Abstract
Enterococcus faecalis and E. faecium are the major pathogens causing community- and healthcare-associated infections, with an ability to acquire resistance to multiple antimicrobials. The present study was conducted to determine the prevalence of virulence factors, drug resistance and its genetic determinants, and clonal lineages of E. faecalis and E. faecium clinical isolates in northern Japan. A total of 480 (426 E. faecalis and 54 E. faecium) isolates collected over a four-month period were analyzed. Three virulence factors promoting bacterial colonization (asa1, efaA, and ace) were more prevalent among E. faecalis (46–59%) than E. faecium, while a similar prevalence of enterococcal surface protein gene (esp) was found in these species. Between E. faecalis and E. faecium, an evident difference was noted for resistance to erythromycin, gentamicin, and levofloxacin and its responsible resistance determinants. Oxazolidinone resistance gene optrA and phenicol exporter gene fexA were identified in an isolate of E. faecalis belonging to ST480 and revealed to be located on a cluster similar to those of isolates reported in other Asian countries. The E. faecalis isolates analyzed were differentiated into 12 STs, among which ST179 and ST16 of clonal complex (CC) 16 were the major lineage. Nearly all the E. faecium isolates were assigned into CC17, which consisted of 10 different sequence types (STs), including a dominant ST17 containing multidrug resistant isolates and ST78 with isolates harboring the hyaluronidase gene (hyl). The present study revealed the genetic profiles of E. faecalis and E. faecium clinical isolates, with the first identification of optrA in ST480 E. faecalis in Japan.
1. Introduction
The genus Enterococcus forms the normal flora of the gastrointestinal tract of humans and animals and is associated with an ability to survive and persist in broader environments. Bacterial species of this genus can be opportunistic or nosocomial pathogens causing a wide range of infections in humans, including urinary tract infections, wound infections, intra-abdominal infections, medical device-associated infections, and endocarditis and bloodstream infections [1]. Among Enterococcus spp., E. faecalis and E. faecium are the most prevalent healthcare-associated pathogens worldwide. While they are intrinsically resistant to multiple antimicrobials classes such as cephalosporins, aminoglycosides, lincosamides, and trimethoprim-sulfamethoxazole, enterococcal species have a remarkable ability to acquire new resistance determinants due to the plasticity of their genome [2,3]. Particularly, vancomycin-resistant enterococci (VRE) that acquired van gene (e.g., vanA) clusters has become one of the major nosocomial bacteria worldwide [2,4].
In recent years, in response to the emergence and rapid spread of VRE, importance of alternative antimicrobial, such as linezolid (LZD), has been increasing [5]. LZD is an oxazolidinone that blocks the bacterial protein synthesis by binding to rRNA on both the 30S and 50S ribosomal subunits [6]. It is one of the last resort antimicrobials for the treatment of infections caused by multidrug-resistant Gram-positive bacteria including VRE and methicillin-resistant Staphylococcus aureus (MRSA) [5,7]. However, linezolid resistance has been occurring in Enterococcus, which poses a new public health concern [8]. Resistance to linezolid has been known to be linked with mutations in the 23S rRNA V domain and the rplC/rplD encoding the 50S ribosomal proteins L3/L4, or with the acquisition of optrA or poxtA encoding for an ATP-binding cassette (ABC)-F protein, or cfr encoding 23S rRNA methyltransferase [9]. Among the three resistance genes, optrA is considered to be primarily responsible for the LZD-resistant enterococci in isolates from humans through the protection of the bacterial ribosome, a target of oxazolidinones [9,10].
Pathogenesis of enterococci is attributed to a variety of virulence factors, which can be divided into two groups: secreted virulence factors such as cytolysin (cylA), gelatinase (gelE), and hyaluronidase (hyl) and cell surface virulence factors such as aggregation substances (asa1), enterococcal surface protein (esp), endocarditis antigen (efaA), and collagen-binding protein (ace), providing important role in biofilm formation, adhesion to host cells, and invasion and facilitation of disease progression [11]. Together with antimicrobial resistance, to monitor the prevalence of virulence factors among Enterococcus isolates is significant to understand their clinical impact in a specific region or medical facility, and to consider control measures [12].
In Japan, only limited information is available for the molecular epidemiological characteristics of clinical enterococcal isolates with respect to the prevalence of genotypes, virulence factors, and antimicrobial resistance profile [13,14,15,16]. The prevalence of VRE- and LZD-resistant enterococci appears to be extremely low according to the latest national nosocomial infection surveillance [17]. However, sporadic regional outbreaks of VRE have been reported recently [18,19]. Furthermore, the ratio of oral LZD use to total LZD use increased in the past decade [20], and detection of LZD-resistant Enterococcus due to cfr/optrA or mutation in 23S rRNA in sporadic patients has been reported [21,22,23]. Nevertheless, the prevalence of LZD resistance among clinical isolates remains to be determined.
In the present study, we investigated the prevalence of virulence determinants and antibiotic resistance, and clonal diversity in clinical isolates of E. faecalis and E. faecium in Hokkaido, the northern main island of Japan. With comprehensive information of the bacteriological profiles of enterococcal isolates, we reported here the identification of an LZD-nonsusceptible E. faecalis harboring optrA-fexA, genotyped as ST480 for the first time in Japanese clinical isolates.
2. Results
2.1. Bacterial Isolates
A total of 480 enterococci isolates comprising 426 E. faecalis and 54 E. faecium were analyzed. These isolates were collected consecutively, for a four-month period starting from May 2022 in Sapporo Mirai Laboratory, Co., Ltd., (Sapporo, Japan) where various clinical specimens were submitted from hospitals and clinics in Hokkaido prefecture for microbiological examination. The age range of patients was 8 months to 101 years, while the sex ratio (female/male) was 1.4 (282/198). Sixty-five percent of isolates (311/480) were derived from outpatients. The source of specimens was diverse, with urine being the most common (78.5%, n = 377), followed by vaginal discharge (13.8%, n = 66) and miscellaneous specimens (7.7%, n = 37), including blood (n = 8), pus (n = 7), bile (n = 5), sputum (n = 3), IVH tube (n = 3), wound (n = 2), catheter tip (n = 2), drain fluid (n = 1), urethral stent (n = 1), oral cavity (n = 1), umbilical swab (n = 1), pleural fluid (n = 1), semen (n = 1), and central venous catheter (n = 1). Only one isolate per patient was included in this study.
2.2. Prevalence of Virulence Factors
Three virulence factor genes that promote bacterial colonization (asa1, efaA, and ace) were more prevalent among E. faecalis (46–59%) than E. faecium (Table 1). Enterococcal surface protein gene (esp) was virtually the sole cell surface virulence factor detected in E. faecium, while showing a similar prevalence to E. faecalis. Among extracellularly secreting virulence factors, gelatinase gene (gelE) was the most common in E. faecalis (58%), followed by the cytolysin gene (cylA), whereas in E. faecium, only the hyaluronidase gene (hyl) was detected at low rate (11%).

Table 1.
Prevalence of virulence factors in clinical isolates of E. faecalis (n = 426) and E. faecium (n = 54).
2.3. Prevalence and Profile of Antimicrobial Resistance and Resistance Determinants
The resistance rates to thirteen antimicrobials, resistance determinants, and their profiles of E. faecalis and E. faecium clinical isolates are shown in Table 2 and Table 3. The profiles of antimicrobial resistance and prevalence of resistance determinants were considerably different between E. faecalis and E. faecium. Higher resistance rates and resistance to more antimicrobials were found in E. faecium. Levofloxacin (LVX) resistance was identified in nearly all isolates of E. faecium (96.3%), in contrast to a significantly lower rate (6.8%) in E. faecalis. Although mutations in the quinolone-resistance determining region (QRDR) of both GyrA and ParC were detected in most of the LVX-resistant isolates, mutations in ParC were different between the species. S82R was found in only E. faecium, accounting for 46% of LVX-resistant isolates, while almost all E. faecalis had the S82I mutation. The majority of E. faecium isolates (88.9%) and nearly half of E. faecalis isolates showed resistance to erythromycin (ERY), associated with erm(B) at a higher prevalence in E. faecalis, and msrC in only E. faecium. High-level gentamicin resistance (GEN-HLR) was noted in 13–19% in both species, while the responsible aminoglycoside modifying enzyme (AME) gene, aac(6′)-Ie-aph(2″)-Ia showed a higher detection rate (23%) than GEN-HLR in E. faecalis. Among E. faecalis, 119 isolates (28%) harbored any one or more of the four AME genes, among which copresence of aac(6′)-Ie-aph(2″)-Ia and aph(3′)-IIIa, with or without other AME genes was the most commonly found (60.5%, 72/119).

Table 2.
Prevalence of antimicrobial resistance and resistance genes in clinical isolates of E. faecalis (n = 426) and E. faecium (n = 54).

Table 3.
Profiles of antimicrobial resistance determinants in E. faecalis and E. faecium showing resistance to erythromycin, gentamicin, and levofloxacin.
All the isolates were susceptible to vancomycin (VAN), daptomycin (DAP), and teicoplanin (TEC). However, non-susceptibility to LZD (MIC, 4 mg/L) was detected in one isolate of E. faecalis (ID, ES443), which harbored optrA, representing a detection rate of 0.23% (1/426) in E. faecalis. This isolate also had fexA and showed resistance to chloramphenicol (CHL) and florfenicol (FFC), (MIC, 16 mg/L and 64 mg/L, respectively), while being susceptible to tedizolid (TDZ) (MIC, <0.25 mg/L).
2.4. Characterization of optrA Gene and Its Cluster
Nucleotide sequence of a region surrounding optrA, i.e., optrA cluster comprising approximately 15 kb, was determined for the LZD-nonsusceptible E. faecalis isolate ES443 and compared with the published sequences (Figure 1). The optrA gene sequence was identical to that of the prototype gene reported for pE349 [24]. The sequences of this region containing fexA-optrA and other genes with identical orientations in E. faecalis strains were explored by BLAST search. The whole region of ES443 showed the highest identity to that of E. faecalis strain EFS17 (from pork in South Korea) (99.86%), JF3A-223 (from pig in South Korea) (99.84%), AT40a (from pet food in Switzerland) (99.83%), QZ076 (from chicken in China) (99.69%), SJ82 (from urine of human in Bangladesh), A101 (from feces of human in China) (99.73%), and so forth. Slight sequence diversity to these strains was found in the downstream region of optrA (e.g., RNase J and efrA). In contrast, RNase J gene of ES443 was identical to that of E. faecalis strain AKSZ-211 (from environment in China) and WE0851 (from urine of human in the UK).
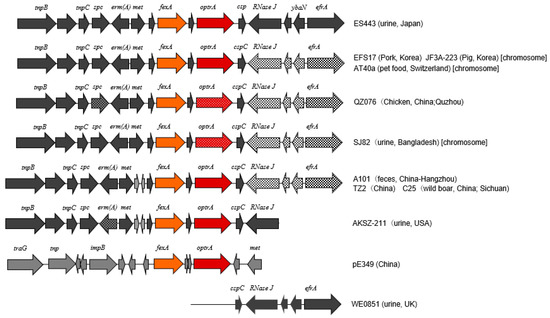
Figure 1.
Schematic representation of the genetic background of optrA in E. faecalis isolate ES443 (uppermost) and the genetic organization or components similar to those of ES443 in other strains reported previously [25] or those available in the GenBank database. Prototype of the fexA–optrA cluster in the pE349 of E. faecalis strain E349 [24] is shown second from the bottom. Arrows indicate the transcription direction of genes. Different textures in arrows denote divergent sequences of genes. Gene names are shown above arrows, and the strain names are indicated on the right.
2.5. Genotypes of Isolates with Different Characteristics
Sequence type (ST) based on multilocus sequence typing (MLST) scheme was determined for all the E. faecium isolates (n = 54) and selected representative E. faecalis isolates (n = 31). E. faecalis isolates, which were derived from various specimens, and those that showed different antimicrobial resistance profiles were chosen for MLST (Table 4 and Table 5). Two novel STs of E. faecalis (ST1296, ST1305) and three novel STs of E. faecium (ST2263, ST2264, ST2267) were identified in the present study.

Table 4.
Genotypes, virulence factors and antimicrobial resistance profile of selected E. faecalis isolates (n = 31).

Table 5.
Genotypes, virulence factors and antimicrobial resistance profile of all the E. faecium isolates (n = 54).
Twelve STs were identified for the E. faecalis isolates analyzed. Among them, STs belonging to clonal complex (CC) 16 were the most common (48%; 11 and 4 isolates of ST179 and ST16, respectively), followed by ST1296 (n = 5), ST28 (n = 2), and ST40 (n = 2) (Table 4). The 12 STs of E. faecalis were phylogenetically diverse and unrelated, except for CC16 (ST16 and ST179), ST480 and ST895, and ST741 and ST1296 (Figure 2a). Most of ST179 E. faecalis isolates (9 among 11 isolates) had 4–6 virulence factors. The prevalence of virulence factors in other STs was variable. Isolates of the prevalent CC16 lineage were generally susceptible to most antimicrobials, except for a few isolates (ES14 and ES116). In contrast, multiple resistance to ERY, LVX, and GEN (high-level) was found in ST4, ST16, and ST1296 isolates. LZD-nonsusceptible isolate ES443 belonged to ST480 and also showed resistance to ERY and LVX, harboring various resistance genes with QRDR mutations and virulence factors asa1, efaA, and esp. In this isolate, cfr was not identified, and no mutation was observed in the 23S rRNA gene (V domain) and L3- and L4-encoding genes. ES443 was derived from urine of an outpatient, while no information was available for administration of LZD for treatment of this patient. Although ST480 was identified in only this isolate, among the STs found in the present study, ST895, a single-locus variant of ST480, was found in an isolate.
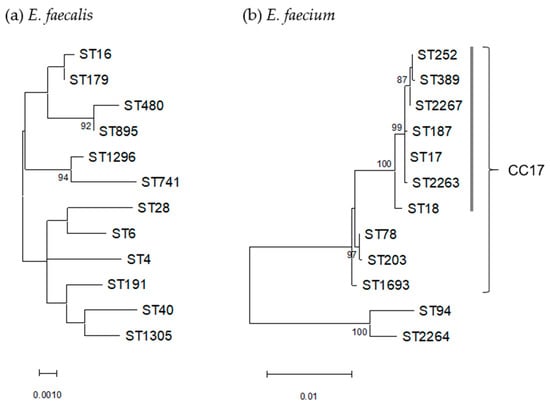
Figure 2.
Phylogenetic trees based on concatenated sequences of seven MLST loci of 12 STs each of E. faecalis (a) and E. faecium (b). Dendrogram was constructed by maximum-likelihood method with the MEGA11 program and statistically supported by bootstrapping with 1000 replicates, and genetic distances were calculated by the Kimura two-parameter model. Variation scale is shown at the bottom. Percent bootstrap support is indicated by the values at each node (the values <80 are omitted). A cluster of E. faecium containing ST17 within CC17 is shown by a vertical bar on the right.
Nearly all the E. faecium (96%, n = 52) were assigned to CC17, which consisted of 10 different STs (ST17, ST18, ST78, ST187, ST203, ST252, ST389, ST1693, ST2263, and ST2267), with ST17 being dominant (59%, n = 32) (Table 5). Among the 10 STs of CC17, two most common types, ST17 and ST78, were genetically distinct, with ST17 forming a main cluster with seven STs (Figure 2b). ST78, a single-locus variant of ST17 (n = 10), contained three isolates positive for the hyaluronidase gene (hyl). In contrast, this gene was not detected in ST17 isolates, while esp was identified in 66% of ST17. Multiple resistance to ERY, LVX, RIF, and GEN (high level) was found in six isolates belonging to ST17 (four isolates), ST252 and ST2267 (one isolate each). In contrast, isolates of non-CC17 lineage (ST94 and ST2264) were susceptible to most of antimicrobials including LVX, without virulence factors being examined.
3. Discussion
The present study revealed the comprehensive status of the antimicrobial resistance, virulence factors, and genotypes of current E. faecalis and E. faecium clinical isolates in northern Japan. Though the prevalence of individual virulence factors in clinical isolates has not yet been sufficiently studied to date, their incidence in Enterococcus species appears to be considerably different depending on origin (human, animal, foodstuff; samples were from infections or healthy individuals) [26,27,28,29,30,31,32,33,34]. The isolates in our study were derived from clinical specimens, mostly from urinary tract infections, and a higher prevalence of asa1 and gelE (approx. 60%) was noted in E. faecalis, with other factors, ace, cylA, esp, being detected in 30–50% of isolates. A similarly high rate of asa1 and gelE was described for isolates from food (fish, milk) and animal [29,32,33], and dominance of gelE was shown for those from infections in humans [30,31] and ruminants [34]. In contrast, ace was ubiquitously distributed to E. faecalis from ocular infections in Japan [30] and those from patients, healthy individuals, and the environment in Italy [27]. A difference in the prevalence of esp depending on country was also shown [31]. Thus, it is suggested that the prevalence of ace and esp in E. faecalis may be diverse by region as well as infection type, in contrast to the universal distribution of asa1 and gelE. On the other hand, E. faecium isolates in our study carried esp (approx. 50%) and hyl (11%) as the main virulence factors, with the absence of ace and gelE. Similarly, esp was the most prevalent in clinical E. faecium isolates in Italy and the UK, while hyl was more common in the UK among VAN-resistant isolates [26]. esp, which was prevalent in almost half of the clinical isolates of both enterococcal species in our study, is a surface protein associated with biofilm formation through amyloid-like aggregation [35,36], and hyl is considered a factor to facilitate intestinal colonization of the bacterial cell involved in the occurrence of infections [37,38]. An increasing prevalence of esp and hyl in E. faecium associated with VAN resistance has been described in European countries [12,39,40], and its global spread is a concern [38]. Therefore, the monitoring of esp and hyl may be of significance for clinical isolates of E. faecalis and E. faecium, though the prevalence of hyl and VAN resistance is still low in Japan.
The prevalence of antimicrobial resistance observed in the present study seems to be comparable to that from national surveillance [17], without detection of isolates resistant to VAN and TEC. Detection rates of GEN-HLR/aac(6′)-Ie-aph(2″)-Ia in E. faecalis (13% and 23%, respectively) were lower than our previous study in northern Japan (1997–2007) [13] and Tokyo (2010) [16], suggesting the decrease in GEN-resistance due to infrequent use of aminoglycosides. A higher proportion of aac(6′)-Ie-aph(2″)-Ia than GEN-HLR is suggested to be ascribable to the low expression level of aac(6′)-Ie-aph(2″)-Ia or the presence of its psedogene [41]. Despite the fact that there have only been a limited numbers of isolates studied, a high prevalence of GEN-HLR and resistance to ERY seems to be persisting in E. faecium [13,14,16]. Though msrC, which encodes the efflux pump of macrolide, was detected in E. faecium at a high rate [14,42], the present study showed a somewhat lower rate (56%), which may suggest that it is not intrinsic to this enterococcal species, as indicated previously [43].
In the present study, a high resistance rate to LVX was noted for E. faecium (96%), as observed in our previous study [15], being significantly higher than E. faecalis (7%). In both species, mutations in QRDR of both GyrA and ParC were detected in most of the LVX-resistant isolates (90% in E. faecalis; 98% in E. faecium), while a lower rate was shown in the previous study (72% in E. faecium) [15]. The occurrence of mutations in both GyrA and ParC have been described as being related to increased MIC, rather than the presence of a single mutation in either of the proteins [15]. Therefore, the present study indicates further progress of quinolone resistance, particularly in E. faecium. In Japan, the proportion of quinolone consumption among all antimicrobial classes is relatively high, with increasing tendency [44], which may be one of the causes spreading quinolone resistance in enterococcus.
In the present study, it was remarkable that an LZD-nonsusceptible E. faecalis isolate harboring optrA was isolated; optrA encodes the ATP-binding cassette (ABC)-F protein, which protects ribosome to confer oxazolidinone resistance [8,45]. Among enterococci, nonsusceptibility to LZD (MIC of 4 mg/L) is prevalent globally at a low rate (<0.38%); a dominant resistance determinant in E. faecalis is optrA [25]. Nevertheless, a remarkably high prevalence of optrA-positive E. faecalis/E. faecium clinical isolates (1–4%) has been noted in China [8,24,46,47,48,49]. In Japan, the prevalence of optrA has not yet been clear, though genomic analysis has been reported for two strains (ST634, ST729) from infected patients [22,23]. In the present study, the prevalence of optrA-positive E. faecalis could be presumed to be 0.2% (1/426), which may be comparable to Austria (0.2%) [50] and Spain (0.7%) [51]. Genotypes (ST) of optrA-positive E. faecalis distributed globally have been classified into various STs, including some major STs, i.e., ST16, ST116, ST256, ST476 ST480, ST766, ST775 [24,48,52,53]. ST480, identified in the present study for isolate ES443, has been described as one of the major optrA-positive clones in China, Germany, and Ireland [24,54,55], and has also been described in many reports in China and Korea [47,56,57,58,59,60], and European and Latin American countries [10,25,51,52,61]. Our present study is the first identification of optrA-positive ST480 E. faecalis in Japan, and thus may indicate its global dissemination.
The OptrA amino acid sequence of isolate ES443 was identical to that of the wild type [24,62], and the nucleotide sequence of the optrA-fexA cluster was similar to chromosomal elements found in isolates distributed to Asian countries (Figure 1). The structure of the optrA-fexA cluster in ES443 was genetically close to those reported in Tn6674 in chromosome [50,63], while it was distinct from that in plasmid [24,56]. Though the medical history of the patient was not available, ES443 was isolated from the urine of an outpatient as a sole pathogen, showing low MIC to LZD; accordingly, this isolate is not likely to be selected by the use of oxazolidinone. Presumably, other antimicrobials such as LVX or ERY might cause the selective persistence of the optrA-harboring strain that might be distributed in the community, because the presence of this gene among healthy populations has been documented [62,64]. Similar views of selection by non-oxazolidinone antibiotics have been described previously for LZD-resistant E. faecalis in China [58].
The E. faecalis isolates analyzed in the present study belonged to various STs, among which the dominant ST16 and ST179 (CC16) were also common among isolates with GEN-HLR in Japan [16]. A newly identified ST1296 was the second most common, following CC16, and comprised heterologous strains, including an isolate (ES94) with multiple virulence factors and drug resistance. Furthermore, two isolates in our study were identified as ST28, which had multiple virulence factors. This ST had been referred to as a high-risk multidrug resistant strain with a potential public health concern in India [65]. Accordingly, the prevalence of ST1296 and ST28, as well as CC16 E. faecalis, should be carefully monitored in Japan.
In contrast, the E. faecium isolates in the present study were substantially homogenous, belonging to CC17, which has been known as being responsible for hospital-associated infections, acquiring antimicrobial resistance [66]. Though ST17 was dominant in CC17 in our study, hyl was not detected in ST17, but it is commonly present in ST78 (3 positives among 10 isolates). ST78, a single-locus variant of ST17, is described as one of the main genotypes of VRE, particularly those with VanA type in Germany and China [67,68], posing a potential to emerge as a successful clone. Although VanA type VRE is still rare in Japan, attention should be paid to the prevalence and antimicrobial resistance of ST78 E. faecium.
The present study revealed the antimicrobial resistance and genetic traits of E. faecalis and E. faecium that are relevant to potential public health concerns in northern Japan. The obtained findings will contribute to the focus on the important points for further epidemiological surveillance and infection control measures.
4. Materials and Methods
4.1. Clinical Isolates and Species Identification
Clinical specimens submitted to Sapporo Mirai Laboratory, Co., Ltd. were initially cultured on Sheep Blood Agar plates (Nissui Pharmaceutical, Co., Tokyo, Japan), and occasionally on Columbia CA Sheep Blood Agar plates (Kohjin Bio, Co., Tokyo, Japan) to promote bacterial growth. Species identification was performed by MALDI-TOF mass spectrometry using MALDI Biotyper (BRUKER). All the isolates were confirmed as E. faecalis and E. faecium by the PCR targeting species-specific sequence of PBP5 genes, as described previously [69]. For some isolates that could not be identified by the PCR, the species was confirmed by the determination of the 16S rRNA gene sequence through direct sequencing with the PCR product amplified by specific primers [70]. Individual isolates were stored in Microbank (Pro-Lab Diagnostics, Richmond Hill, ON, Canada) at −80 °C, and were recovered when they were analyzed.
4.2. Antibiotic Susceptibility Testing
Susceptibility to penicillin (PEN), ampicillin (AMP), ampicillin-sulbactam (SAM), imipenem (IPM), high level gentamicin resistance (GEN-HLR), minocycline (MIN), erythromycin (ERY), levofloxacin (LVX), linezolid (LZD), rifampicin (RIF), daptomycin (DAP), teicoplanin (TEC), and vancomycin (VAN) was measured by broth microdilution test, using Dry Plate Eiken DP42 (Eiken, Tokyo, Japan). Antimicrobials and their concentrations (mg/L) used were as follows: PEN (0.12–8), AMP (0.25–8), SAM (1/2–8/16), IPM (0.25–8), GEN-HLR (500), MIN (1–8), ERY (0.25–4), LVX (0.25–4), LZD (0.5–4), RIF (0.5–2), DAP (0.25–4), TEC (0.5–16), VAN (0.5–16). For the LZD non-susceptible isolate (ES443), the MIC of chloramphenicol (CHL), florfenicol (FFC), and tedizolid (TDZ) were determined by the broth microdilution method. Susceptibility/resistance was judged according to the break points mentioned in the CLSI and EUCAST guidelines [71,72]. For CHL and FFC, the MIC breakpoints for susceptibility interpretation was performed as described previously [73].
4.3. Detection of Virulence Factors Genes
For all the isolates, the following virulence factor genes were detected by PCR using previously reported primers and conditions [26,27]: aggregation substance (asa1), collagen-binding protein (ace), virulence factor associated with infective endocarditis (efaA), enterococcal surface protein associated with biofilm production (esp), gelatinase (gelE), cytolysin (cylA), and hyaluronidase (hyl).
4.4. Detection of Drug Resistance Genes
The presence of the following drug resistance genes was examined by uniplex or multiplex PCR assays by primers and conditions, as described previously [41,74,75]: beta-lactamase gene, blaZ; aminoglycoside modifying enzymes (AME) genes, aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, ant(4′)-Ia, aph(2″)-Id/Ie, and ant(9)-Ia; macrolide resistance genes, erm(A), erm(B), erm(C), erm(T), msrA, msrB, and msrC; lincosamide resistance genes, lnuA, lnuB, lsaA, and mefA/E; vancomycin resistance genes, vanA, vanB, vanD, and vanM; oxazolidinone and phenicol resistance gene, optrA, poxtA, and cfr; and the phenicol exporter gene, fexA. Nucleotide sequences of the quinolone resistance-determining region (QRDR) of GyrA and ParC were determined by PCR and direct sequencing to detect mutations [15].
4.5. Genetic Determinants of Oxazolidinone Resistance Isolate
One isolate (ES443) exhibiting non-susceptibility to linezolid (MIC = 4 mg/L) was further analyzed for the mutation in 23S rRNA and L3- and L4-encoding genes, as described previously [41]. The nucleotide sequence of the fexA–optrA gene cluster was determined by PCR and direct sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an automated DNA sequencer (ABI PRISM 3100). The primers used for sequencing are shown in Table S1 (Supplementary Materials). The multiple alignment of nucleotide/amino acid sequences determined in the present study and those retrieved from the GenBank database was performed by Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 10 December 2022), which was also used for the calculation of sequence identity.
4.6. Multilocus Sequence Typing (MLST), Phylogenetic Analysis
For all the E. faecium isolates (n = 54) and selected E. faecalis isolates (n = 31) having different drug resistance profiles and derived from various specimen, the sequence type (ST) based on the MLST schemes [76,77] were identified using the web-based genotyping tool PubMLST (https://pubmlst.org/efaecium/, accessed on 31 October 2022) and (https://pubmlst.org/efaecalis/, accessed on 31 October 2022), respectively. The MLST data were further assigned to the clonal complex (CC) by BURST analysis available in the PubMLST website. To analyze the genetic relatedness of STs identified for E. faecalis and E. faecium, MEGA11 software (https://megasoftware.net/home, accessed on 28 December 2022) was used to construct the phylogenetic dendrograms of concatenated sequences of seven MLST loci.
4.7. GenBank Accession Number
The nucleotide sequences of a genetic cluster, including fexA–optrA, was deposited in the GenBank database under the accession number OP795985.
4.8. Statistical Analysis
The difference in the prevalence of virulence factors and antimicrobial resistance/resistance determinants between E. faecalis and E. faecium was statistically analyzed by Fisher′s exact test using the js-STAR XR ver.1.1.9 software (https://www.kisnet.or.jp/nappa/software/star/index.htm, accessed on 31 December 2022). A p-value < 0.01 was considered statistically significant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12010108/s1, Table S1: Primers used for sequencing of optrA-fexA cluster identified in this study.
Author Contributions
Conceptualization, M.S.A. and N.K.; methodology, M.S.A. and N.K.; investigation, M.S.A. and N.U.; data curation, M.S.A. and N.K.; resources, M.K., M.H., N.O., K.K., N.T. and M.I.; writing—original draft preparation, M.S.A.; writing—review and editing, M.S.A. and N.K.; supervision and project administration, N.K.; funding acquisition, M.S.A. and N.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS (Japan Society for the Promotion of Science) KAKENHI Grant Number JP20H03933, and JP21K10401.
Institutional Review Board Statement
Ethical review and approval were waived for this study because no human participants were directly involved. We analyzed isolates that had been routinely cultured from clinical specimens from hospitals and clinics sent to the Sapporo Mirai Laboratory, Co., Ltd., Hokkaido, Japan.
Informed Consent Statement
All the patients′ data were kept anonymous and patient consent was waived in this study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gilmore, M.S.; Clewell, D.B.; Ike, Y.; Shankar, N. (Eds.) Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–569. [Google Scholar] [CrossRef] [PubMed]
- Sparo, M.; Delpech, G.; Allende, N.G. Impact on Public Health of the Spread of High-Level Resistance to Gentamicin and Vancomycin in Enterococci. Front. Microbiol. 2018, 9, 3073. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.; Coque, T.M.; Hammerum, A.M.; Hope, R.; Hryniewicz, W.; Johnson, A.; Klare, I.; Kristinsson, K.G.; Leclercq, R.; Lester, C.H.; et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Eurosurveillance 2008, 13, 19046. [Google Scholar] [CrossRef] [PubMed]
- Ament, P.W.; Jamshed, N.; Horne, J.P. Linezolid: Its role in the treatment of gram-positive, drug-resistant bacterial infections. Am. Fam. Physician 2002, 65, 663. [Google Scholar] [PubMed]
- Hashemian, S.M.; Farhadi, T.; Ganjparvar, M. Linezolid: A review of its properties, function, and use in critical care. Drug Des. Dev. Ther. 2018, 12, 1759–1767. [Google Scholar] [CrossRef]
- Lentino, J.R.; Narita, M.; Yu, V.L. New antimicrobial agents as therapy for resistant gram-positive cocci. Eur. J. Clin. Microbiol. 2007, 27, 3–15. [Google Scholar] [CrossRef]
- Bi, R.; Qin, T.; Fan, W.; Ma, P.; Gu, B. The emerging problem of linezolid-resistant enterococci. J. Glob. Antimicrob. Resist. 2018, 13, 11–19. [Google Scholar] [CrossRef]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Updat. 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Freitas, A.R.; Tedim, A.P.; Novais, C.; Lanza, V.F.; Peixe, L. Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb. Genom. 2020, 6, e000350. [Google Scholar] [CrossRef]
- Geraldes, C.; Tavares, L.; Gil, S.; Oliveira, M. Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment. Antibiotics 2022, 11, 857. [Google Scholar] [CrossRef]
- Billström, H.; Lund, B.; Sullivan, A.; Nord, C.E. Virulence and antimicrobial resistance in clinical Enterococcus faecium. Int. J. Antimicrob. Agents 2008, 32, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Kobayashi, N.; Quiñones, D.; Nagashima, S.; Uehara, N.; Watanabe, N. Genetic diversity of enterococci harboring the high-level gentamicin resistance gene aac(6′)-Ie-aph(2″)-Ia or aph(2″)-Ie in a Japanese hospital. Microb. Drug Resist. 2009, 15, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Isogai, N.; Urushibara, N.; Kawaguchiya, M.; Ghosh, S.; Suzaki, K.; Watanabe, N.; Quiñones, D.; Kobayashi, N. Characterization of Enterococcus faecium with macrolide resistance and reduced susceptibility to quinupristin/dalfopristin in a Japanese hospital: Detection of extensive diversity in erm(B)-regulator regions. Microb. Drug Resist. 2013, 19, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Urushibara, N.; Suzaki, K.; Kawaguchiya, M.; Aung, M.S.; Shinagawa, M.; Takahashi, S.; Kobayashi, N. Contribution of Type II Topoisomerase Mutations to Fluoroquinolone Resistance in Enterococcus faecium from Japanese Clinical Setting. Microb. Drug Resist. 2018, 24, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Harada, S.; Shibue, Y.; Aoki, K.; Ishii, Y.; Tateda, K. Prevalence of High-Level Aminoglycoside Resistance and Genes Encoding Aminoglycoside-Modifying Enzymes in Enterococcus faecalis and Enterococcus faecium Isolated in a University Hospital in Tokyo. Jpn. J. Infect. Dis. 2020, 73, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Japan Nosocomial Infections Surveillance (JANIS). Annual Open Report 2019 (All Facilities). Clinical Laboratory Division. Available online: https://janis.mhlw.go.jp/english/report/open_report/2019/3/1/ken_Open_Report_Eng_201900_clsi2012.pdf (accessed on 20 December 2022).
- Matsushima, A.; Takakura, S.; Yamamoto, M.; Matsumura, Y.; Shirano, M.; Nagao, M.; Ito, Y.; Iinuma, Y.; Shimizu, T.; Fujita, N.; et al. Regional spread and control of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis in Kyoto, Japan. Eur. J. Clin. Microbiol. 2011, 31, 1095–1100. [Google Scholar] [CrossRef]
- Saito, N.; Kitazawa, J.; Horiuchi, H.; Yamamoto, T.; Kimura, M.; Inoue, F.; Matsui, M.; Minakawa, S.; Itoga, M.; Tsuchiya, J.; et al. Interhospital transmission of vancomycin-resistant Enterococcus faecium in Aomori, Japan. Antimicrob. Resist. Infect. Control 2022, 11, 99. [Google Scholar] [CrossRef]
- Goto, R.; Inose, R.; Kusama, Y.; Kawabe, A.; Ishii, S.; Ebisui, A.; Ishikane, M.; Yagi, T.; Ohmagari, N.; Muraki, Y. Trends of the Use of Anti-methicillin-Resistant Staphylococcus aureus Agents in Japan Based on Sales Data from 2006 to 2015. Biol. Pharm. Bull. 2020, 43, 1906–1910. [Google Scholar] [CrossRef]
- Nihonyanagi, S.; Adachi, Y.; Onuki, T.; Nakazaki, N.; Hirata, Y.; Fujiki, K.; Takayama, Y.; Kanoh, Y.; Bandoh, Y.; Dantsuji, Y.; et al. Emergence of linezolid-resistant Enterococcus faecalis strains from two inpatients in a pediatric ward. Kansenshogaku Zasshi 2012, 86, 555–562. [Google Scholar] [CrossRef]
- Kuroda, M.; Sekizuka, T.; Matsui, H.; Suzuki, K.; Seki, H.; Saito, M.; Hanaki, H. Complete Genome Sequence and Characterization of Linezolid-Resistant Enterococcus faecalis Clinical Isolate KUB3006 Carrying a cfr(B)-Transposon on Its Chromosome and optrA-Plasmid. Front. Microbiol. 2018, 9, 2576. [Google Scholar] [CrossRef]
- Iimura, M.; Hayashi, W.; Arai, E.; Natori, T.; Horiuchi, K.; Matsumoto, G.; Tanaka, H.; Soga, E.; Nagano, Y.; Arakawa, Y.; et al. Identification of a multiresistant mosaic plasmid carrying a new segment of IS1216E-flanked optrA with integrated Tn551-ermB element in linezolid-resistant Enterococcus faecalis human isolate. J. Glob. Antimicrob. Resist. 2020, 22, 697–699. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, L.M.; Castanheira, M.; Flamm, R.K.; Mendes, R.E. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: Results from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 2018, 73, 2314–2322. [Google Scholar] [CrossRef] [PubMed]
- Vankerckhoven, V.; Van Autgaerden, T.; Vael, C.; Lammens, C.; Chapelle, S.; Rossi, R.; Jabes, D.; Goossens, H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 2004, 42, 4473–4479. [Google Scholar] [CrossRef]
- Creti, R.; Imperi, M.; Bertuccini, L.; Fabretti, F.; Orefici, G.; Di Rosa, R.; Baldassarri, L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 2004, 53, 13–20. [Google Scholar] [CrossRef]
- Mannu, L.; Paba, A.; Daga, E.; Comunian, R.; Zanetti, S.; Duprè, I.; Sechi, L. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int. J. Food Microbiol. 2003, 88, 291–304. [Google Scholar] [CrossRef]
- Hammad, A.M.; Shimamoto, T.; Shimamoto, T. Genetic characterization of antibiotic resistance and virulence factors in Enterococcus spp. from Japanese retail ready-to-eat raw fish. Food Microbiol. 2014, 38, 62–66. [Google Scholar] [CrossRef]
- Todokoro, D.; Suzuki, T.; Kobayakawa, S.; Tomita, H.; Ohashi, Y.; Akiyama, H. Postoperative Enterococcus faecalis endophthalmitis: Virulence factors leading to poor visual outcome. Jpn. J. Ophthalmol. 2017, 61, 408–414. [Google Scholar] [CrossRef]
- Lins, R.X.; Junior, H.R.; Wilson, M.; Lewis, M.A.O.; Fidel, R.A.S.; Williams, D. Comparison of genotypes, antimicrobial resistance and virulence profiles of oral and non oral Enterococcus faecalis from Brazil, Japan and the United Kingdom. J. Dent. 2019, 84, 49–54. [Google Scholar] [CrossRef]
- El-Zamkan, M.A.; Mohamed, H.M.A. Antimicrobial resistance, virulence genes and biofilm formation in Enterococcus species isolated from milk of sheep and goat with subclinical mastitis. PLoS ONE 2021, 16, e0259584. [Google Scholar] [CrossRef]
- Zarzecka, U.; Zadernowska, A.; Chajęcka-Wierzchowska, W. Effects of osmotic and high pressure stress on expression of virulence factors among Enterococcus spp. isolated from food of animal origin. Food Microbiol. 2022, 102, 103900. [Google Scholar] [CrossRef] [PubMed]
- Smoglica, C.; Vergara, A.; Angelucci, S.; Festino, A.R.; Antonucci, A.; Marsilio, F.; Di Francesco, C.E. Evidence of Linezolid Resistance and Virulence Factors in Enterococcus spp. Isolates from Wild and Domestic Ruminants, Italy. Antibiotics 2022, 11, 223. [Google Scholar] [CrossRef] [PubMed]
- Tendolkar, P.M.; Baghdayan, A.S.; Gilmore, M.S.; Shankar, N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 2004, 72, 6032–6039. [Google Scholar] [CrossRef]
- Taglialegna, A.; Matilla-Cuenca, L.; Dorado-Morales, P.; Navarro, S.; Ventura, S.; Garnett, J.A.; Lasa, I.; Valle, J. The biofilm-associated surface protein Esp of Enterococcus faecalis forms amyloid-like fibers. NPJ Biofilms Microbiomes 2020, 6, 15. [Google Scholar] [CrossRef]
- Rice, L.B.; Carias, L.; Rudin, S.; Vael, C.; Goossens, H.; Konstabel, C.; Klare, I.; Nallapareddy, S.R.; Huang, W.; Murray, B.E. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 2003, 187, 508–512. [Google Scholar] [CrossRef]
- Freitas, A.R.; Tedim, A.P.; Novais, C.; Ruiz-Garbajosa, P.; Werner, G.; Laverde-Gomez, J.A.; Cantón, R.; Peixe, L.; Baquero, F.; Coque, T.M. Global spread of the hylEfm colonization-virulence gene in megaplasmids of the Enterococcus faecium CC17 polyclonal subcluster. Antimicrob. Agents Chemother. 2010, 54, 2660–2665. [Google Scholar] [CrossRef] [PubMed]
- Klare, I.; Konstabel, C.; Mueller-Bertling, S.; Werner, G.; Strommenger, B.; Kettlitz, C.; Borgmann, S.; Schulte, B.; Jonas, D.; Serr, A.; et al. Spread of ampicillin/vancomycin-resistant Enterococcus faecium of the epidemic-virulent clonal complex-17 carrying the genes esp and hyl in German hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Mato, R.; Almeida, F.; Pires, R.; Rodrigues, P.; Ferreira, T.; Sanches, I.S. Assessment of high-level gentamicin and glycopeptide-resistant Enterococcus faecalis and E. faecium clonal structure in a Portuguese hospital over a 3-year period. Eur. J. Clin. Microbiol. 2009, 28, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Aung, M.S.; Paul, S.K.; Ahmed, S.; Haque, N.; Khan, E.R.; Barman, T.K.; Islam, A.; Abedin, S.; Sultana, C.; et al. Drug Resistance Determinants in Clinical Isolates of Enterococcus faecalis in Bangladesh: Identification of Oxazolidinone Resistance Gene optrA in ST59 and ST902 Lineages. Microorganisms 2020, 8, 1240. [Google Scholar] [CrossRef]
- Singh, K.V.; Malathum, K.; Murray, B.E. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, Is associated with an increase in macrolide susceptibility. Antimicrob. Agents Chemother. 2001, 45, 263–266. [Google Scholar] [CrossRef]
- Werner, G.; Hildebrandt, B.; Witte, W. The newly described msrC gene is not equally distributed among All Isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 2001, 45, 3672–3673. [Google Scholar] [CrossRef] [PubMed]
- Muraki, Y.; Yagi, T.; Tsuji, Y.; Nishimura, N.; Tanabe, M.; Niwa, T.; Watanabe, T.; Fujimoto, S.; Takayama, K.; Murakami, N.; et al. Japanese antimicrobial consumption surveillance: First report on oral and parenteral antimicrobial consumption in Japan (2009–2013). J. Glob. Antimicrob. Resist. 2016, 7, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Zhang, W.; Du, X.-D.; Krüger, H.; Feßler, A.T.; Ma, S.; Zhu, Y.; Wu, C.; Shen, J.; Wang, Y. Mobile Oxazolidinone Resistance Genes in Gram-Positive and Gram-Negative Bacteria. Clin. Microbiol. Rev. 2021, 34, e00188-20. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Zou, J.; Zhao, J.; Tang, Y.; Yuan, Y.; Yang, B.; Huang, J.; Xia, P.; Xia, Y. Emergence of optrA-Mediated Linezolid Resistance in Enterococcus faecium: A Molecular Investigation in a Tertiary Hospital of Southwest China from 2014–2018. Infect. Drug Resist. 2022, 15, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Wang, Y.; Schwarz, S.; Lv, H.; Li, Y.; Liao, K.; Yu, S.; Zhao, K.; Gu, D.; Wang, X.; et al. Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010–2014. Clin. Microbiol. Infect. 2015, 21, 1095.e1–1095.e4. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, Y.; Lv, Y.; Wang, S.; Song, Y.; Li, Y.; Liu, J.; Xue, F.; Yang, W.; Zhang, J. Nationwide Surveillance of Novel Oxazolidinone Resistance Gene optrA in Enterococcus Isolates in China from 2004 to 2014. Antimicrob. Agents Chemother. 2016, 60, 7490–7493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dong, G.; Li, J.; Chen, L.; Liu, H.; Bi, W.; Lu, H.; Zhou, T. A high incidence and coexistence of multiresistance genes cfr and optrA among linezolid-resistant enterococci isolated from a teaching hospital in Wenzhou, China. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1441–1448. [Google Scholar] [CrossRef]
- Kerschner, H.; Rosel, A.C.; Hartl, R.; Hyden, P.; Stoeger, A.; Ruppitsch, W.; Allerberger, F.; Apfalter, P. Oxazolidinone Resistance Mediated by optrA in Clinical Enterococcus faecalis Isolates in Upper Austria: First Report and Characterization by Whole Genome Sequencing. Microb. Drug Resist. 2021, 27, 685–690. [Google Scholar] [CrossRef]
- Rodríguez-Lucas, C.; Fernández, J.; Vázquez, X.; de Toro, M.; Ladero, V.; Fuster, C.; Rodicio, R.; Rodicio, M.R. Detection of the optrA Gene Among Polyclonal Linezolid-Susceptible Isolates of Enterococcus faecalis Recovered from Community Patients. Microb. Drug Resist. 2022, 28. [Google Scholar] [CrossRef]
- Mendes, R.E.; Deshpande, L.; Streit, J.M.; Sader, H.S.; Castanheira, M.; Hogan, P.A.; Flamm, R.K. ZAAPS programme results for 2016: An activity and spectrum analysis of linezolid using clinical isolates from medical centres in 42 countries. J. Antimicrob. Chemother. 2018, 73, 1880–1887. [Google Scholar] [CrossRef]
- Chen, M.; Pan, H.; Lou, Y.; Wu, Z.; Zhang, J.; Huang, Y.; Yu, W.; Qiu, Y. Epidemiological characteristics and genetic structure of linezolid-resistant Enterococcus faecalis. Infect. Drug Resist. 2018, 11, 2397–2409. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.K.; Fleige, C.; Lange, D.; Klare, I.; Werner, G. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int. J. Antimicrob. Agents 2018, 52, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Egan, S.; Shore, A.C.; O′Connell, B.; Brennan, G.I.; Coleman, D.C. Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: High prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J. Antimicrob. Chemother. 2020, 75, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Shen, Y.; Schwarz, S.; Cai, J.; Lv, Y.; Li, J.; Feßler, A.T.; Zhang, R.; Wu, C.; Shen, J.; et al. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 2016, 71, 1466–1473. [Google Scholar] [CrossRef]
- Zhou, W.; Gao, S.; Xu, H.; Zhang, Z.; Chen, F.; Shen, H.; Zhang, C. Distribution of the optrA gene in Enterococcus isolates at a tertiary care hospital in China. J. Glob. Antimicrob. Resist. 2019, 17, 180–186. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Yin, Y.; Li, S.; Zhang, Y.; Wang, Q.; Wang, H. Molecular characteristics of oxazolidinone resistance in enterococci from a multicenter study in China. BMC Microbiol. 2019, 19, 162. [Google Scholar] [CrossRef]
- Park, K.; Jeong, Y.S.; Chang, J.; Sung, H.; Kim, M.N. Emergence of optrA-Mediated Linezolid-Nonsusceptible Enterococcus faecalis in a Tertiary Care Hospital. Ann. Lab. Med. 2020, 40, 321–325. [Google Scholar] [CrossRef]
- Morroni, G.; Brenciani, A.; Simoni, S.; Vignaroli, C.; Mingoia, M.; Giovanetti, E. Commentary: Nationwide Surveillance of Novel Oxazolidinone Resistance Gene optrA in Enterococcus Isolates in China from 2004 to 2014. Front. Microbiol. 2017, 8, 1631. [Google Scholar] [CrossRef]
- Argudín, M.A.; Youzaga, S.; Dodémont, M.; Heinrichs, A.; Roisin, S.; Deplano, A.; Nonhoff, C.; Hallin, M. Detection of optrA-positive enterococci clinical isolates in Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 985–987. [Google Scholar] [CrossRef]
- Cai, J.; Schwarz, S.; Chi, D.; Wang, Z.; Zhang, R.; Wang, Y. Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin. Microbiol. Infect. 2019, 25, 630.e1–630.e6. [Google Scholar] [CrossRef]
- Elghaieb, H.; Tedim, A.P.; Abbassi, M.S.; Novais, C.; Duarte, B.; Hassen, A.; Peixe, L.; Freitas, A.R. From farm to fork: Identical clones and Tn6674-like elements in linezolid-resistant Enterococcus faecalis from food-producing animals and retail meat. J. Antimicrob. Chemother. 2019, 75, 30–35. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.; Biggel, M.; Zurfluh, K.; Treier, A.; Stephan, R. Faecal carriage of enterococci harbouring oxazolidinone resistance genes among healthy humans in the community in Switzerland. J. Antimicrob. Chemother. 2022, 77, 2779–2783. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.; Dhawan, B.; Vishnubhatla, S.; Kapil, A.; Das, B.; Sood, S. Emergence of high-risk multidrug-resistant Enterococcus faecalis CC2 (ST181) and CC87 (ST28) causing healthcare-associated infections in India. Infect. Genet. Evol. 2020, 85, 104519. [Google Scholar] [CrossRef]
- Lee, T.; Pang, S.; Abraham, S.; Coombs, G.W. Antimicrobial-resistant CC17 Enterococcus faecium: The past, the present and the future. J. Glob. Antimicrob. Resist. 2018, 16, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Eisenberger, D.; Tuschak, C.; Werner, M.; Bogdan, C.; Bollinger, T.; Hossain, H.; Friedrich, P.; Hussein, Z.; Pöhlmann, C.; Würstl, B.; et al. Whole-genome analysis of vancomycin-resistant Enterococcus faecium causing nosocomial outbreaks suggests the occurrence of few endemic clonal lineages in Bavaria, Germany. J. Antimicrob. Chemother. 2020, 75, 1398–1404. [Google Scholar] [CrossRef]
- Sun, L.; Xu, J.; Wang, W.; He, F. Emergence of vanA-Type Vancomycin-Resistant Enterococcus faecium ST 78 Strain with a rep2-Type Plasmid Carrying a Tn1546-Like Element Isolated from a Urinary Tract Infection in China. Infect. Drug Resist. 2020, 13, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Alam, M.; Nishimoto, Y.; Urasawa, S.; Uehara, N.; Watanabe, N. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol. Infect. 2001, 126, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Mahbub Alam, M.; Kobayashi, N.; Ishino, M.; Sumi, A.; Kobayashi, K.; Uehara, N.; Watanabe, N. Detection of a novel aph(2″) allele (aph [2″]-Ie) conferring high-level gentamicin resistance and a spectinomycin resistance gene ant(9)-Ia (aad 9) in clinical isolates of enterococci. Microb. Drug Resist. 2005, 11, 239–247. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; M100-Ed32; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. 2021. Available online: http://www.eucast.org (accessed on 23 June 2022).
- Tamang, M.D.; Moon, D.C.; Kim, S.-R.; Kang, H.Y.; Lee, K.; Nam, H.-M.; Jang, G.-C.; Lee, H.-S.; Jung, S.-C.; Lim, S.-K. Detection of novel oxazolidinone and phenicol resistance gene optrA in enterococcal isolates from food animals and animal carcasses. Vet. Microbiol. 2017, 201, 252–256. [Google Scholar] [CrossRef]
- Aung, M.; Urushibara, N.; Kawaguchiya, M.; Hirose, M.; Ike, M.; Ito, M.; Kobayashi, N. Distribution of Virulence Factors and Resistance Determinants in Three Genotypes of Staphylococcus argenteus Clinical Isolates in Japan. Pathogens 2021, 10, 163. [Google Scholar] [CrossRef]
- Nomura, T.; Hashimoto, Y.; Kurushima, J.; Hirakawa, H.; Tanimoto, K.; Zheng, B.; Ruan, G.; Xue, F.; Liu, J.; Hisatsune, J.; et al. New colony multiplex PCR assays for the detection and discrimination of vancomycin-resistant enterococcal species. J. Microbiol. Methods 2018, 145, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Garbajosa, P.; Bonten, M.J.M.; Robinson, D.A.; Top, J.; Nallapareddy, S.R.; Torres, C.; Coque, T.M.; Cantón, R.; Baquero, F.; Murray, B.E.; et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 2006, 44, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Homan, W.L.; Tribe, D.; Poznanski, S.; Li, M.; Hogg, G.; Spalburg, E.; Van Embden, J.D.; Willems, R.J. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 2002, 40, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).