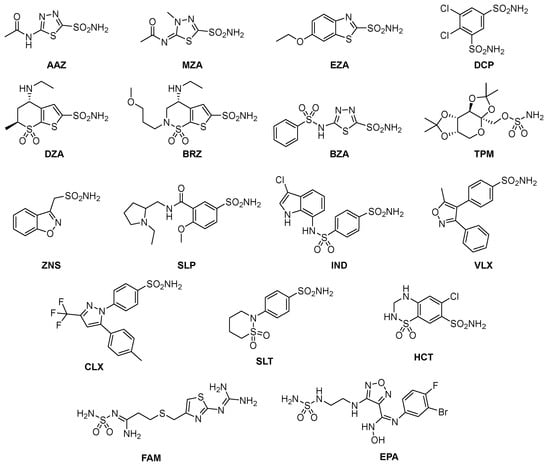
Figure 5.
Structure of clinically used sulfonamide/sulfamate/sulfamide derivatives AAZ-HCT.
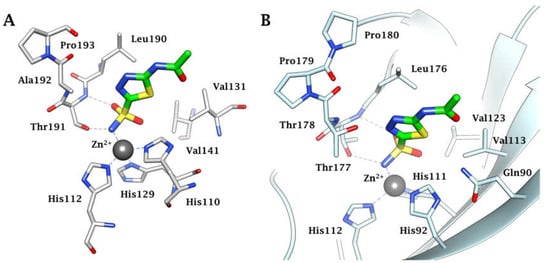
Figure 6.
(A) Active site view of the α-CA of Helicobacter pylori in adduct with AAZ (4YGF). (B) Active site ribbon view of the α-CA of N. gonorrhoeae in adduct with AAZ (8DYQ). H-bonds are represented as black dashed lines.
3. Where Are We NOW with the Inhibition of the Bacterial CAs
In the last decade, several in vitro experiments were conducted employing the CAIs outlined in the preceding paragraph to inhibit the four bacterial CA classes (α, β, γ, and ι). Many of these studies were focused on bacterial CAs derived from pathogenic bacteria, such as Mycobacterium tuberculosis, Vibrio cholerae, Francisella tularensis, Burkholderia pseudomallei, Porphyromonas gingivalis, Legionella pneumophila, Clostridium perfringens, Mammaliccosu sciuri, etc. [36,82,83,84,85]. Most sulfonamide CAIs exert potent inhibition on most recombinant CAs belonging to the bacteria mentioned above [34,86,87,88,89,90]. The most interesting aspect was that, some of these CAIs, including acetazolamide and methazolamide, significantly limit the growth of bacteria in cell cultures [91]. In this context, some experimental shreds of evidence prove that the inhibition of bacteria CAs can be potentially used to combat the resistance of many pathogens to the existing antimicrobial drugs.
For example, ethoxzolamide (EZA), an authorized diuretic and carbonic anhydrase inhibitor, kills Helicobacter pylori in vitro, suggesting it could be turned into an anti-H. pylori medication [92].
The influence of the selective CA inhibitor AAZ on the bacterial lifecycle was tested by analyzing the growth of E. coli and its consumption of glucose, added as the only carbon source to the bacterial culture media [93]. Carbon sources are required for biosynthetic activities and metabolism is directly correlated with the rate at which carbon sources are used. The FDA-approved carbonic anhydrase AAZ was able to interfere with E. coli growth and glucose uptake at 31.2 μg/mL. Intriguingly, AAZ resulted in a good inhibitor of the two recombinant E. coli CAs, β-CA (CynT2) and γ-CA (EcoCAγ), with a KI of 227 and 248 nM, respectively [94,95]. AAZ prevents sugar consumption due to its inhibitory action on bacterial CAs, which are directly engaged in providing CO2/HCO3− required for the bacterial metabolic need [93].
It has been reproposed the FDA-approved carbonic anhydrase drug AAZ could be used to design potent antienterococcal agents. The authors, modifying the AAZ scaffold, arrived at two leads possessing improved potency against clinical vancomycin-resistant enterococci (VRE) strains [96]. The classical AAZ showed a MIC of 2 μg/mL, while the two leads had a MIC = 0.007 μg/mL and 1 μg/mL, respectively. DZA, another classical CAIs, resulted in MIC values of 1–8 μg/mL against a panel of clinical VRE isolates [97]. Based on the results of homology modeling and molecular dynamics simulations, the authors demonstrated that the α and γ CAs encoded by the Vancomycin-Resistant Enterococcus are the intracellular targets of the compounds [96,98]. In addition, AAZ fared better than linezolid (a standard drug for VRE infections) when tested in two in vivo VRE mouse models –murine colonization–reduction and VRE septicemia [99].
Finally, AAZ inhibited the growth of the Gram-negative bacterium Neisseria gonorrhoeae both in the in vitro as well as in vivo mouse model of a gonococcal genital tract infection [100,101]. Recently, Portela et al. [102] demonstrated that sulfonamide pretreatment has a positive outcome on the strength of dentin and the prevention of Streptococcus mutans colonization in teeth treated with two potent bacterial CA sulfonamide inhibitors [102].
It is also interesting to note that over the last few years, there has been a significant focus on developing non-classical CAIs for treating multiple diseases due to the prevalence of sulfonamide allergies among the general population as well as their use as potential new antibacterials [103]. The classes of non-classical inhibitors that show strong potential as lead compounds for isoform-specific drug design include phenols, polyamines, carboxylic acids, and coumarins and their derivatives. These compounds can anchor to the zinc-bound water/hydroxide ion or bind outside the active site to block substrate entry, exhibiting atypical binding mechanisms of the classical sulfonamide CAIs [103].
4. Conclusions
The well-known zinc-binding groups (ZBGs), such as the primary sulfonamide (-SO2NH2), primary sulfamate (-OSO2NH2), and sulfamide (-NHSO2NH2), which are present in the structure of several clinically-approved drugs and an increasing number of investigational medicines, have been proven to affect the inhibition of the CAs encoded by pathogenic bacteria [78,79,80,81]. Intriguingly, various types of these nonantibiotic sulfonamides, many of which offer pharmacologic applications as antiglaucoma, antiobesity, antitumor, or diuretic, resulted in potent inhibitors (kI in the nanomolar range) of such CAs. These findings prompt scientists to consider the CAIs as a new approach to fight antibiotic resistance developed by bacteria versus the common FDA-approved antibiotics. Unlike common antibiotics, the CAIs impair the growth of pathogenic bacteria through a novel mechanism of action: perturbating/depleting the intracellular levels of CO2 and HCO3−, which are necessary to microbial biosynthetic pathways. In addition, the antibacterial growth due to the CA inhibition is strongly supported by the fact that the slow non-catalytic CO2 hydration/dehydration reaction is unable to restore these levels. These attractive facts led to the writing of fascinating manuscripts that discuss the repurposing of drugs such as ethoxzolamide (EZA), acetazolamide (AZA), and dorzolamide (DZA) as treatments for interfering with the life cycle of E. coli, Helicobacter pylori, Neisseria gonorrhoeae, and vancomycin-resistant enterococci.
In conclusion, the licensed sulfonamides/sulfamates/sulfamides acting as CAIs exhibit antibacterial properties in their native or slightly modified scaffolds [97], hypothesizing that the CAIs can be potentially employed either by themselves or in conjunction with an antibiotic or even as “antibiotic adjuvants” to increase the effectiveness of certain antibiotics.
Author Contributions
Supervision, C.T.S. and C.C.; Writing—original draft, C.C.; Writing—review and editing, A.N., C.T.S. and C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Italian Ministry of University and Research, project FISR2019_04819 BacCAD (to C.T.S. and C.C.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to Valentina Brasiello for her assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Doolan, J.A.; Williams, G.T.; Hilton, K.L.F.; Chaudhari, R.; Fossey, J.S.; Goult, B.T.; Hiscock, J.R. Advancements in antimicrobial nanoscale materials and self-assembling systems. Chem. Soc. Rev. 2022, 51, 8696–8755. [Google Scholar] [CrossRef]
- Wagenlehner, F.M.E.; Dittmar, F. Re: Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Eur. Urol. 2022, 82, 658–670. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J. 2022, 20, e07209. [Google Scholar]
- Reardon, S. Resistance to last-ditch antibiotic has spread farther than anticipated. Nature 2017. [Google Scholar] [CrossRef]
- Uruen, C.; Garcia, C.; Fraile, L.; Tommassen, J.; Arenas, J. How Streptococcus suis escapes antibiotic treatments. Vet. Res. 2022, 53, 91–123. [Google Scholar] [CrossRef]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Oz, T.; Guvenek, A.; Yildiz, S.; Karaboga, E.; Tamer, Y.T.; Mumcuyan, N.; Ozan, V.B.; Senturk, G.H.; Cokol, M.; Yeh, P.; et al. Strength of Selection Pressure Is an Important Parameter Contributing to the Complexity of Antibiotic Resistance Evolution. Mol. Biol. Evol 2014, 31, 2387–2401. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Sulis, G.; Pai, M.; Gandra, S. Comment on: Global consumption of antimicrobials: Impact of the WHO Global Action Plan on Antimicrobial Resistance and 2019 coronavirus pandemic (COVID-19). J. Antimicrob. Chemother. 2022, 77, 2891–2892. [Google Scholar] [CrossRef]
- Iwu, C.D.; Patrick, S.M. An insight into the implementation of the global action plan on antimicrobial resistance in the WHO African region: A roadmap for action. Int. J. Antimicrob. Agents 2021, 58, 106411–106417. [Google Scholar] [CrossRef]
- Cycon, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment-Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338–382. [Google Scholar] [CrossRef]
- Pusparajah, P.; Letchumanan, V.; Goh, B.H.; McGaw, L.J. Editorial: Novel Approaches to the Treatment of Multidrug-Resistant Bacteria. Front. Pharmacol. 2022, 13, 972935–972937. [Google Scholar] [CrossRef]
- da Silva, T.H.; Hachigian, T.Z.; Lee, J.; King, M.D. Using computers to ESKAPE the antibiotic resistance crisis. Drug Discov. Today 2022, 27, 456–470. [Google Scholar] [CrossRef]
- Nataraj, B.H.; Mallappa, R.H. Antibiotic Resistance Crisis: An Update on Antagonistic Interactions between Probiotics and Methicillin-Resistant Staphylococcus aureus (MRSA). Curr. Microbiol. 2021, 78, 2194–2211. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Hansen, M.P.; Hoffmann, T.C.; McCullough, A.R.; van Driel, M.L.; Del Mar, C.B. Antibiotic Resistance: What are the Opportunities for Primary Care in Alleviating the Crisis? Front. Public Health 2015, 3, 35–41. [Google Scholar] [CrossRef]
- Sun, D.X.; Gao, W.; Hu, H.X.; Zhou, S.M. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Bronstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Krishnamurthy, N.; Grimshaw, A.A.; Axson, S.A.; Choe, S.H.; Miller, J.E. Drug repurposing: A systematic review on root causes, barriers and facilitators. BMC Health Serv. Res. 2022, 22, 970. [Google Scholar] [CrossRef]
- Mitchell, A.P. Fungal CO2 sensing: A breath of fresh air. Curr. Biol. 2005, 15, R934–R9366. [Google Scholar] [CrossRef]
- Michenkova, M.; Taki, S.; Blosser, M.C.; Hwang, H.J.; Kowatz, T.; Moss, F.J.; Occhipinti, R.; Qin, X.; Sen, S.; Shinn, E.; et al. Carbon dioxide transport across membranes. Interface Focus 2021, 11, 20200090–20200107. [Google Scholar] [CrossRef]
- Cummins, E.P.; Selfridge, A.C.; Sporn, P.H.; Sznajder, J.I.; Taylor, C.T. Carbon dioxide-sensing in organisms and its implications for human disease. Cell Mol. Life Sci. 2014, 71, 831–845. [Google Scholar] [CrossRef]
- Shimamura, T.; Watanabe, S.; Sasaki, S. Enhancement of enterotoxin production by carbon dioxide in Vibrio cholerae. Infect. Immun. 1985, 49, 455–456. [Google Scholar] [CrossRef]
- Lotlikar, S.R.; Hnatusko, S.; Dickenson, N.E.; Choudhari, S.P.; Picking, W.L.; Patrauchan, M.A. Three functional beta-carbonic anhydrases in Pseudomonas aeruginosa PAO1: Role in survival in ambient air. Microbiology 2013, 159 Pt 8, 1748–1759. [Google Scholar] [CrossRef]
- Supuran, C.T.; Capasso, C. A Highlight on the Inhibition of Fungal Carbonic Anhydrases as Drug Targets for the Antifungal Armamentarium. Int. J. Mol. Sci. 2021, 22, 4324. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin. Ther. Targets 2015, 19, 1689–1704. [Google Scholar] [CrossRef]
- Campestre, C.; De Luca, V.; Carradori, S.; Grande, R.; Carginale, V.; Scaloni, A.; Supuran, C.T.; Capasso, C. Carbonic Anhydrases: New Perspectives on Protein Functional Role and Inhibition in Helicobacter pylori. Front. Microbiol. 2021, 12, 629163–629174. [Google Scholar] [CrossRef]
- Supuran, C.T.; Capasso, C. An Overview of the Bacterial Carbonic Anhydrases. Metabolites 2017, 7, 56. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzym. Inhib. Med. Chem. 2015, 30, 325–332. [Google Scholar] [CrossRef]
- Nocentini, A.; Supuran, C.T.; Capasso, C. An overview on the recently discovered iota-carbonic anhydrases. J. Enzym. Inhib. Med. Chem. 2021, 36, 1988–1995. [Google Scholar] [CrossRef]
- Supuran, C.T.; Capasso, C. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J. Enzym. Inhib. Med. Chem. 2016, 31, 1254–1260. [Google Scholar] [CrossRef]
- Supuran, C.T.; Capasso, C. Biomedical applications of prokaryotic carbonic anhydrases. Expert Opin. Ther. Pat. 2018, 28, 745–754. [Google Scholar] [CrossRef]
- Annunziato, G.; Angeli, A.; D’Alba, F.; Bruno, A.; Pieroni, M.; Vullo, D.; De Luca, V.; Capasso, C.; Supuran, C.T.; Costantino, G. Discovery of New Potential Anti-Infective Compounds Based on Carbonic Anhydrase Inhibitors by Rational Target-Focused Repurposing Approaches. ChemMedChem 2016, 11, 1904–1914. [Google Scholar] [CrossRef]
- Ozensoy Guler, O.; Capasso, C.; Supuran, C.T. A magnificent enzyme superfamily: Carbonic anhydrases, their purification and characterization. J. Enzym. Inhib. Med. Chem. 2016, 31, 689–694. [Google Scholar] [CrossRef]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; Ferraroni, M.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Sulfonamide inhibition studies of the beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorganic Med. Chem. 2016, 24, 1115–1120. [Google Scholar] [CrossRef]
- Del Prete, S.; De Luca, V.; De Simone, G.; Supuran, C.T.; Capasso, C. Cloning, expression and purification of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. J. Enzym. Inhib. Med. Chem. 2016, 31, 54–59. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. An Overview of the Carbonic Anhydrases from Two Pathogens of the Oral Cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr. Top. Med. Chem. 2016, 16, 2359–2368. [Google Scholar] [CrossRef]
- Del Prete, S.; Nocentini, A.; Supuran, C.T.; Capasso, C. Bacterial iota-carbonic anhydrase: A new active class of carbonic anhydrase identified in the genome of the Gram-negative bacterium Burkholderia territorii. J. Enzym. Inhib. Med. Chem. 2020, 35, 1060–1068. [Google Scholar] [CrossRef]
- Pinard, M.A.; Lotlikar, S.R.; Boone, C.D.; Vullo, D.; Supuran, C.T.; Patrauchan, M.A.; McKenna, R. Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorganic Med. Chem. 2015, 23, 4831–4838. [Google Scholar] [CrossRef]
- Ferraroni, M.; Del Prete, S.; Vullo, D.; Capasso, C.; Supuran, C.T. Crystal structure and kinetic studies of a tetrameric type II beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71 Pt 12, 2449–2456. [Google Scholar] [CrossRef]
- De Simone, G.; Monti, S.M.; Alterio, V.; Buonanno, M.; De Luca, V.; Rossi, M.; Carginale, V.; Supuran, C.T.; Capasso, C.; Di Fiore, A. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorganic Med. Chem. Lett. 2015, 25, 2002–2006. [Google Scholar] [CrossRef]
- Zolnowska, B.; Slawinski, J.; Pogorzelska, A.; Chojnacki, J.; Vullo, D.; Supuran, C.T. Carbonic anhydrase inhibitors. Synthesis, and molecular structure of novel series N-substituted N′-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)guanidines and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Eur. J. Med. Chem. 2014, 71, 135–147. [Google Scholar]
- De Luca, L.; Ferro, S.; Damiano, F.M.; Supuran, C.T.; Vullo, D.; Chimirri, A.; Gitto, R. Structure-based screening for the discovery of new carbonic anhydrase VII inhibitors. Eur. J. Med. Chem. 2014, 71, 105–111. [Google Scholar] [CrossRef]
- Di Fiore, A.; Capasso, C.; De Luca, V.; Monti, S.M.; Carginale, V.; Supuran, C.T.; Scozzafava, A.; Pedone, C.; Rossi, M.; De Simone, G. X-ray structure of the first ‘extremo-alpha-carbonic anhydrase’, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69 Pt 6, 1150–1159. [Google Scholar] [CrossRef]
- Supuran, C.T. Structure-based drug discovery of carbonic anhydrase inhibitors. J. Enzym. Inhib. Med. Chem. 2012, 27, 759–772. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic anhydrases—An overview. Curr. Pharm. Des. 2008, 14, 603–614. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Senda, M.; Fukuda, K.; Yu, H.Y.; Ishida, M.; Taira, M.; Kinbara, K.; Senda, T. Characterization of a novel type of carbonic anhydrase that acts without metal cofactors. BMC Biol. 2021, 19, 105. [Google Scholar] [CrossRef]
- Supuran, C.T.; Capasso, C. Carbonic Anhydrase from Porphyromonas Gingivalis as a Drug Target. Pathogens 2017, 6, 30. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. An Overview of the Selectivity and Efficiency of the Bacterial Carbonic Anhydrase Inhibitors. Curr. Med. Chem. 2015, 22, 2130–2139. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. Sulfa and trimethoprim-like drugs—Antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J. Enzym. Inhib. Med. Chem. 2014, 29, 379–387. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. Anti-infective carbonic anhydrase inhibitors: A patent and literature review. Expert Opin. Ther. Pat. 2013, 23, 693–704. [Google Scholar] [CrossRef] [PubMed]
- James, P.; Isupov, M.N.; Sayer, C.; Saneei, V.; Berg, S.; Lioliou, M.; Kotlar, H.K.; Littlechild, J.A. The structure of a tetrameric alpha-carbonic anhydrase from Thermovibrio ammonificans reveals a core formed around intermolecular disulfides that contribute to its thermostability. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70 Pt 10, 2607–2618. [Google Scholar] [CrossRef]
- Huang, S.; Xue, Y.; Sauer-Eriksson, E.; Chirica, L.; Lindskog, S.; Jonsson, B.H. Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide. J. Mol. Biol. 1998, 283, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Kisker, C.; Schindelin, H.; Alber, B.E.; Ferry, J.G.; Rees, D.C. A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 1996, 15, 2323–2330. [Google Scholar] [CrossRef] [PubMed]
- De Simone, G.; Di Fiore, A.; Capasso, C.; Supuran, C.T. The zinc coordination pattern in the eta-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorganic Med. Chem. Lett. 2015, 25, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Petreni, A.; Carginale, V.; Scaloni, A.; Supuran, C.T.; Capasso, C. Effect of amino acids and amines on the activity of the recombinant iota-carbonic anhydrase from the Gram-negative bacterium Burkholderia territorii. J. Enzym. Inhib. Med. Chem. 2021, 36, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Petreni, A.; Nocentini, A.; Scaloni, A.; Supuran, C.T.; Capasso, C. Effect of Sulfonamides and Their Structurally Related Derivatives on the Activity of iota-Carbonic Anhydrase from Burkholderia territorii. Int. J. Mol. Sci. 2021, 22, 571. [Google Scholar] [CrossRef]
- Petreni, A.; De Luca, V.; Scaloni, A.; Nocentini, A.; Capasso, C.; Supuran, C.T. Anion inhibition studies of the Zn(II)-bound iota-carbonic anhydrase from the Gram-negative bacterium Burkholderia territorii. J. Enzym. Inhib. Med. Chem. 2021, 36, 372–376. [Google Scholar] [CrossRef]
- Supuran, C.T.; Capasso, C. Antibacterial carbonic anhydrase inhibitors: An update on the recent literature. Expert Opin. Ther. Pat. 2020, 30, 963–982. [Google Scholar] [CrossRef]
- Kusian, B.; Sultemeyer, D.; Bowien, B. Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO2 concentrations. J. Bacteriol. 2002, 184, 5018–5026. [Google Scholar] [CrossRef] [PubMed]
- Cronk, J.D.; Endrizzi, J.A.; Cronk, M.R.; O’Neill, J.W.; Zhang, K.Y. Crystal structure of E. coli beta-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci. 2001, 10, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Merlin, C.; Masters, M.; McAteer, S.; Coulson, A. Why is carbonic anhydrase essential to Escherichia coli? J. Bacteriol. 2003, 185, 6415–6424. [Google Scholar] [CrossRef] [PubMed]
- Nishimori, I.; Minakuchi, T.; Maresca, A.; Carta, F.; Scozzafava, A.; Supuran, C.T. The beta-carbonic anhydrases from Mycobacterium tuberculosis as drug targets. Curr. Pharm. Des. 2010, 16, 3300–3309. [Google Scholar] [CrossRef] [PubMed]
- Kohler, S.; Ouahrani-Bettache, S.; Winum, J.Y. Brucella suis carbonic anhydrases and their inhibitors: Towards alternative antibiotics? J. Enzym. Inhib. Med. Chem. 2017, 32, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Supuran, C.T. 3D-QSAR CoMFA studies on sulfonamide inhibitors of the Rv3588c beta-carbonic anhydrase from Mycobacterium tuberculosis and design of not yet synthesized new molecules. J. Enzym. Inhib. Med. Chem. 2014, 29, 449–455. [Google Scholar] [CrossRef]
- Ceruso, M.; Vullo, D.; Scozzafava, A.; Supuran, C.T. Sulfonamides incorporating fluorine and 1,3,5-triazine moieties are effective inhibitors of three beta-class carbonic anhydrases from Mycobacterium tuberculosis. J. Enzym. Inhib. Med. Chem. 2014, 29, 686–689. [Google Scholar] [CrossRef]
- Carta, F.; Maresca, A.; Covarrubias, A.S.; Mowbray, S.L.; Jones, T.A.; Supuran, C.T. Carbonic anhydrase inhibitors. Characterization and inhibition studies of the most active beta-carbonic anhydrase from Mycobacterium tuberculosis, Rv3588c. Bioorganic Med. Chem. Lett. 2009, 19, 6649–6654. [Google Scholar] [CrossRef]
- Modak, J.K.; Tikhomirova, A.; Gorrell, R.J.; Rahman, M.M.; Kotsanas, D.; Korman, T.M.; Garcia-Bustos, J.; Kwok, T.; Ferrero, R.L.; Supuran, C.T.; et al. Anti-Helicobacter pylori activity of ethoxzolamide. J. Enzym. Inhib. Med. Chem. 2019, 34, 1660–1667. [Google Scholar] [CrossRef]
- Ronci, M.; Del Prete, S.; Puca, V.; Carradori, S.; Carginale, V.; Muraro, R.; Mincione, G.; Aceto, A.; Sisto, F.; Supuran, C.T.; et al. Identification and characterization of the alpha-CA in the outer membrane vesicles produced by Helicobacter pylori. J. Enzym. Inhib. Med. Chem. 2019, 34, 189–195. [Google Scholar] [CrossRef]
- Buzas, G.M. Helicobacter pylori—2010. Orv. Hetil. 2010, 151, 2003–2010. [Google Scholar] [CrossRef] [PubMed]
- Abuaita, B.H.; Withey, J.H. Bicarbonate Induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 2009, 77, 4111–4120. [Google Scholar] [CrossRef] [PubMed]
- Rollenhagen, C.; Bumann, D. Salmonella enterica highly expressed genes are disease specific. Infect. Immun. 2006, 74, 1649–1660. [Google Scholar] [CrossRef]
- Nishimori, I.; Minakuchi, T.; Vullo, D.; Scozzafava, A.; Supuran, C.T. Inhibition studies of the beta-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium with sulfonamides and sulfamates. Bioorganic Med. Chem. 2011, 19, 5023–5030. [Google Scholar] [CrossRef]
- Vullo, D.; Nishimori, I.; Minakuchi, T.; Scozzafava, A.; Supuran, C.T. Inhibition studies with anions and small molecules of two novel beta-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium. Bioorganic Med. Chem. Lett. 2011, 21, 3591–3595. [Google Scholar] [CrossRef]
- Lotlikar, S.R.; Kayastha, B.B.; Vullo, D.; Khanam, S.S.; Braga Reygan, E.; Murray, A.B.; McKenna, R.; Supuran, C.T.; Patrauchan, M.A. Pseudomonas aeruginosa β-carbonic anhydrase, psCA1, is required for calcium deposition and contributes to virulence. Cell Calcium. 2019, 84, 102080–102095. [Google Scholar] [CrossRef] [PubMed]
- Guragain, M.; King, M.M.; Williamson, K.S.; Pérez-Osorio, A.C.; Akiyama, T.; Khanam, S.; Patrauchan, M.A.; Franklin, M.J. The Pseudomonas aeruginosa PAO1 Two-Component Regulator CarSR Regulates Calcium Homeostasis and Calcium-Induced Virulence Factor Production through Its Regulatory Targets CarO and CarP. J. Bacteriol. 2016, 198, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin. Drug Discov. 2017, 12, 61–88. [Google Scholar] [CrossRef]
- Supuran, C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016, 473, 2023–2032. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic anhydrase inhibition and the management of neuropathic pain. Expert Rev. Neurother 2016, 16, 961–968. [Google Scholar] [CrossRef]
- Supuran, C.T. Drug interaction considerations in the therapeutic use of carbonic anhydrase inhibitors. Expert Opin. Drug Metab. Toxicol. 2016, 12, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Sulfonamide inhibition profiles of the beta-carbonic anhydrase from the pathogenic bacterium Francisella tularensis responsible of the febrile illness tularemia. Bioorganic Med. Chem. 2017, 25, 3555–3561. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Del Prete, S.; Di Fonzo, P.; Carginale, V.; Donald, W.A.; Supuran, C.T.; Capasso, C. Comparison of the Sulfonamide Inhibition Profiles of the beta- and gamma-Carbonic Anhydrases from the Pathogenic Bacterium Burkholderia pseudomallei. Molecules 2017, 22, 421. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Comparison of the sulfonamide inhibition profiles of the alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorganic Med. Chem. Lett. 2016, 26, 1941–1946. [Google Scholar] [CrossRef]
- Dedeoglu, N.; DeLuca, V.; Isik, S.; Yildirim, H.; Kockar, F.; Capasso, C.; Supuran, C.T. Sulfonamide inhibition study of the beta-class carbonic anhydrase from the caries producing pathogen Streptococcus mutans. Bioorganic Med. Chem. Lett. 2015, 25, 2291–2297. [Google Scholar] [CrossRef]
- Cau, Y.; Mori, M.; Supuran, C.T.; Botta, M. Mycobacterial carbonic anhydrase inhibition with phenolic acids and esters: Kinetic and computational investigations. Org. Biomol. Chem. 2016, 14, 8322–8330. [Google Scholar] [CrossRef]
- Modak, J.K.; Liu, Y.C.; Supuran, C.T.; Roujeinikova, A. Structure-Activity Relationship for Sulfonamide Inhibition of Helicobacter pylori alpha-Carbonic Anhydrase. J. Med. Cheml 2016, 59, 11098–11109. [Google Scholar] [CrossRef]
- Supuran, C.T. Bortezomib inhibits bacterial and fungal beta-carbonic anhydrases. Bioorganic Med. Chem 2016, 24, 4406–4409. [Google Scholar] [CrossRef]
- Supuran, C.T. Legionella pneumophila Carbonic Anhydrases: Underexplored Antibacterial Drug Targets. Pathogens 2016, 5, 44. [Google Scholar] [CrossRef]
- Vullo, D.; Kumar, R.S.S.; Scozzafava, A.; Ferry, J.G.; Supuran, C.T. Sulphonamide inhibition studies of the beta-carbonic anhydrase from the bacterial pathogen Clostridium perfringens. J. Enzym. Inhib. Med. Chem. 2018, 33, 31–36. [Google Scholar] [CrossRef]
- Shahidzadeh, R.; Opekun, A.; Shiotani, A.; Graham, D.Y. Effect of the carbonic anhydrase inhibitor, acetazolamide, on Helicobacter pylori infection in vivo: A pilot study. Helicobacter 2005, 10, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Buzas, G.M. Helicobacter pylori—2021. Orv. Hetil. 2021, 162, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Carginale, V.; Supuran, C.T.; Capasso, C. The gram-negative bacterium Escherichia coli as a model for testing the effect of carbonic anhydrase inhibition on bacterial growth. J. Enzym. Inhib. Med. Chem. 2022, 37, 2092–2098. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Bua, S.; Supuran, C.T.; Capasso, C. Escherichia coli gamma-carbonic anhydrase: Characterisation and effects of simple aromatic/heterocyclic sulphonamide inhibitors. J. Enzym. Inhib. Med. Chem. 2020, 35, 1545–1554. [Google Scholar] [CrossRef]
- Del Prete, S.; De Luca, V.; Bua, S.; Nocentini, A.; Carginale, V.; Supuran, C.T.; Capasso, C. The Effect of Substituted Benzene-Sulfonamides and Clinically Licensed Drugs on the Catalytic Activity of CynT2, a Carbonic Anhydrase Crucial for Escherichia coli Life Cycle. Int. J. Mol. Sci. 2020, 21, 4175. [Google Scholar] [CrossRef]
- Kaur, J.; Cao, X.; Abutaleb, N.S.; Elkashif, A.; Graboski, A.L.; Krabill, A.D.; AbdelKhalek, A.H.; An, W.; Bhardwaj, A.; Seleem, M.N.; et al. Optimization of Acetazolamide-Based Scaffold as Potent Inhibitors of Vancomycin-Resistant Enterococcus. J. Med. Chem. 2020, 63, 9540–9562. [Google Scholar] [CrossRef]
- Abutaleb, N.S.; Elhassanny, A.E.M.; Flaherty, D.P.; Seleem, M.N. In vitro and in vivo activities of the carbonic anhydrase inhibitor, dorzolamide, against vancomycin-resistant enterococci. Peerj 2021, 9, e11059. [Google Scholar] [CrossRef]
- An, W.W.; Holly, K.J.; Nocentini, A.; Imhoff, R.D.; Hewitt, C.S.; Abutaleb, N.S.; Cao, X.F.; Seleem, M.N.; Supuran, C.T.; Flaherty, D.P. Structure-activity relationship studies for inhibitors for vancomycin-resistant Enterococcus and human carbonic anhydrases. J. Enzym. Inhib. Med. Chem. 2022, 37, 1838–1844. [Google Scholar] [CrossRef]
- Abutaleb, N.S.; Elkashif, A.; Flaherty, D.P.; Seleem, M.N. In Vivo Antibacterial Activity of Acetazolamide. Antimicrob. Agents Chemother. 2021, 65, 65–70. [Google Scholar] [CrossRef]
- Abutaleb, N.S.; Elhassanny, A.E.M.; Seleem, M.N. In vivo efficacy of acetazolamide in a mouse model of Neisseria gonorrhoeae infection. Microb. Pathog. 2022, 164, 105454–105458. [Google Scholar] [CrossRef]
- Hewitt, C.S.; Abutaleb, N.S.; Elhassanny, A.E.M.; Nocentini, A.; Cao, X.F.; Amos, D.P.; Youse, M.S.; Holly, K.J.; Marapaka, A.K.; An, W.W.; et al. Structure-Activity Relationship Studies of Acetazolamide-Based Carbonic Anhydrase Inhibitors with Activity against Neisseria gonorrhoeae. ACS Infect. Dis. 2021, 7, 1969–1984. [Google Scholar] [CrossRef] [PubMed]
- Portela, M.B.; Barboza, C.M.; da Silva, E.M.; de Moraes, D.C.; Simão, R.A.; de Souza, C.R.; Cardoso, V.D.S.; Ferreira-Pereira, A.; Vermelho, A.B.; Supuran, C.T. Dentine biomodification by sulphonamides pre-treatment: Bond strength, proteolytic inhibition, and antimicrobial activity. J. Enzym. Inhib. Med. Chem. 2023, 38, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Lomelino, C.L.; Supuran, C.T.; McKenna, R. Non-Classical Inhibition of Carbonic Anhydrase. Int. J. Mol. Sci. 2016, 17, 1150. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).