Anti-Biofilm and Antibacterial Activities of Cycas media R. Br Secondary Metabolites: In Silico, In Vitro, and In Vivo Approaches
Abstract
:1. Introduction
2. Materials and Methods
2.1. General
2.2. Plant, Extraction, and Isolation
2.3. Bacterial Isolates
2.4. Animals
2.5. Molecular Docking
2.6. In Vitro Antibacterial Activity
2.6.1. Antibiotic Susceptibility Profile of the Tested Isolates
2.6.2. Susceptibility Testing of GINK and SOTE
2.6.3. Impact of SOTE on the Membrane Depolarization
2.6.4. Studying the Effect of SOTE on the Bacterial Morphology
2.6.5. Anti-Biofilm Activity of SOTE
2.6.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.7. In Vivo Antibacterial Activity
2.7.1. Experimental Model
2.7.2. Liver Function Tests
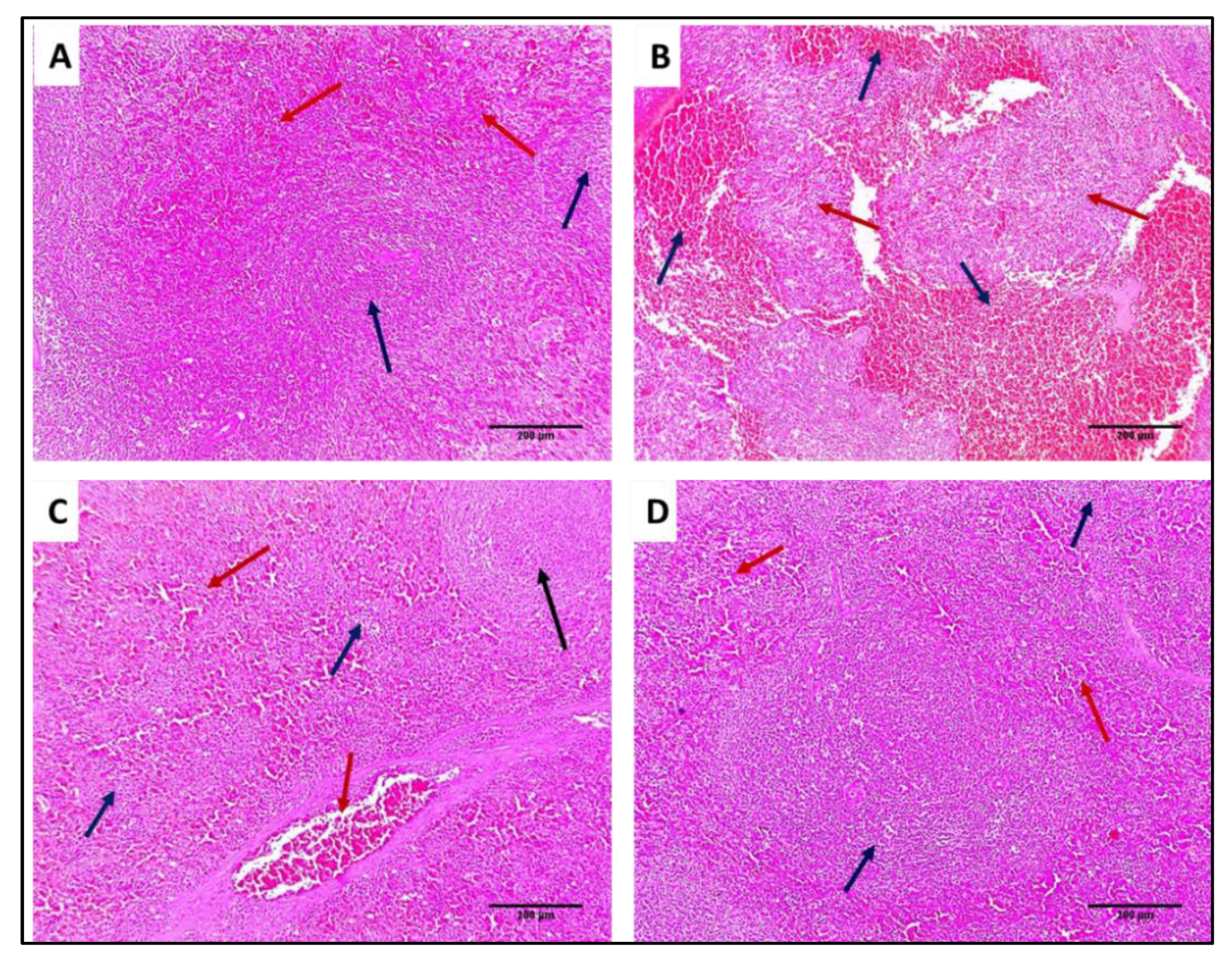
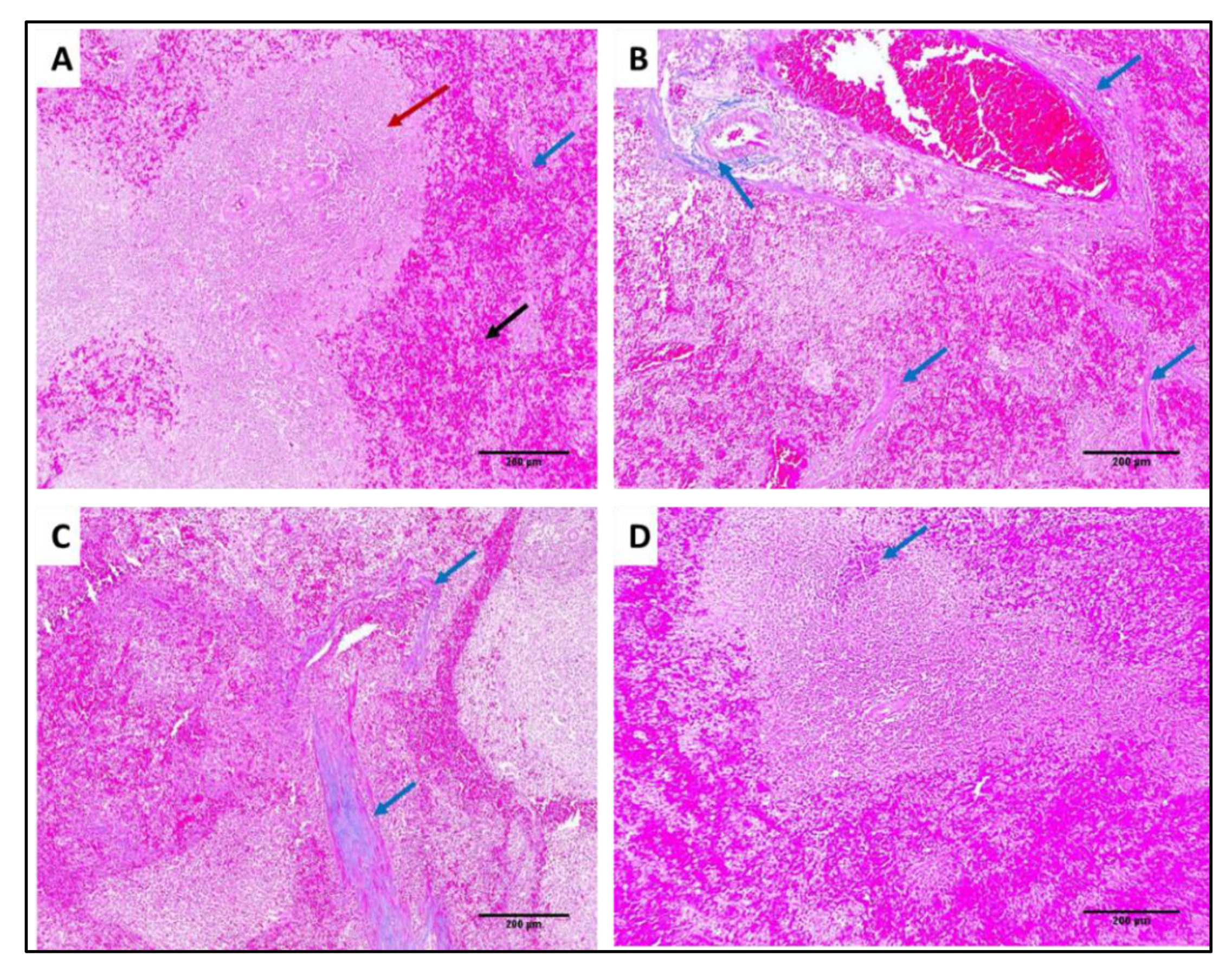
2.7.3. Histological Assessment
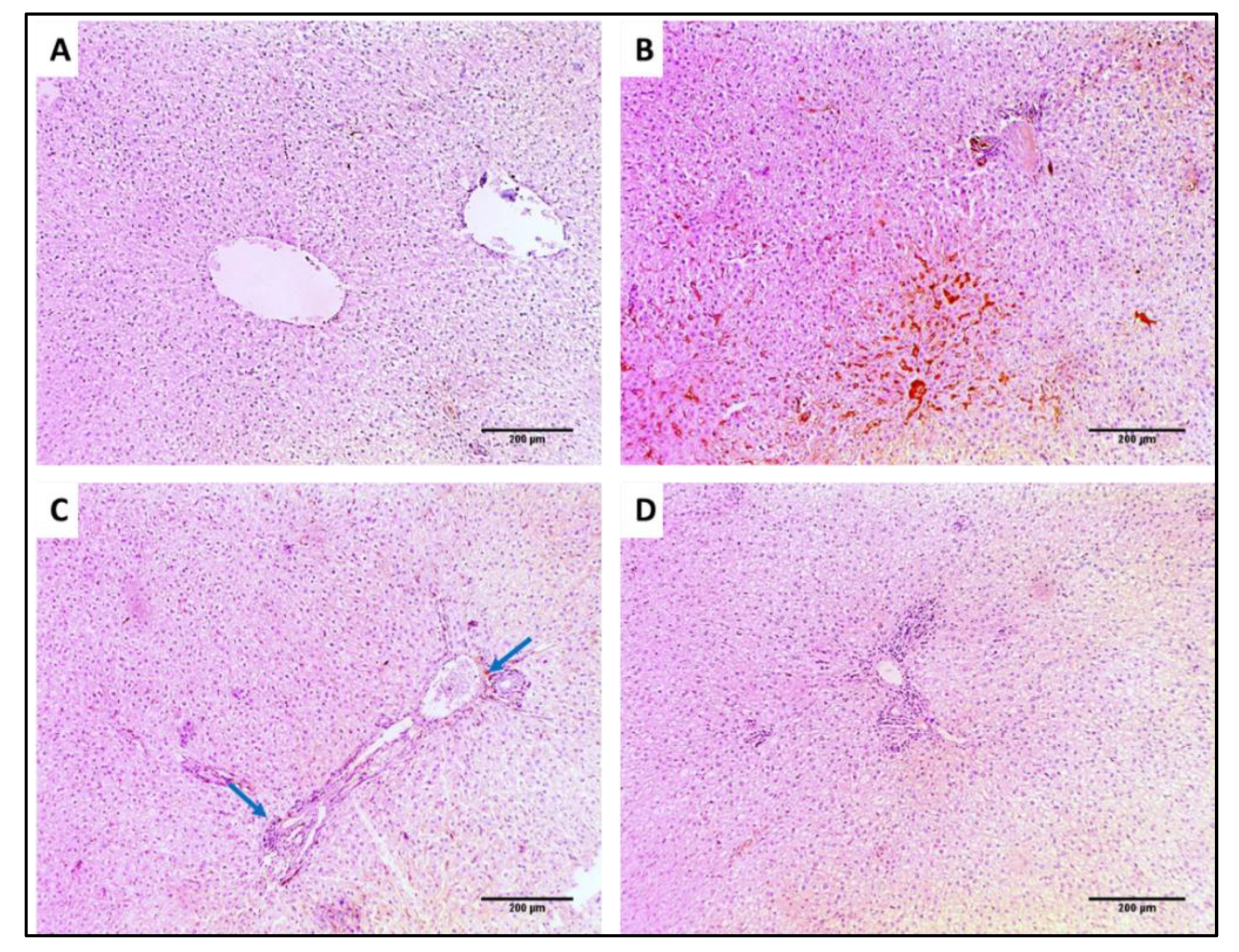
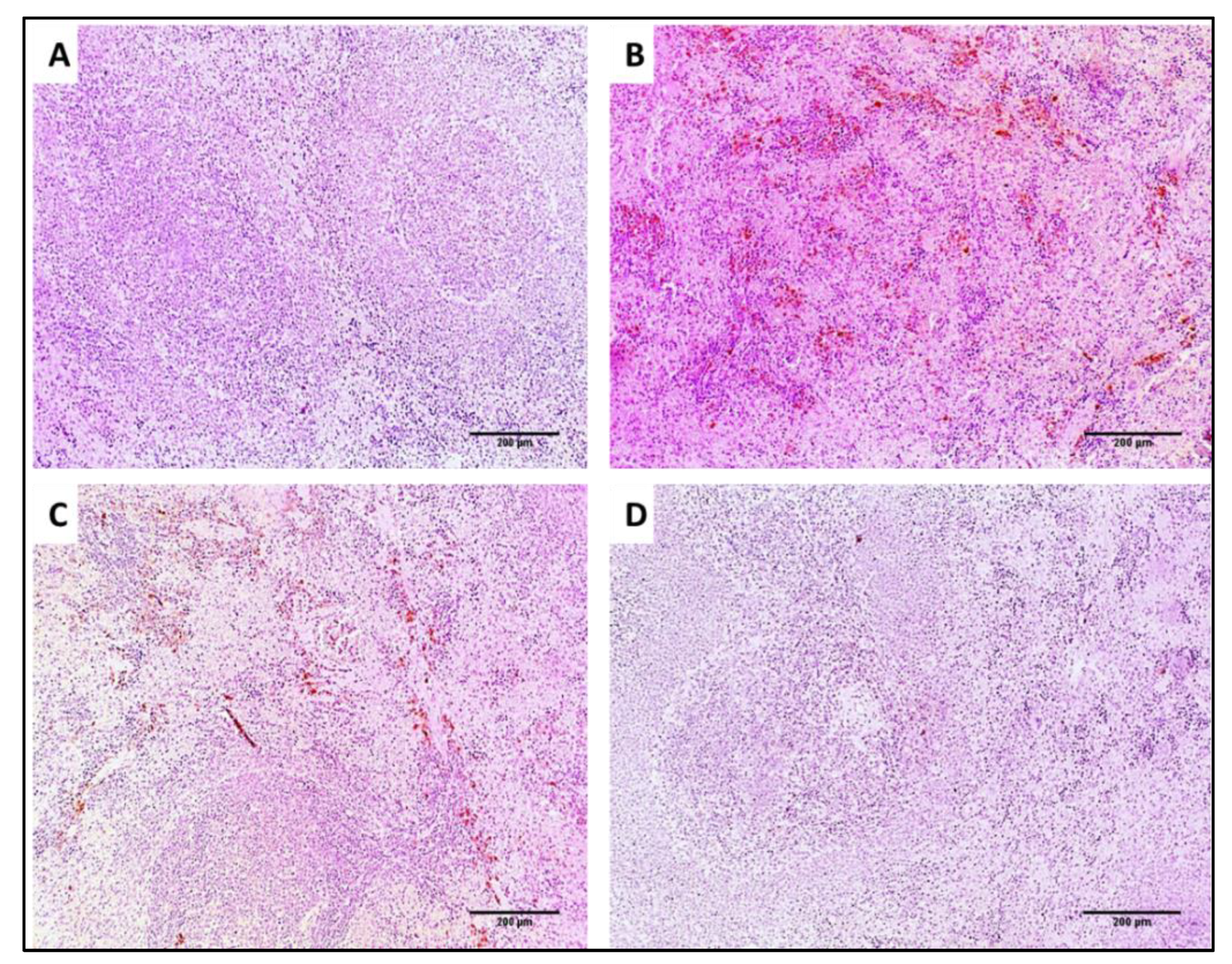
2.7.4. Immunohistochemistry
2.7.5. Measuring the Inflammatory Markers Using ELISA
2.8. Statistical Analysis
3. Results
3.1. Structure Elucidation of the Isolated Compounds
3.2. Molecular Docking Studies
3.3. In Vitro Antibacterial Activity
3.3.1. Membrane Depolarization
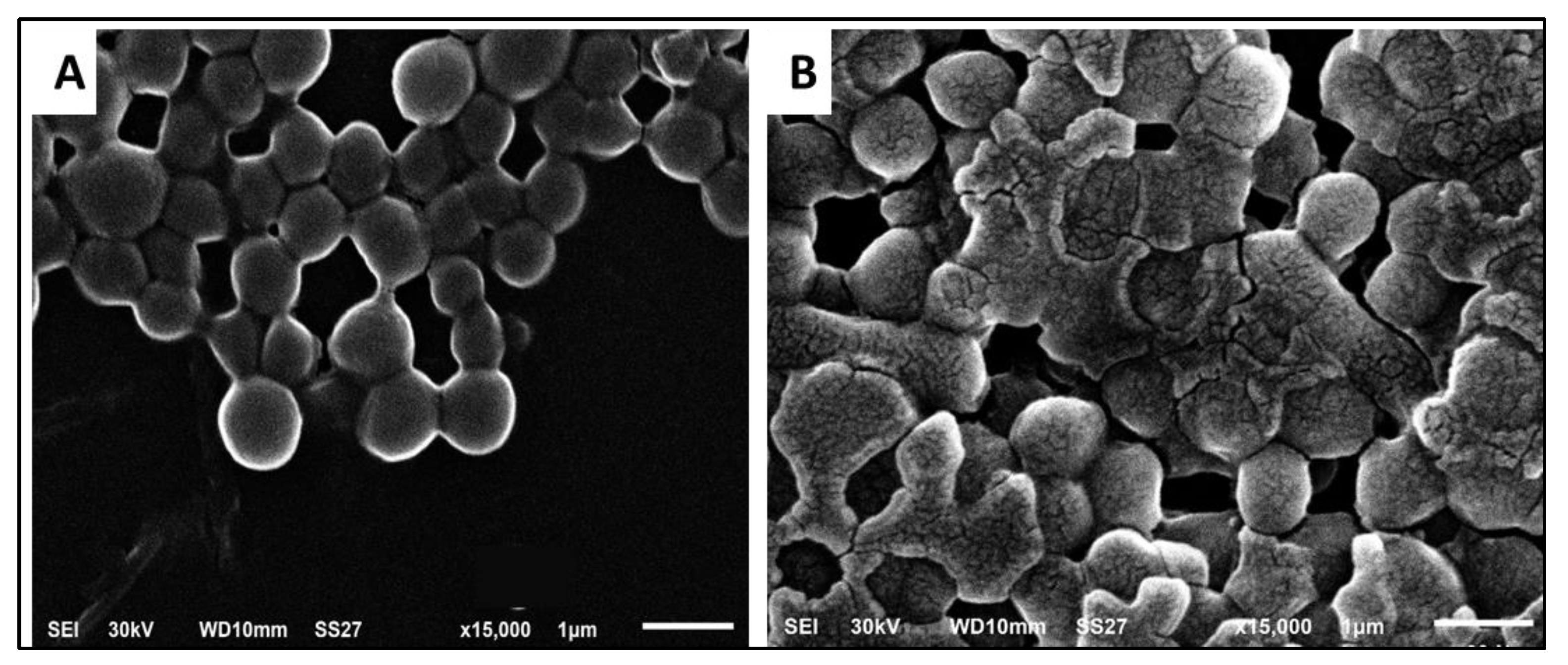
3.3.2. Bacterial Morphology
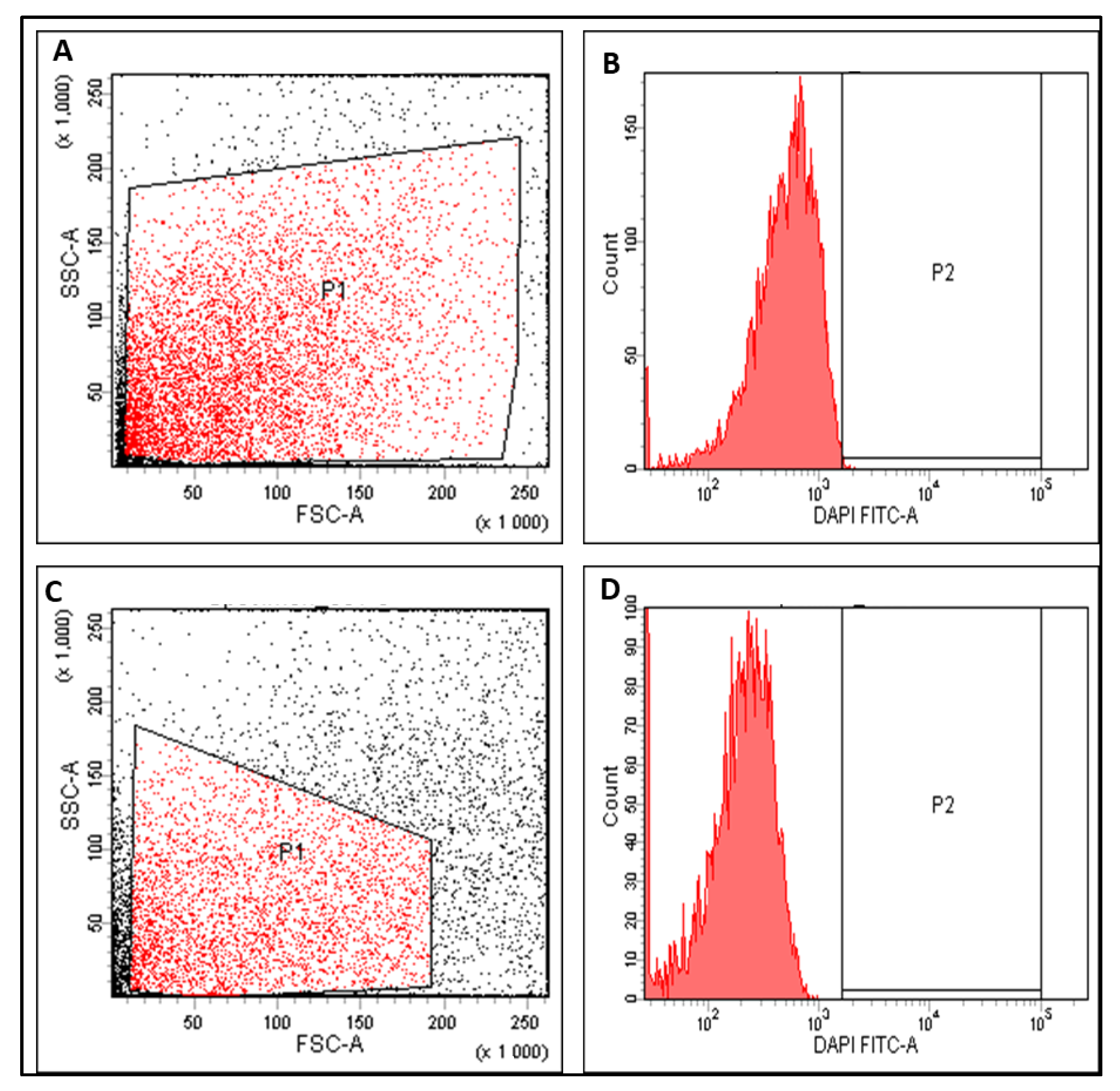
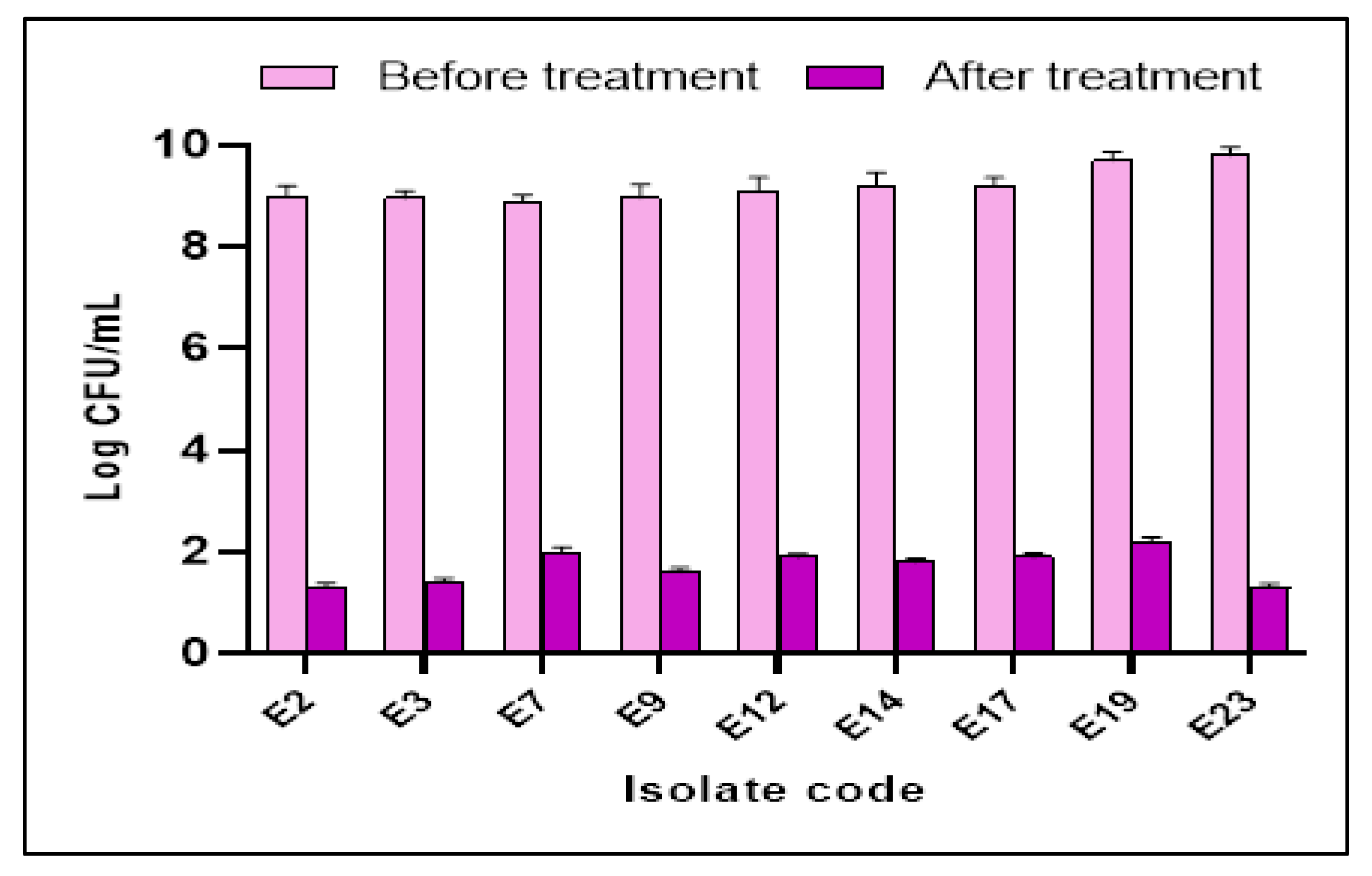
3.3.3. Anti-Biofilm Activity
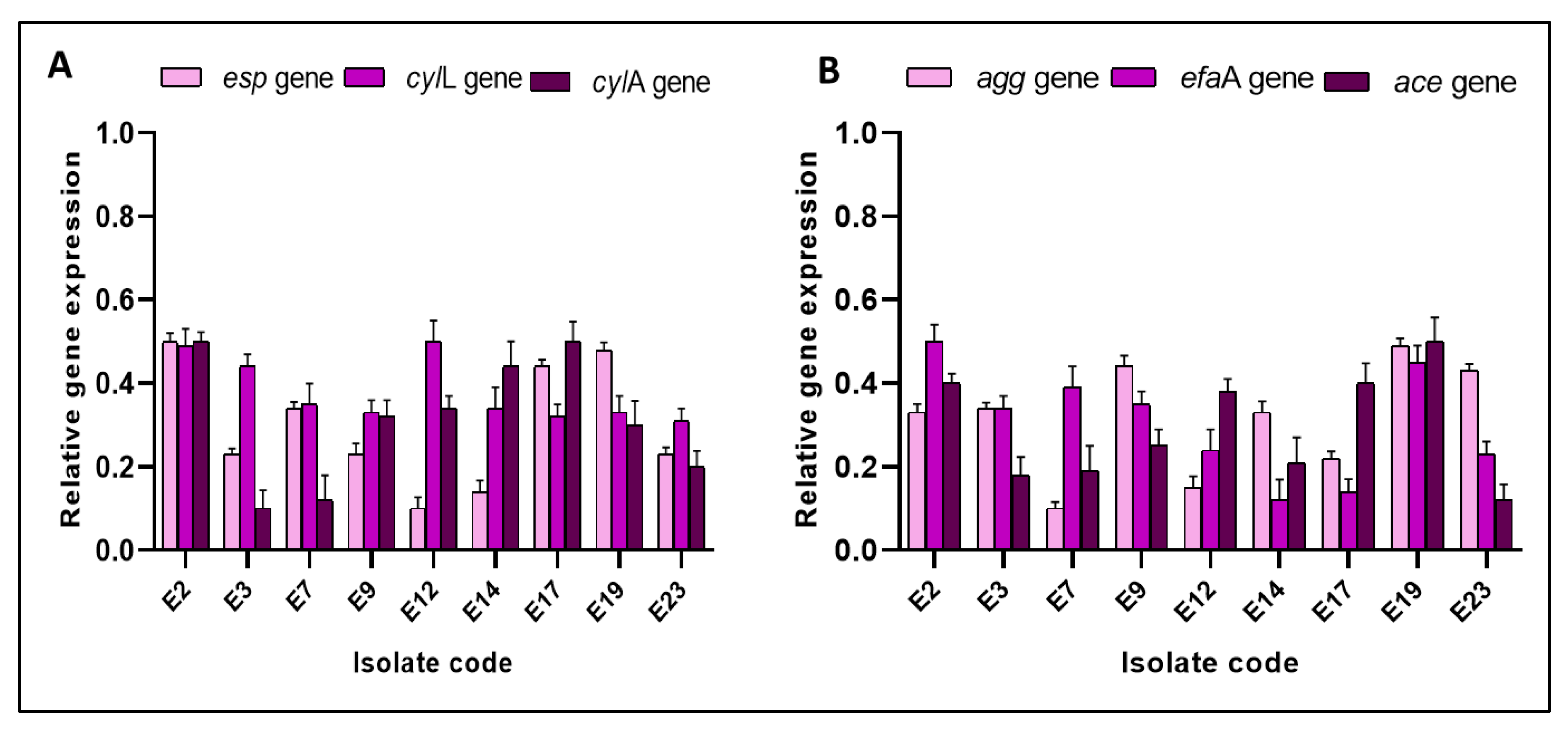
3.3.4. qRT-PCR
3.4. In Vivo Antibacterial Activity
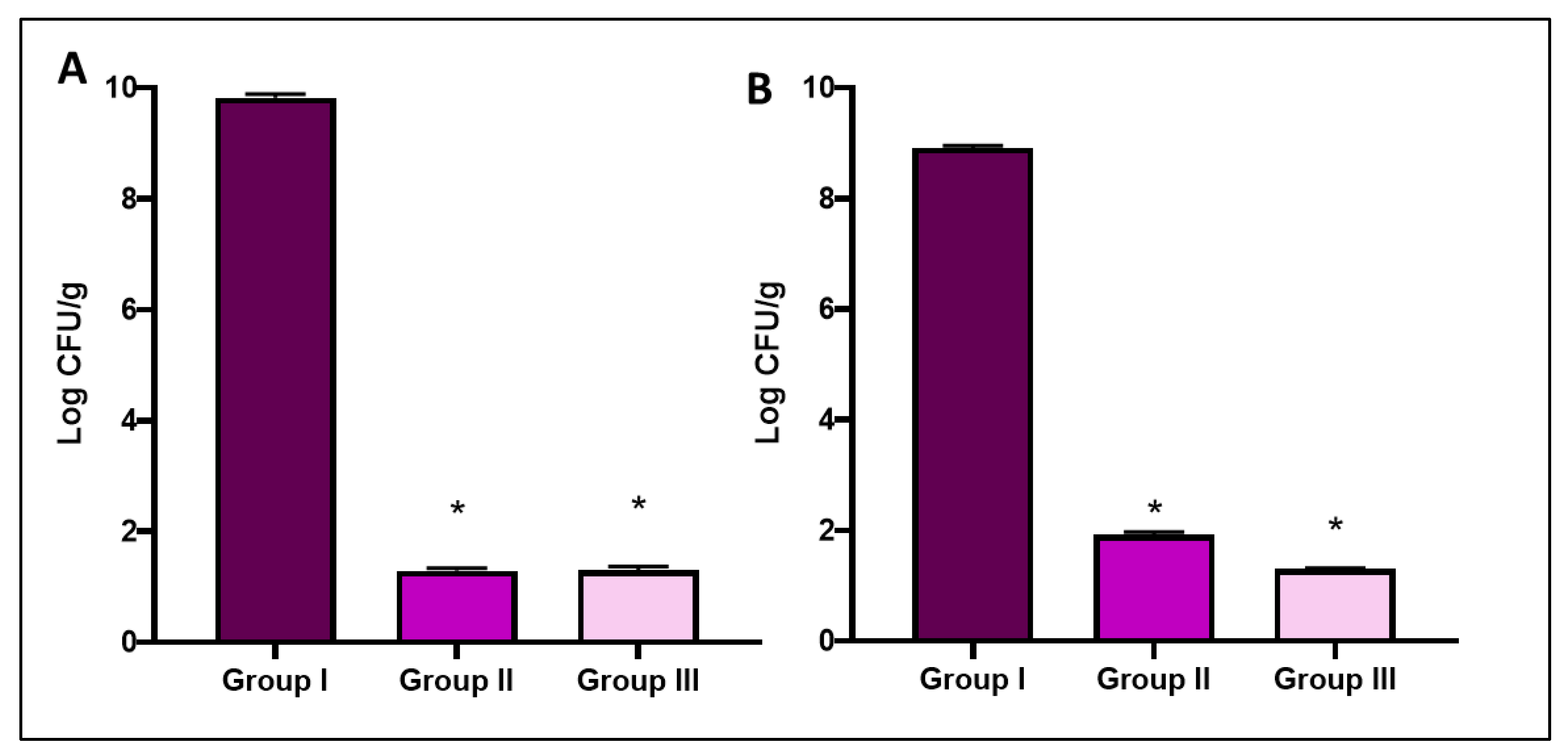
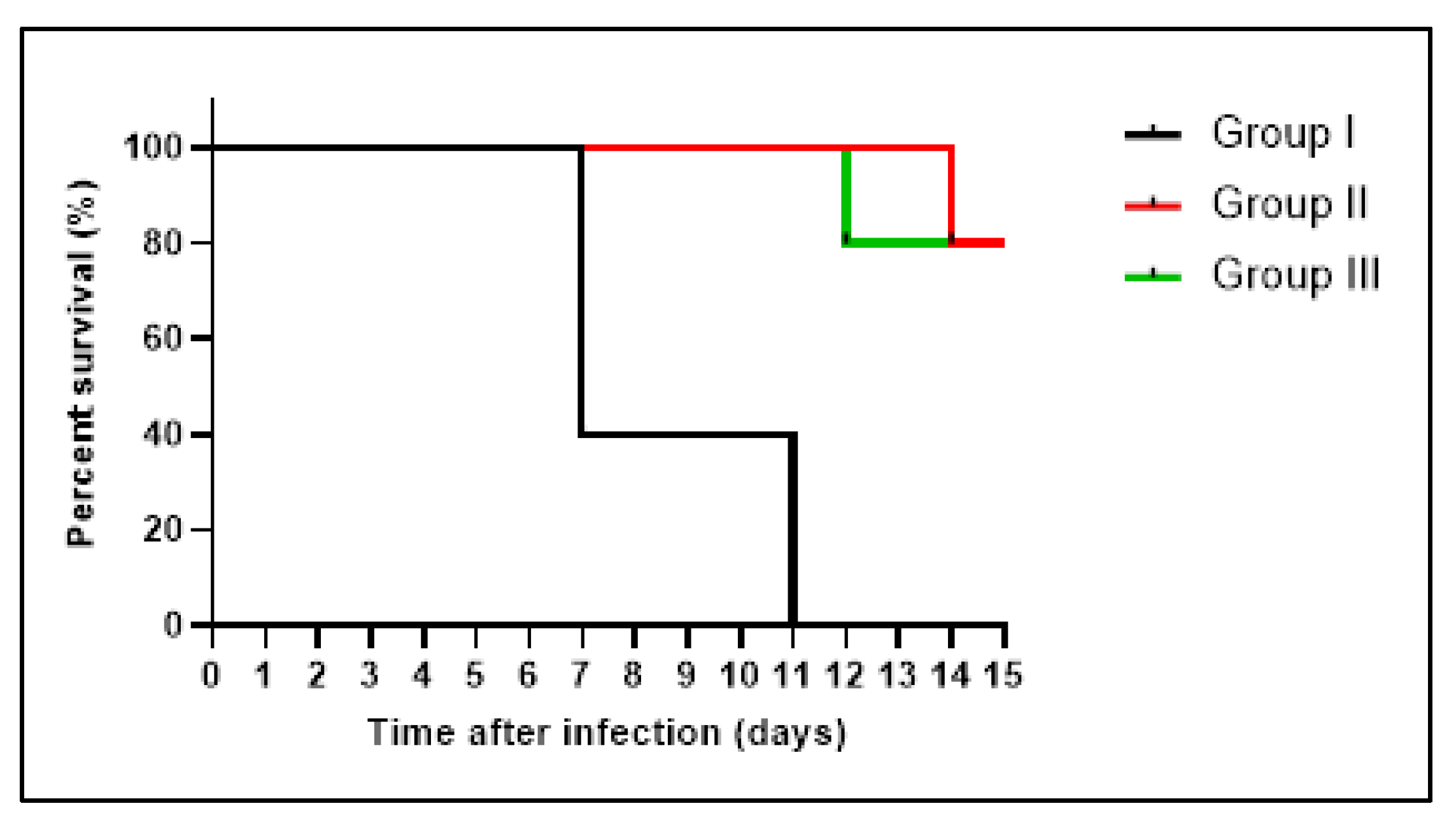
3.4.1. Bacterial Burden and Survival Curve
Liver Function Tests
3.4.2. Histological Studies
3.4.3. Immunohistochemistry
3.4.4. Inflammation Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fiore, E.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of enterococci. Microbiol. Spectr. 2019, 7, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Chen, J.; Sun, X.; Xu, G.; Li, P.; Deng, Q.; Yu, Z.; Chen, Z.; Zheng, J. The Antibacterial and Antibiofilm Activity of Telithromycin Against Enterococcus spp. Isolated From Patients in China. Front. Microbiol. 2021, 11, 616797. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.-C.; Chen, W.-C.; Chang, Y.-D.; Chen, J.-N.; Liu, F.-Y.; Huang, Y.-S.; You, C.-X.; Wu, E.H. Exposure to One Antibiotic Leads to Acquisition of Resistance to Another Antibiotic via Quorum Sensing Mechanisms. Front. Microbiol. 2021, 11, 580466. [Google Scholar] [CrossRef] [PubMed]
- Popović, N.; Dinić, M.; Tolinački, M.; Mihajlović, S.; Terzić-Vidojević, A.; Bojić, S.; Djokić, J.; Golić, N.; Veljović, K. New insight into biofilm formation ability, the presence of virulence genes and probiotic potential of Enterococcus sp. dairy isolates. Front. Microbiol. 2018, 9, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Chassagne, F.; Samarakoon, T.; Porras, G.; Lyles, J.T.; Dettweiler, M.; Marquez, L.; Salam, A.M.; Shabih, S.; Farrokhi, D.R.; Quave, C.L. A systematic review of plants with antibacterial activities: A taxonomic and phylogenetic perspective. Front. Pharmacol. 2021, 11, 2069. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, S.; Nagalingum, N.S.; Chiang, Y.-C.; Lindstrom, A.J.; Gong, X. Phylogeny of the gymnosperm genus Cycas L. (Cycadaceae) as inferred from plastid and nuclear loci based on a large-scale sampling: Evolutionary relationships and taxonomical implications. Mol. Phylogenetics Evol. 2018, 127, 87–97. [Google Scholar] [CrossRef]
- Negm, W.A.; Ibrahim, A.E.-R.S.; El-Seoud, K.A.; Attia, G.I.; Ragab, A.E. A new cytotoxic and antioxidant Amentoflavone Monoglucoside from Cycas revoluta Thunb growing in Egypt. J. Pharm. Sci. Res. 2016, 8, 343. [Google Scholar]
- Negm, W.A.; El-Aasr, M.; Kamer, A.A.; Elekhnawy, E. Investigation of the Antibacterial Activity and Efflux Pump Inhibitory Effect of Cycas thouarsii R. Br. Extract against Klebsiella pneumoniae Clinical Isolates. Pharmaceuticals 2021, 14, 756. [Google Scholar] [CrossRef]
- Ornduff, R. Size classes, reproductive behavior, and insect associates of Cycas media (Cycadaceae) in Australia. Bot. Gaz. 1991, 152, 203–207. [Google Scholar] [CrossRef]
- Facklam, R.; Collins, M. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 1989, 27, 731–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumpel, S.; Razeto, A.; Pillar, C.M.; Vijayan, V.; Taylor, A.; Giller, K.; Gilmore, M.S.; Becker, S.; Zweckstetter, M. Structure and DNA-binding properties of the cytolysin regulator CylR2 from Enterococcus faecalis. EMBO J. 2004, 23, 3632–3642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnuraj, K.; Narayana, S.V.L. Crystal structure of ACE19, the collagen binding subdomain of Enterococus faecalis surface protein ACE. Proteins Struct. Funct. Bioinform. 2007, 69, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.; Jiang, K.; Camacho, M.I.; Jonna, V.R.; Hofer, A.; Westerlund, F.; Christie, P.J.; Berntsson, R.P.-A. PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction. Mol. Microbiol. 2018, 109, 291–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegelman, L.; Zhang, L.; Tezcan, A.; Ghosh, P. Enterococcal surface Protein, Partial N-Terminal Region. Available online: https://www.rcsb.org/structure/6ori (accessed on 24 February 2022).
- Swiss-Model (A0A7H0FPW4) Enterococcus Faecalis Gelatinase. Available online: https://swissmodel.expasy.org/repository/uniprot/A0A7H0FPW4 (accessed on 3 January 2022).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- DeRoo, S.S.; Pudalov, N.J.; Fu, L.Y. Planning for a COVID-19 vaccination program. Jama 2020, 323, 2458–2459. [Google Scholar] [CrossRef]
- Seleem, N.M.; Abd El Latif, H.K.; Shaldam, M.A.; El-Ganiny, A. Drugs with new lease of life as quorum sensing inhibitors: For combating MDR Acinetobacter baumannii infections. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1687–1702. [Google Scholar] [CrossRef]
- Hagens, A.; İnkaya, A.Ç.; Yildirak, K.; Sancar, M.; van der Schans, J.; Acar Sancar, A.; Ünal, S.; Postma, M.; Yeğenoğlu, S. COVID-19 Vaccination Scenarios: A Cost-Effectiveness Analysis for Turkey. Vaccines 2021, 9, 399. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Modeling 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Patel, J.B.; Cockerill, F.; Bradford, P.A. Performance standards for antimicrobial susceptibility testing: Twenty-fifth informational supplement. M100 2015, 35, 29–50. [Google Scholar]
- Almukainzi, M.; El-Masry, T.A.; Negm, W.A.; Elekhnawy, E.; Saleh, A.; Sayed, A.E.; Khattab, M.A.; Abdelkader, D.H. Gentiopicroside PLGA Nanospheres: Fabrication, in vitro Characterization, Antimicrobial Action, and in vivo Effect for Enhancing Wound Healing in Diabetic Rats. Int. J. Nanomed. 2022, 17, 1203–1225. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, B.; Negm, W.A.; Elekhnawy, E.; El-Masry, T.A.; Elharty, M.E.; Saleh, A.; Abdelkader, D.H.; Mokhtar, F.A. Antibacterial activity of nano zinc oxide green-synthesised from Gardenia thailandica triveng. Leaves against Pseudomonas aeruginosa clinical isolates: In vitro and in vivo study. Artif. Cells Nanomed. Biotechnol. 2022, 50, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Elmongy, E.I.; Negm, W.A.; Elekhnawy, E.; El-Masry, T.A.; Attallah, N.G.; Altwaijry, N.; Batiha, G.E.-S.; El-Sherbeni, S.A. Antidiarrheal and Antibacterial Activities of Monterey Cypress Phytochemicals: In Vivo and In Vitro Approach. Molecules 2022, 27, 346. [Google Scholar] [CrossRef]
- Alotaibi, B.; Mokhtar, F.A.; El-Masry, T.A.; Elekhnawy, E.; Mostafa, S.A.; Abdelkader, D.H.; Elharty, M.E.; Saleh, A.; Negm, W.A. Antimicrobial Activity of Brassica rapa L. Flowers Extract on Gastrointestinal Tract Infections and Antiulcer Potential Against Indomethacin-Induced Gastric Ulcer in Rats Supported by Metabolomics Profiling. J. Inflamm. Res. 2021, 14, 7411–7430. [Google Scholar] [CrossRef]
- Abdelaziz, A.; Sonbol, F.; Elbanna, T.; El-Ekhnawy, E. Exposure to sublethal concentrations of benzalkonium chloride induces antimicrobial resistance and cellular changes in Klebsiellae pneumoniae clinical isolates. Microb. Drug Resist. 2019, 25, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Elekhnawy, E.A.; Sonbol, F.I.; Elbanna, T.E.; Abdelaziz, A.A. Evaluation of the impact of adaptation of Klebsiella pneumoniae clinical isolates to benzalkonium chloride on biofilm formation. Egypt. J. Med. Hum. Genet. 2021, 22, 51. [Google Scholar] [CrossRef]
- Elekhnawy, E.; Negm, W.A.; El-Aasr, M.; Kamer, A.A.; Alqarni, M.; Batiha, G.E.-S.; Obaidullah, A.J.; Fawzy, H.M. Histological assessment, anti-quorum sensing, and anti-biofilm activities of Dioon spinulosum extract: In vitro and in vivo approach. Sci. Rep. 2022, 12, 180. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Beshiru, A. Antimicrobial resistance, virulence determinants, and biofilm formation of Enterococcus species from ready-to-eat seafood. Front. Microbiol. 2019, 10, 728. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Alqahtani, M.J.; Elekhnawy, E.; Negm, W.A.; Mahgoub, S.; Hussein, I.A. Encephalartos villosus Lem. Displays a Strong In Vivo and In Vitro Antifungal Potential against Candida glabrata Clinical Isolates. J. Fungi 2022, 8, 521. [Google Scholar] [CrossRef]
- Hasan Khudhair, D.; Al-Gareeb, A.I.; Al-kuraishy, H.; El-Kadem, A.H.; Elekhnawy, E.; Negm, W.A.; Saber, S.; Cavalu, S.; Tirla, A.; Alotaibi, S.S.; et al. Combination of vitamin C and curcumin safeguards against methotrexate-induced acute liver injury in mice by synergistic antioxidant effects. Front. Med. 2022, 9, 866343. [Google Scholar] [CrossRef] [PubMed]
- Attallah, N.G.; El-Sherbeni, S.A.; El-Kadem, A.H.; Elekhnawy, E.; El-Masry, T.A.; Elmongy, E.I.; Altwaijry, N.; Negm, W.A. Elucidation of the Metabolite Profile of Yucca gigantea and Assessment of its Cytotoxic, Antimicrobial, and Anti-Inflammatory Activities. Molecules 2022, 27, 1329. [Google Scholar] [CrossRef]
- Negm, W.A.; El-Kadem, A.H.; Elekhnawy, E.; Attallah, N.G.; Al-Hamoud, G.A.; El-Masry, T.A.; Zayed, A. Wound-Healing Potential of Rhoifolin-Rich Fraction Isolated from Sanguisorba officinalis Roots Supported by Enhancing Re-Epithelization, Angiogenesis, Anti-Inflammatory, and Antimicrobial Effects. Pharmaceuticals 2022, 15, 178. [Google Scholar] [CrossRef] [PubMed]
- Negm, W.A.; Abo El-Seoud, K.A.; Kabbash, A.; Kassab, A.A.; El-Aasr, M. Hepatoprotective, cytotoxic, antimicrobial and antioxidant activities of Dioon spinulosum leaves Dyer Ex Eichler and its isolated secondary metabolites. Nat. Prod. Res. 2020, 35, 5166–5176. [Google Scholar] [CrossRef]
- Moawad, A.; Hetta, M.; Zjawiony, J.K.; Jacob, M.R.; Hifnawy, M.; Marais, J.P.; Ferreira, D. Phytochemical investigation of Cycas circinalis and Cycas revoluta leaflets: Moderately active antibacterial biflavonoids. Planta Med. 2010, 76, 796–802. [Google Scholar] [CrossRef] [Green Version]
- Tendolkar, P.M.; Baghdayan, A.S.; Gilmore, M.S.; Shankar, N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 2004, 72, 6032–6039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, V.C.; Thurlow, L.R.; Boyle, D.; Hancock, L.E. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 2008, 190, 5690–5698. [Google Scholar] [CrossRef] [Green Version]
- Alotaibi, B.; Negm, W.A.; Elekhnawy, E.; El-Masry, T.A.; Elseady, W.S.; Saleh, A.; Alotaibi, K.N.; El-Sherbeni, S.A. Antibacterial, Immunomodulatory, and Lung Protective Effects of Boswelliadalzielii Oleoresin Ethanol Extract in Pulmonary Diseases: In Vitro and In Vivo Studies. Antibiotics 2021, 10, 1444. [Google Scholar] [CrossRef]
- Perera, M.; Dighe, S.N.; Katavic, P.L.; Collet, T.A. Antibacterial Potential of Extracts and Phytoconstituents Isolated from Syncarpia hillii Leaves In Vitro. Plants 2022, 11, 283. [Google Scholar] [CrossRef]
- Markham, K.R.; Sheppard, C.; Geiger, H. 13C NMR studies of some naturally occurring amentoflavone and hinokiflavone biflavonoids. Phytochemistry 1987, 26, 3335–3337. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Alonso-Calleja, C.; García-Fernández, C.; Carballo, J.; Capita, R. Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) for twelve antimicrobials (biocides and antibiotics) in eight strains of Listeria monocytogenes. Biology 2021, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, X.; Hu, Y.; Lei, T.; Liu, T. Sotetsuflavone induces autophagy in non-small cell lung cancer through blocking PI3K/Akt/mTOR signaling pathway in vivo and in vitro. Front. Pharmacol. 2019, 10, 1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Hu, Y.; Yan, Y.; Cheng, Z.; Liu, T. Sotetsuflavone inhibits proliferation and induces apoptosis of A549 cells through ROS-mediated mitochondrial-dependent pathway. BMC Complement. Altern. Med. 2018, 18, 235. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Rasul, A.; Hussain, G.; Shah, M.A.; Zahoor, M.K.; Anwar, H.; Sarfraz, I.; Riaz, A.; Manzoor, M.; Adem, Ş.; et al. Ginkgetin: A natural biflavone with versatile pharmacological activities. Food Chem. Toxicol. 2020, 145, 111642. [Google Scholar] [CrossRef] [PubMed]
- Lian, N.; Tong, J.; Li, W.; Wu, J.; Li, Y. Ginkgetin ameliorates experimental atherosclerosis in rats. Biomed. Pharmacother. 2018, 102, 510–516. [Google Scholar] [CrossRef]
- Šamec, D.; Karalija, E.; Dahija, S.; Hassan, S.T. Biflavonoids: Important Contributions to the Health Benefits of Ginkgo (Ginkgo biloba L.). Plants 2022, 11, 1381. [Google Scholar] [CrossRef]
- Te Winkel, J.; Gray, D.; Seistrup, K.; Hamoen, L.; Strahl, H. Analysis of Antimicrobial-Triggered Membrane Depolarization Using Voltage Sensitive Dyes. Front. Cell Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef] [Green Version]
- Goldbeck, J.C.; Victoria, F.N.; Motta, A.; Savegnago, L.; Jacob, R.G.; Perin, G.; Lenardao, E.J.; da Silva, W.P. Bioactivity and morphological changes of bacterial cells after exposure to 3-(p-chlorophenyl) thio citronellal. LWT-Food Sci. Technol. 2014, 59, 813–819. [Google Scholar] [CrossRef] [Green Version]
- Sathiyamoorthi, E.; Faleye, O.S.; Lee, J.-H.; Raj, V.; Lee, J. Antibacterial and antibiofilm activities of chloroindoles against Vibrio parahaemolyticus. Front. Microbiol. 2021, 12, 2206. [Google Scholar] [CrossRef]
- Raorane, C.J.; Lee, J.-H.; Kim, Y.-G.; Rajasekharan, S.K.; García-Contreras, R.; Lee, J. Antibiofilm and antivirulence efficacies of flavonoids and curcumin against Acinetobacter baumannii. Front. Microbiol. 2019, 10, 990. [Google Scholar] [CrossRef]
- Ge, Y.; Huang, M.; Yao, Y.-M. Biology of interleukin-17 and its pathophysiological significance in sepsis. Front. Immunol. 2020, 11, 1558. [Google Scholar] [CrossRef] [PubMed]
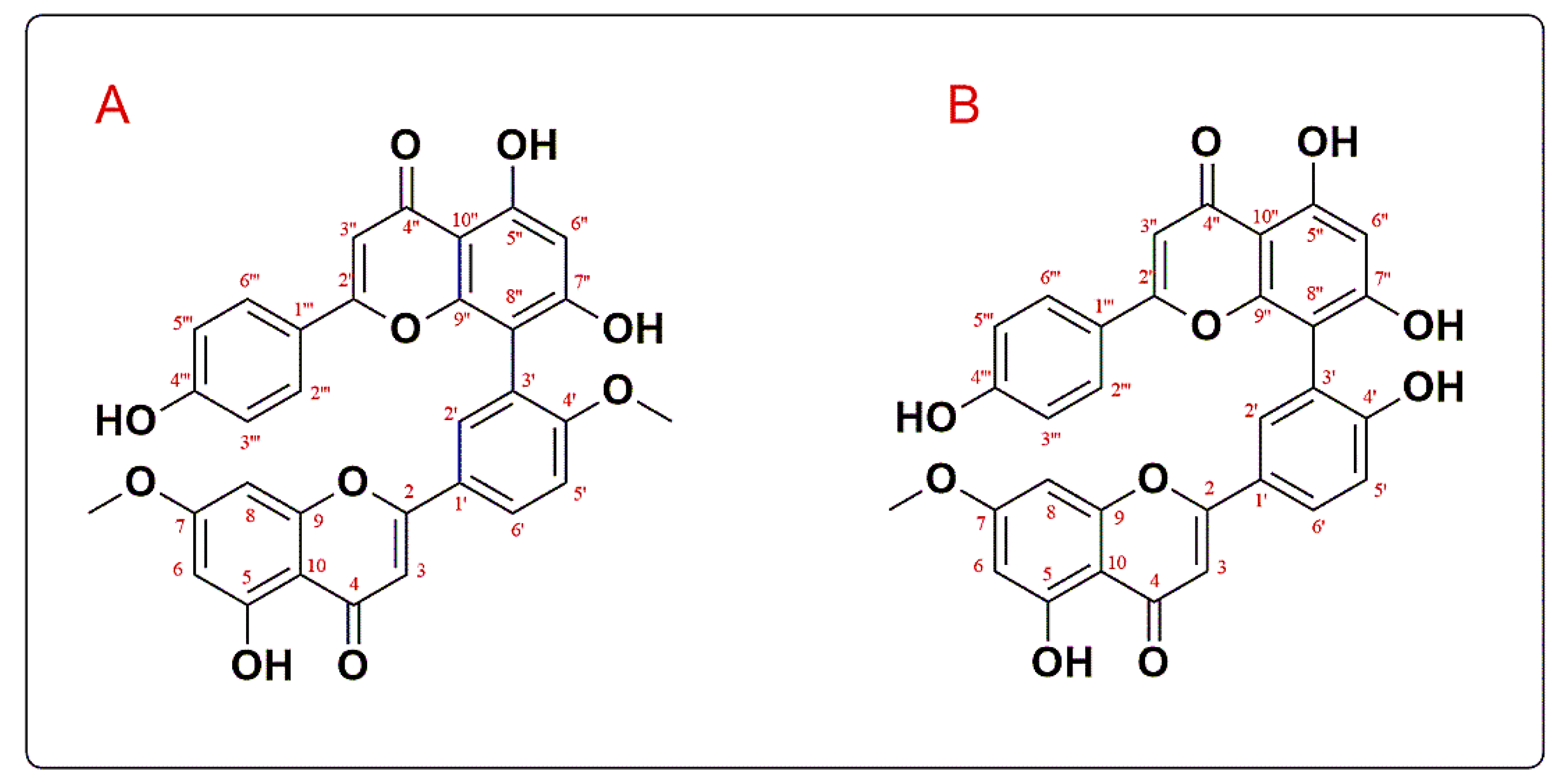
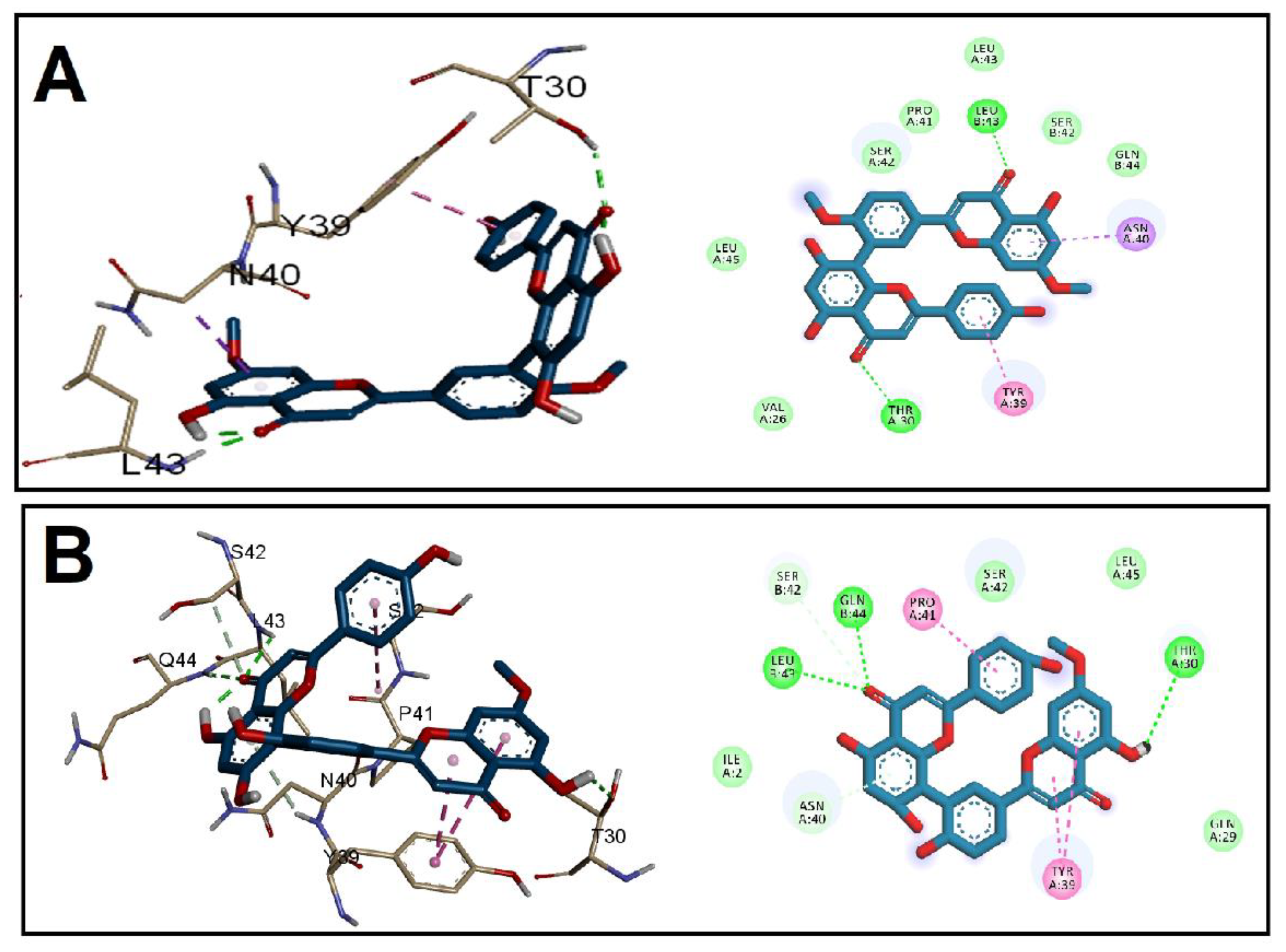
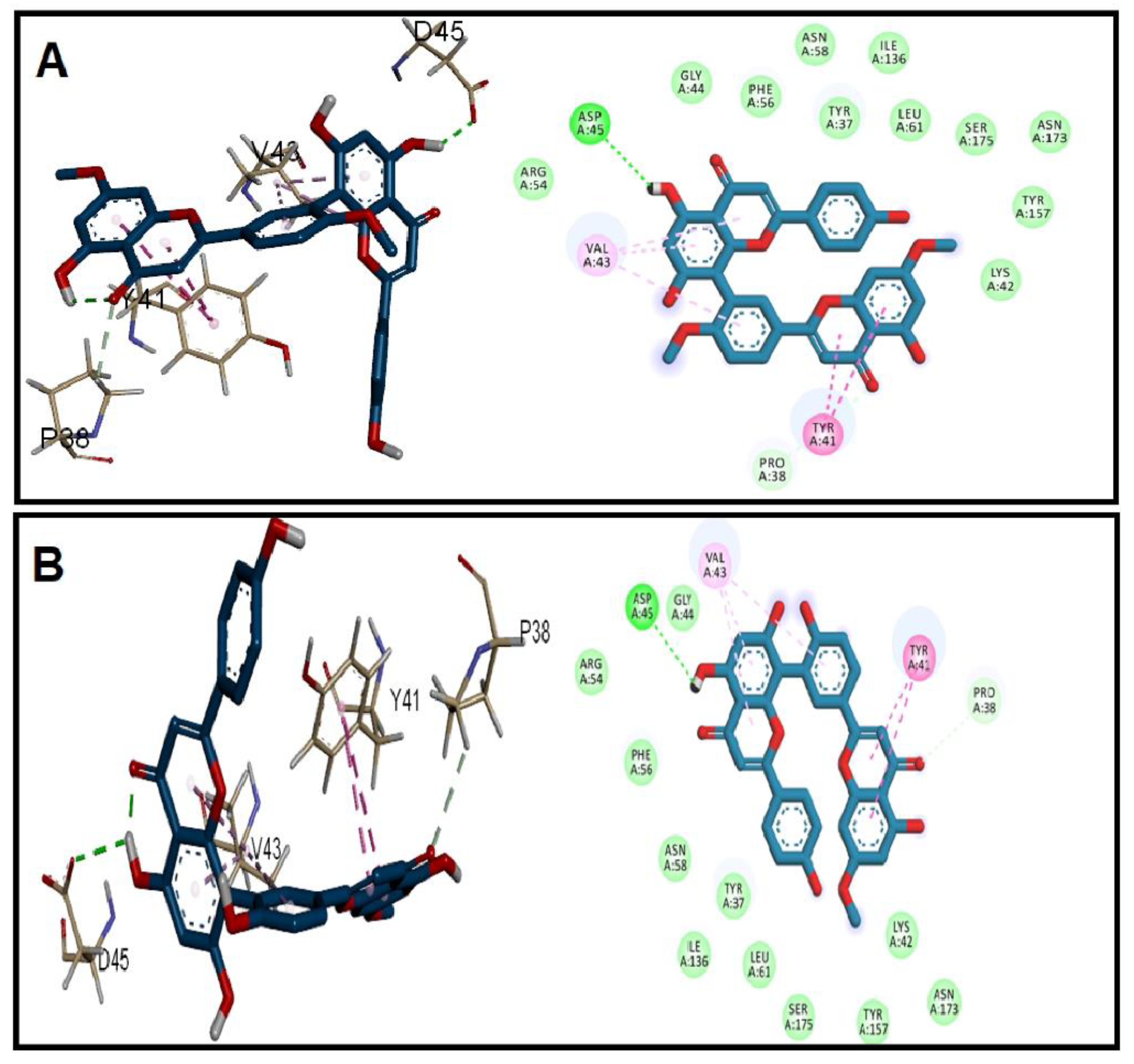
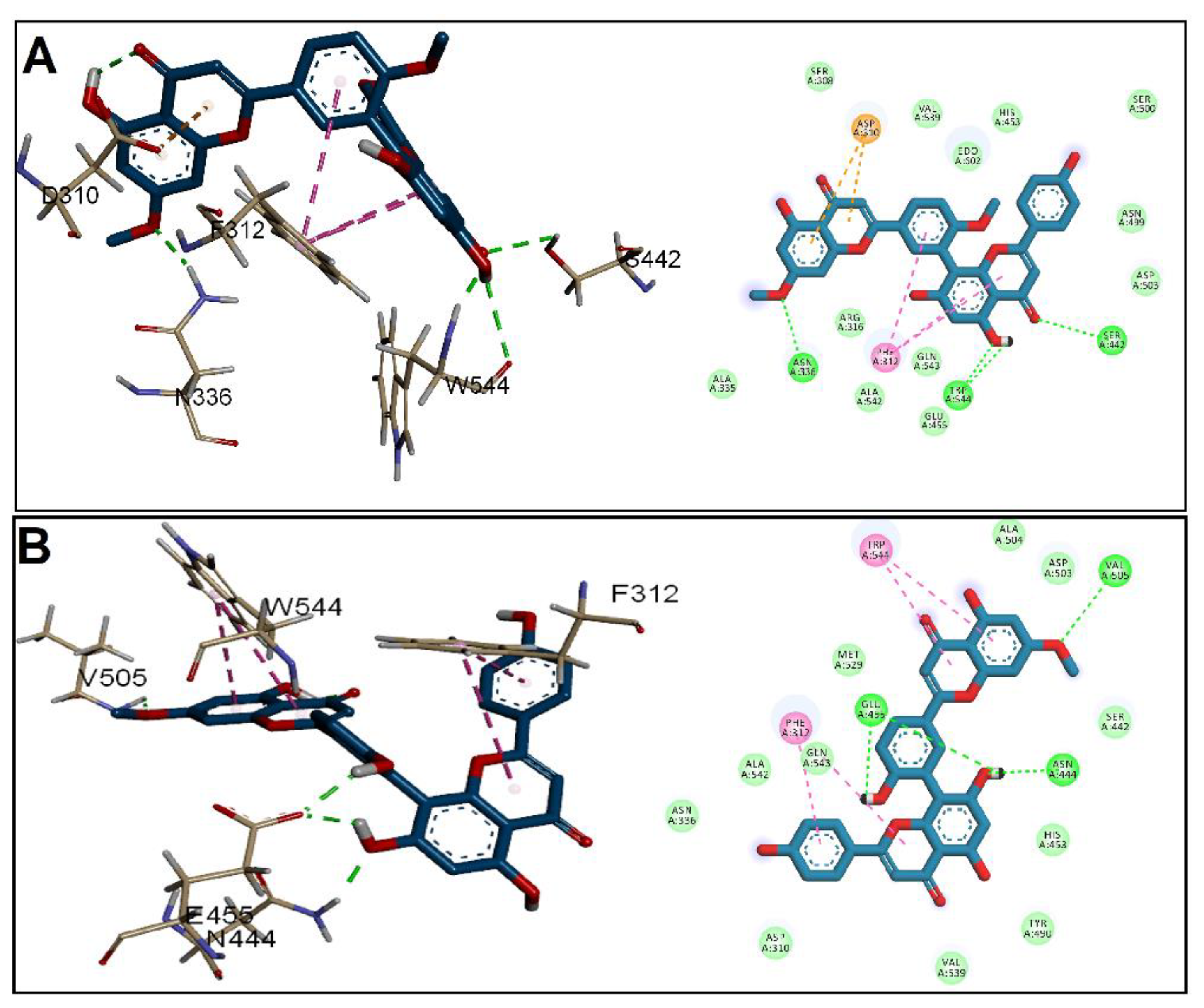
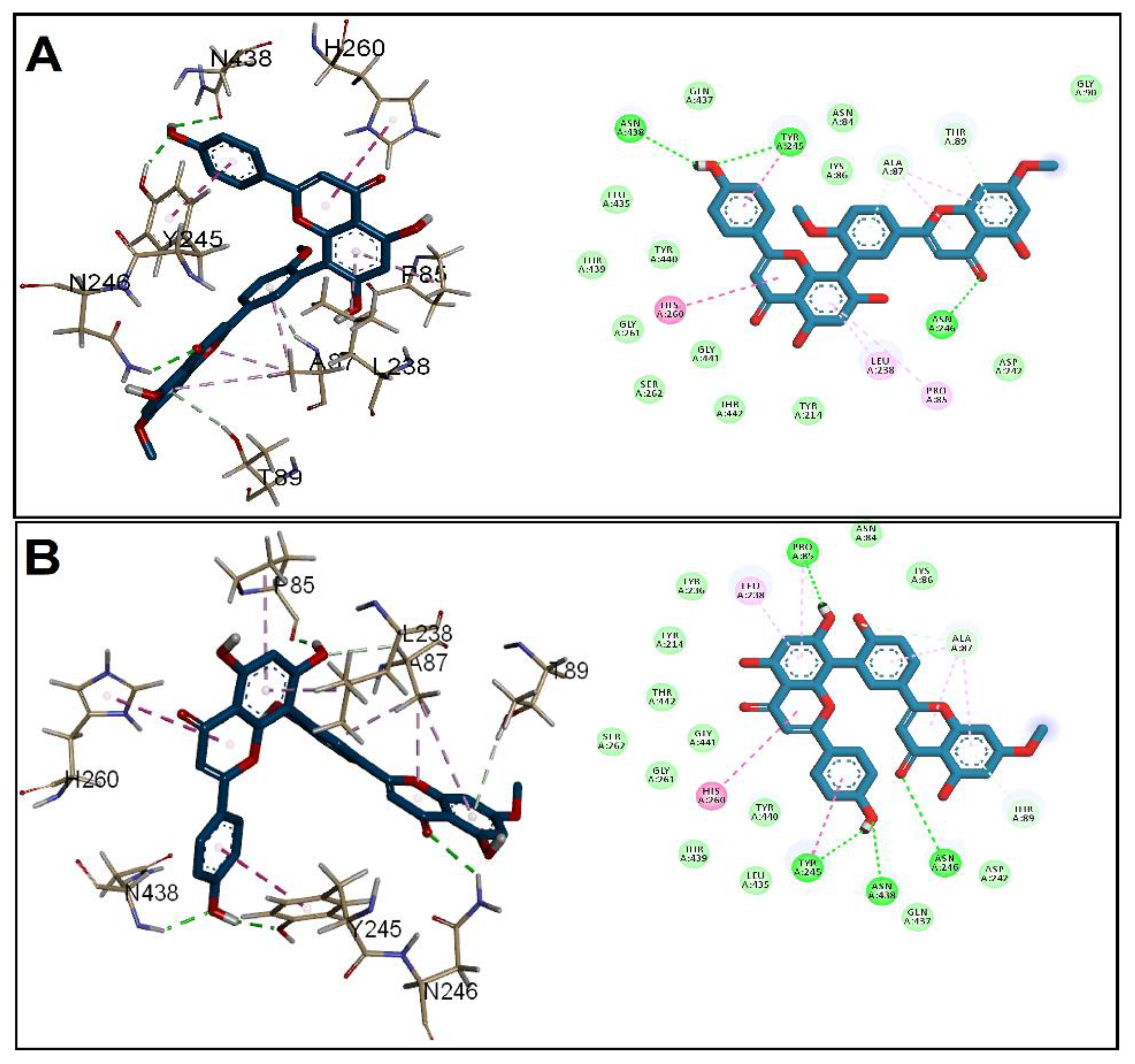
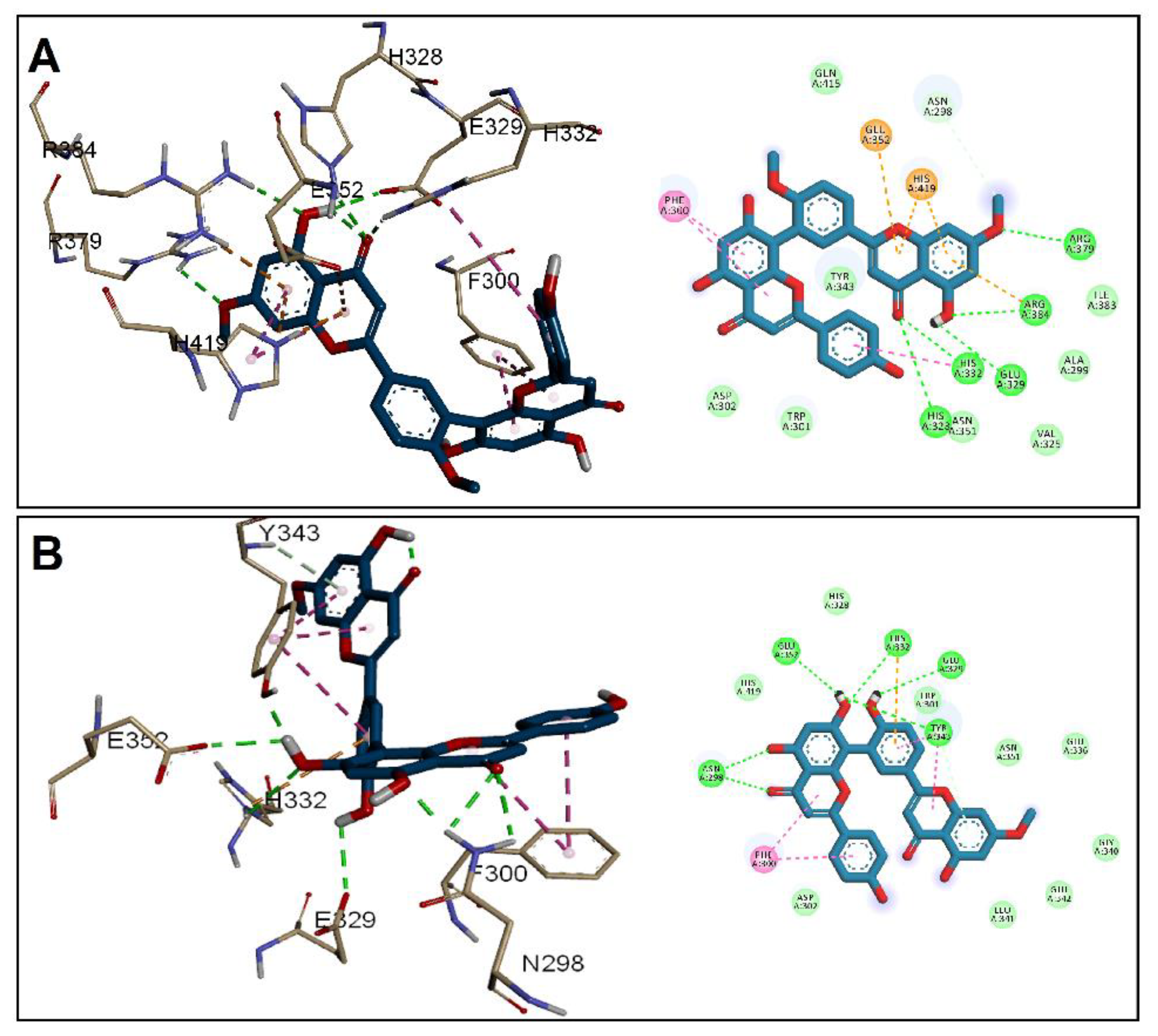


| Receptor | Grid Box (x, y, z) | Affinity (kcal/mol) | ||
|---|---|---|---|---|
| Center | Size | GINK | SOTE | |
| CylR2 | 23.43, −4.93, 12.79 | 20.0, 20.0, 15.0 | −7.4 | −7.5 |
| ACE19 | 3.4, −12.7, 83.3 | 25.9, 24.9, 22.8 | −7.4 | −7.6 |
| PrgB | 36.8, 23.3, 20.6 | 52.8, 66.1, 61.1 | −8.9 | −9.6 |
| EFGE | −0.6, −1.8, 3.1 | 33.7, 34.1, 32.3 | −9.5 | −9.9 |
| ESP | 0.9, −4.0, 0.1 | 41.6, 45.3, 47.3 | −10.7 | −11.1 |
| Compound I (GINK) | Compound II (SOTE) | |||
|---|---|---|---|---|
| δ-H | δ-C | δ-H | δ-C | |
| 2 | 163.7 | 164.2 | ||
| 3 | 6.92 (1H, s) | 103.1 | 6.80 (1H, s) | 102.8 |
| 4 | 181.9 | 181.9 | ||
| 5 | 161.5 | 161.1 | ||
| 6 | 6.19 (1H, d, J = 1.5 Hz) | 98.1 | 6.34 (1H, d, J = 2.5 Hz) | 98.1 |
| 7 | 165.1 | 163.4 | ||
| 8 | 6.49 (1H, d, J = 1.5) | 92.7 | 6.77 (1H, d, J = 2.5) | 92.6 |
| 9 | 157.3 | 157.5 | ||
| 10 | 104.7 | 102.8 | ||
| 1′ | 122.3 | 120.9 | ||
| 2′ | 8.06 (1H, d, J = 2.5) | 128.2 | 8.10 (1H, d, J = 2.5) | 127.6 |
| 3′ | 121. 7 | 121.5 | ||
| 4′ | 160.6 | 160.5 | ||
| 5′ | 7.33 (1H, d, J = 8.5) | 111.7 | 6.98 (1H, d, J = 9) | 117.0 |
| 6′ | 8.18 (1H, dd, J = 2.5, 8.5) | 130.9 | 8.00 (1H, dd, J = 2.5, 9) | 131.5 |
| 2″ | 163.7 | 164.4 | ||
| 3″ | 6.78 (1H, s) | 102.6 | 6.88 (1H, s) | 102.5 |
| 4″ | 182.1 | 182.1 | ||
| 5″ | 160.4 | 160.9 | ||
| 6″ | 6.38 (1H, s) | 98.2 | 6.33 (1H, s) | 99.4 |
| 7″ | 161.6 | 161.9 | ||
| 8″ | 103.7 | 104.7 | ||
| 9″ | 154.5 | 154.5 | ||
| 10″ | 103.6 | 103.1 | ||
| 1″′ | 120.9 | 120.9 | ||
| 2″′ | 7.56 (2H, d, J = 7.5) | 128.0 | 7.57 (2H, d, J = 9) | 128.1 |
| 3″′ | 6.81 (2H, d, J = 7.5) | 115.8 | 6.70 (2H, d, J = 9) | 115.7 |
| 4″′ | 161.1 | 160.2 | ||
| 5″′ | 6.81 (2H, d, J = 7.5) | 115.8 | 7.57 (2H, d, J = 9) | 115.7 |
| 6″′ | 7.56 (2H, d, J = 7.5) | 128.0 | 6.70 (2H, d, J = 9) | 128.1 |
| OCH3 | 3.77, 3.81 | 56.1, 55.9 | 3.79 | 56.1 |
| Biofilm Forming Ability | Number of the Isolates | |
|---|---|---|
| Before Treatment with SOTE | After Treatment with SOTE | |
| Non-biofilm forming | 5 | 10 |
| Weak | 4 | 8 |
| Moderate | 8 | 3 |
| Strong | 6 | 2 |
| Groups | ALT (U/L) | AST (U/L) | Bilirubin (mg/dL) | Total Proteins (g/dL) | Albumin (g/dL) |
|---|---|---|---|---|---|
| Group I | 92.0 ± 2.3 | 135.1 ± 4.3 | 0.56 ± 0.04 | 1.8 ± 0.1 | 2.1 ± 0.3 |
| Group II | 46.2 ± 3.2 * | 70.0 ± 2.5 * | 0.15 ± 0.001 * | 7.0 ± 0.4 * | 4.45 ± 0.3 * |
| Group III | 46.0 ± 2.2 * | 71.2 ±1.2 * | 0.16 ± 0.002 * | 6.9 ± 0.3 * | 4.44 ± 0.2 * |
| Tissues | Liver | Spleen | ||
|---|---|---|---|---|
| Inflammation Markers | IL-1β Level (pg/mL) | IL-6 Level (pg/mL) | IL-1β Level (pg/mL) | IL-6 Level (pg/mL) |
| Group I | 73.3 ± 1.2 | 200.5 ± 5.2 | 71.1 ± 2.0 | 195.4 ±4.3 |
| Group II | 23.2 ± 2.3 * | 100.2 ± 3.4 * | 24.1 ± 1.4 * | 99.1 ± 3.2 * |
| Group III | 22.6 ± 2.3 * | 110.1 ± 4.1 * | 23.9 ± 1.5 * | 98.2 ± 4.3 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attallah, N.G.M.; Al-Fakhrany, O.M.; Elekhnawy, E.; Hussein, I.A.; Shaldam, M.A.; Altwaijry, N.; Alqahtani, M.J.; Negm, W.A. Anti-Biofilm and Antibacterial Activities of Cycas media R. Br Secondary Metabolites: In Silico, In Vitro, and In Vivo Approaches. Antibiotics 2022, 11, 993. https://doi.org/10.3390/antibiotics11080993
Attallah NGM, Al-Fakhrany OM, Elekhnawy E, Hussein IA, Shaldam MA, Altwaijry N, Alqahtani MJ, Negm WA. Anti-Biofilm and Antibacterial Activities of Cycas media R. Br Secondary Metabolites: In Silico, In Vitro, and In Vivo Approaches. Antibiotics. 2022; 11(8):993. https://doi.org/10.3390/antibiotics11080993
Chicago/Turabian StyleAttallah, Nashwah G. M., Omnia Momtaz Al-Fakhrany, Engy Elekhnawy, Ismail A. Hussein, Moataz A. Shaldam, Najla Altwaijry, Moneerah J. Alqahtani, and Walaa A. Negm. 2022. "Anti-Biofilm and Antibacterial Activities of Cycas media R. Br Secondary Metabolites: In Silico, In Vitro, and In Vivo Approaches" Antibiotics 11, no. 8: 993. https://doi.org/10.3390/antibiotics11080993
APA StyleAttallah, N. G. M., Al-Fakhrany, O. M., Elekhnawy, E., Hussein, I. A., Shaldam, M. A., Altwaijry, N., Alqahtani, M. J., & Negm, W. A. (2022). Anti-Biofilm and Antibacterial Activities of Cycas media R. Br Secondary Metabolites: In Silico, In Vitro, and In Vivo Approaches. Antibiotics, 11(8), 993. https://doi.org/10.3390/antibiotics11080993










