Blood Culture Headspace Gas Analysis Enables Early Detection of Escherichia coli Bacteremia in an Animal Model of Sepsis
Abstract
1. Introduction
2. Results
2.1. Grade of Inflammation
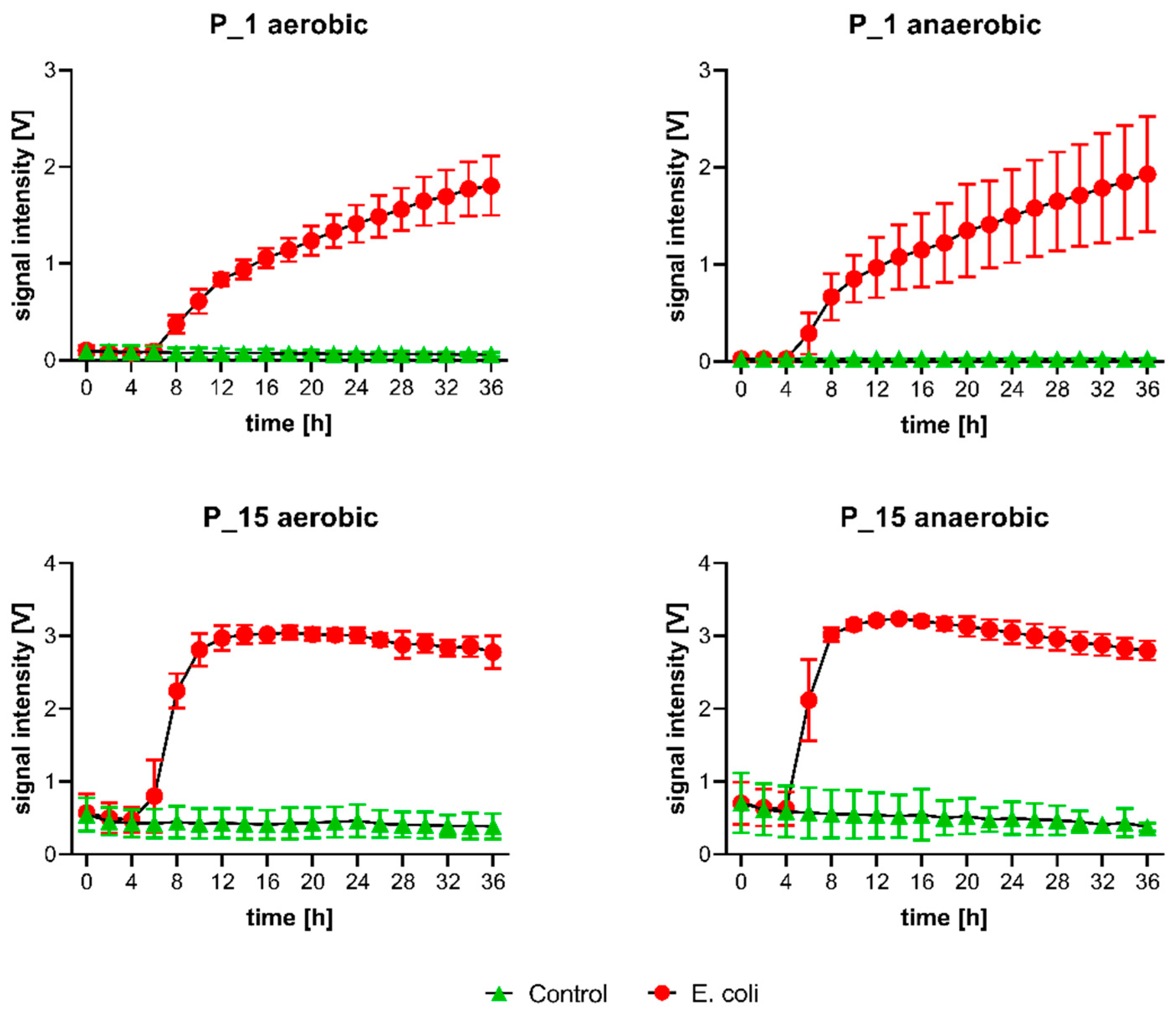
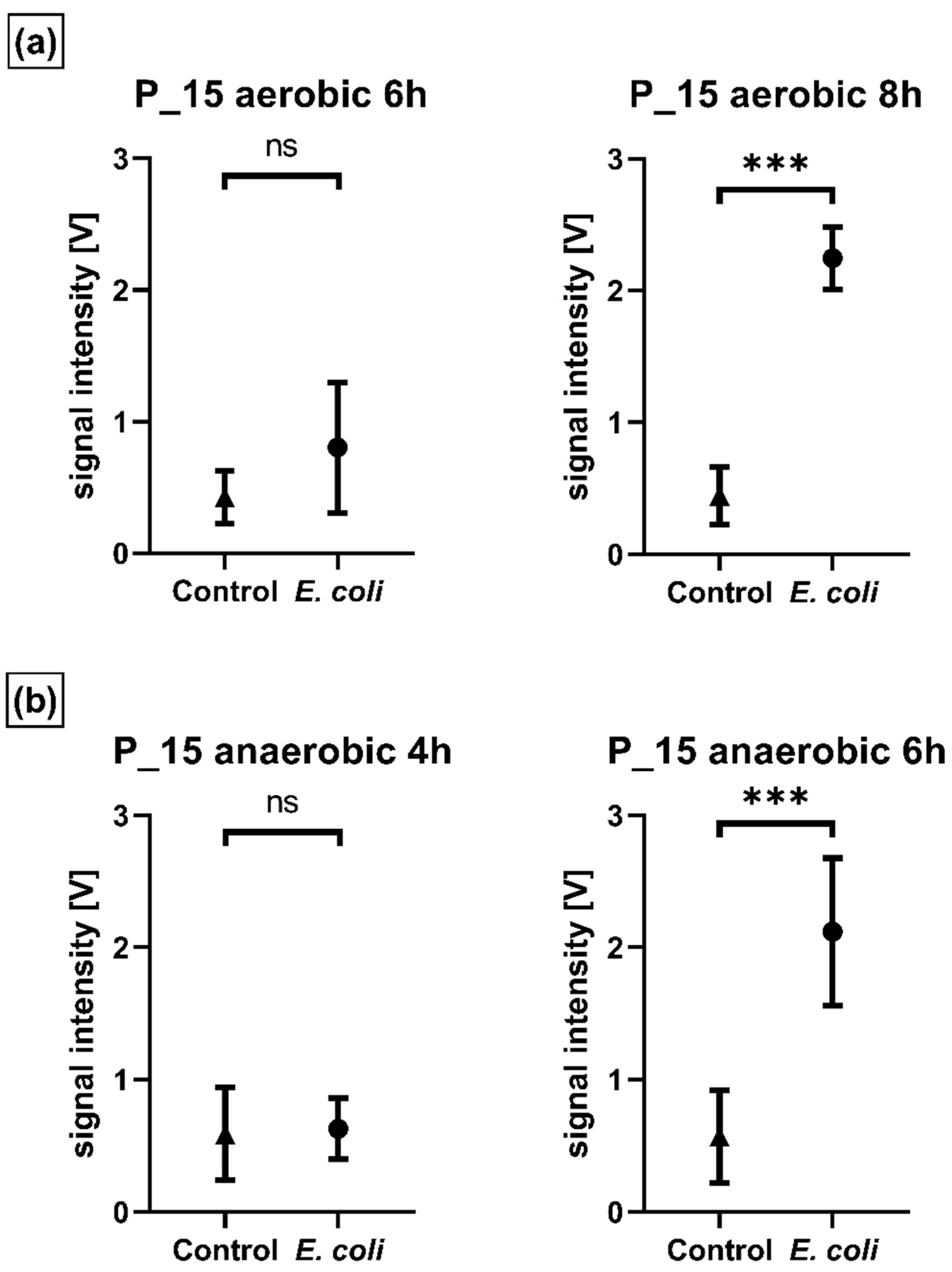
2.2. Volatile Background/mVOCs
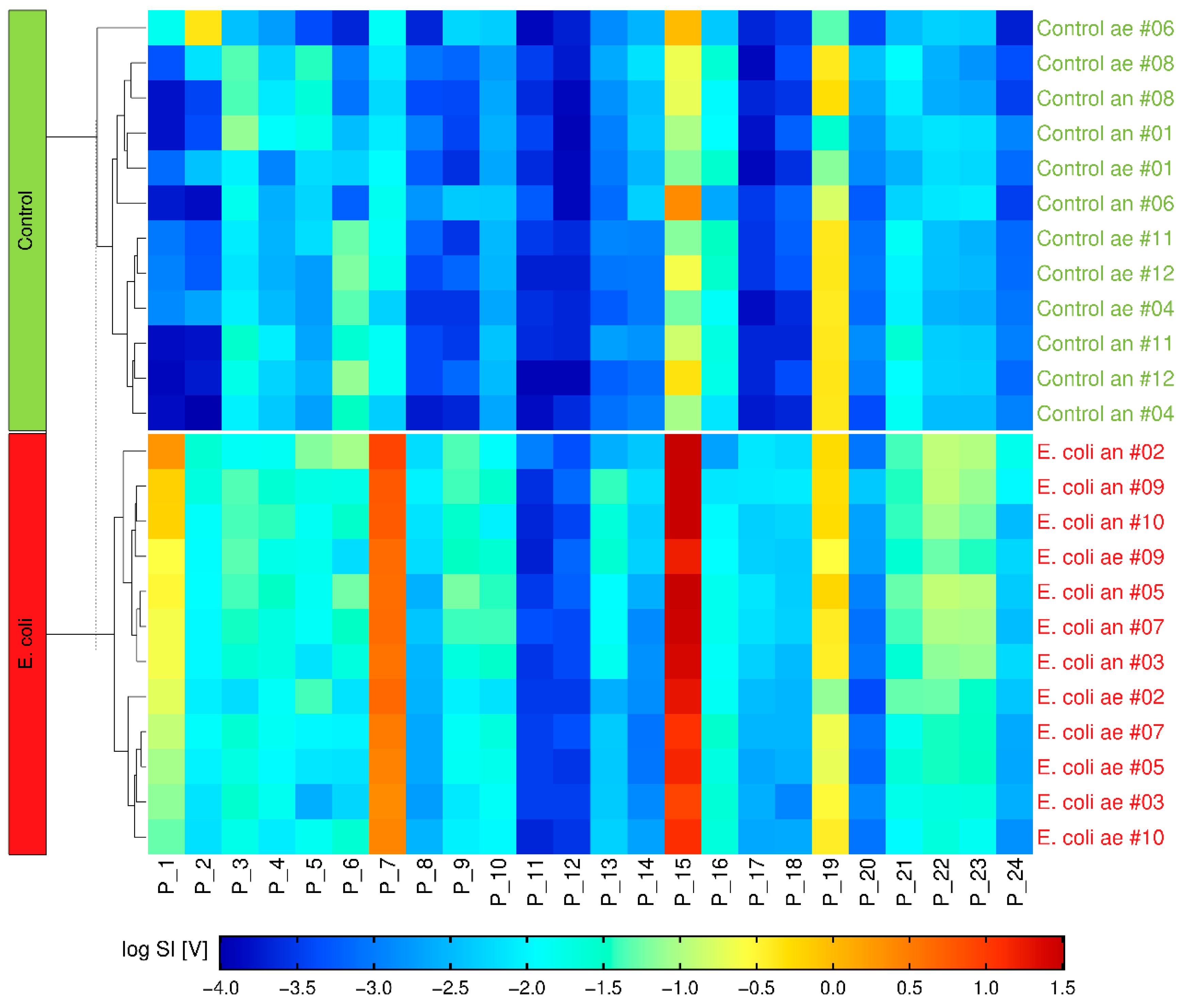
2.3. Multivariate Analysis
2.4. Microbiological Reference
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Rabbit Model of Sepsis
4.3. Culture Conditions
4.4. Gas Chromatography-Ion Mobility Spectrometer
4.5. Automated Sampling
4.6. GC-IMS Measurements
4.7. MALDI-TOF MS
4.8. Laboratory Analyses
4.9. Data Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Tabak, Y.P.; Vankeepuram, L.; Ye, G.; Jeffers, K.; Gupta, V.; Murray, P.R. Blood Culture Turnaround Time in U.S. Acute Care Hospitals and Implications for Laboratory Process Optimization. J. Clin. Microbiol. 2018, 56, e00500–e00518. [Google Scholar] [CrossRef] [PubMed]
- Goff, D.A.; Jankowski, C.; Tenover, F.C. Using rapid diagnostic tests to optimize antimicrobial selection in antimicrobial stewardship programs. Pharmacotherapy 2012, 32, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Peker, N.; Couto, N.; Sinha, B.; Rossen, J.W. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: Recent developments in molecular approaches. Clin. Microbiol. Infect. 2018, 24, 944–955. [Google Scholar] [CrossRef]
- Warhurst, G.; Dunn, G.; Chadwick, P.; Blackwood, B.; McAuley, D.; Perkins, G.D.; McMullan, R.; Gates, S.; Bentley, A.; Young, D.; et al. Rapid detection of health-care-associated bloodstream infection in critical care using multipathogen real-time polymerase chain reaction technology: A diagnostic accuracy study and systematic review. Health Technol. Assess. 2015, 19, 1–142. [Google Scholar] [CrossRef]
- Schubert, S.; Weinert, K.; Wagner, C.; Gunzl, B.; Wieser, A.; Maier, T.; Kostrzewa, M. Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J. Mol. Diagn. 2011, 13, 701–706. [Google Scholar] [CrossRef]
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-Care Testing for Infectious Diseases: Past, Present, and Future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef]
- Reali, S.; Najib, E.Y.; Treuerné Balázs, K.E.; Chern Hui Tan, A.; Váradi, L.; Hibbs, D.E.; Groundwater, P.W. Novel diagnostics for point-of-care bacterial detection and identification. RSC Adv. 2019, 9, 21486–21497. [Google Scholar] [CrossRef]
- Schulz, S.; Dickschat, J.S. Bacterial volatiles: The smell of small organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef]
- Zoller, H.F.; Clark, W.M. The production of volatile fatty acids by bacteria of the dysentery group. J. Gen. Physiol. 1921, 3, 325–330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lemfack, M.C.; Gohlke, B.O.; Toguem, S.M.T.; Preissner, S.; Piechulla, B.; Preissner, R. mVOC 2.0: A database of microbial volatiles. Nucleic Acids Res. 2018, 46, D1261–D1265. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, C.L.; Taba, B.; Queralto, N.; Lukaszewski, R.A.; Martino, R.A.; Rhodes, P.A.; Lim, S.H. The use of colorimetric sensor arrays to discriminate between pathogenic bacteria. PLoS ONE 2013, 8, e62726. [Google Scholar] [CrossRef] [PubMed]
- Ratiu, I.A.; Ligor, T.; Bocos-Bintintan, V.; Buszewski, B. Mass spectrometric techniques for the analysis of volatile organic compounds emitted from bacteria. Bioanalysis 2017, 9, 1069–1092. [Google Scholar] [CrossRef]
- Kunze-Szikszay, N.; Euler, M.; Perl, T. Identification of volatile compounds from bacteria by spectrometric methods in medicine diagnostic and other areas: Current state and perspectives. Appl. Microbiol. Biotechnol. 2021, 105, 6245–6255. [Google Scholar] [CrossRef]
- Eiceman, G.A. Ion Mobility Spectrometry, 3rd ed.; CRC Press: Boca Ranton, FL, USA, 2016. [Google Scholar]
- Vautz, W.; Zimmermann, D.; Hartmann, M.; Baumbach, J.I.; Nolte, J.; Jung, J. Ion mobility spectrometry for food quality and safety. Food Addit. Contam. 2006, 23, 1064–1073. [Google Scholar] [CrossRef]
- Ahmed, W.M.; Lawal, O.; Nijsen, T.M.; Goodacre, R.; Fowler, S.J. Exhaled Volatile Organic Compounds of Infection: A Systematic Review. ACS Infect. Dis. 2017, 3, 695–710. [Google Scholar] [CrossRef]
- Chouinard, C.D.; Wei, M.S.; Beekman, C.R.; Kemperman, R.H.; Yost, R.A. Ion Mobility in Clinical Analysis: Current Progress and Future Perspectives. Clin. Chem. 2016, 62, 124–133. [Google Scholar] [CrossRef]
- Vautz, W.; Franzke, J.; Zampolli, S.; Elmi, I.; Liedtke, S. On the potential of ion mobility spectrometry coupled to GC pre-separation—A tutorial. Anal. Chim. Acta 2018, 1024, 52–64. [Google Scholar] [CrossRef]
- Baumbach, J.I. Ion mobility spectrometry coupled with multi-capillary columns for metabolic profiling of human breath. J. Breath Res. 2009, 3, 034001. [Google Scholar] [CrossRef]
- Jünger, M.; Vautz, W.; Kuhns, M.; Hofmann, L.; Ulbricht, S.; Baumbach, J.I.; Quintel, M.; Perl, T. Ion mobility spectrometry for microbial volatile organic compounds: A new identification tool for human pathogenic bacteria. Appl. Microbiol. Biotechnol. 2012, 93, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Perl, T.; Junger, M.; Vautz, W.; Nolte, J.; Kuhns, M.; Borg-von Zepelin, M.; Quintel, M. Detection of characteristic metabolites of Aspergillus fumigatus and Candida species using ion mobility spectrometry-metabolic profiling by volatile organic compounds. Mycoses 2011, 54, e828–e837. [Google Scholar] [CrossRef] [PubMed]
- Steppert, I.; Schönfelder, J.; Schultz, C.; Kuhlmeier, D. Rapid in vitro differentiation of bacteria by ion mobility spectrometry. Appl. Microbiol. Biotechnol. 2021, 105, 4297–4307. [Google Scholar] [CrossRef] [PubMed]
- Drees, C.; Vautz, W.; Liedtke, S.; Rosin, C.; Althoff, K.; Lippmann, M.; Zimmermann, S.; Legler, T.J.; Yildiz, D.; Perl, T.; et al. GC-IMS headspace analyses allow early recognition of bacterial growth and rapid pathogen differentiation in standard blood cultures. Appl. Microbiol. Biotechnol. 2019, 103, 9091–9101. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zeng, L.; Li, M.; Yan, B.; Gao, D.; Zhou, B.; Lu, W.; He, Q. Use of GC-IMS for detection of volatile organic compounds to identify mixed bacterial culture medium. AMB Express 2022, 12, 31. [Google Scholar] [CrossRef]
- O’Hara, M.; Mayhew, C.A. A preliminary comparison of volatile organic compounds in the headspace of cultures of Staphylococcus aureus grown in nutrient, dextrose and brain heart bovine broths measured using a proton transfer reaction mass spectrometer. J. Breath Res. 2009, 3, 027001. [Google Scholar] [CrossRef]
- Maddula, S.; Blank, L.M.; Schmid, A.; Baumbach, J.I. Detection of volatile metabolites of Escherichia coli by multi capillary column coupled ion mobility spectrometry. Anal. Bioanal. Chem. 2009, 394, 791–800. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Shi, H.; Tang, C.; Zhang, R. Characteristics of volatile organic compounds produced from five pathogenic bacteria by headspace-solid phase micro-extraction/gas chromatography-mass spectrometry. J. Basic Microbiol. 2017, 57, 228–237. [Google Scholar] [CrossRef]
- Thorpe, T.C.; Wilson, M.L.; Turner, J.E.; DiGuiseppi, J.L.; Willert, M.; Mirrett, S.; Reller, L.B. BacT/Alert: An automated colorimetric microbial detection system. J. Clin. Microbiol. 1990, 28, 1608–1612. [Google Scholar] [CrossRef]
- Scotter, J.M.; Allardyce, R.A.; Langford, V.S.; Hill, A.; Murdoch, D.R. The rapid evaluation of bacterial growth in blood cultures by selected ion flow tube-mass spectrometry (SIFT-MS) and comparison with the BacT/ALERT automated blood culture system. J. Microbiol. Methods 2006, 65, 628–631. [Google Scholar] [CrossRef]
- Kuil, S.D.; Hidad, S.; Schneeberger, C.; Singh, P.; Rhodes, P.; de Jong, M.D.; Visser, C.E. Susceptibility Testing by Volatile Organic Compound Detection Direct from Positive Blood Cultures: A Proof-of-Principle Laboratory Study. Antibiotics 2022, 11, 705. [Google Scholar] [CrossRef] [PubMed]
- Hewett, K.; Drabińska, N.; White, P.; Avison, M.B.; Persad, R.; Ratcliffe, N.; Costello, B.L. Towards the Identification of Antibiotic-Resistant Bacteria Causing Urinary Tract Infections Using Volatile Organic Compounds Analysis-A Pilot Study. Antibiotics 2020, 9, 797. [Google Scholar] [CrossRef] [PubMed]
- Kunze-Szikszay, N.; Walliser, K.; Luther, J.; Cambiaghi, B.; Reupke, V.; Dullin, C.; Vautz, W.; Bremmer, F.; Telgheder, U.; Zscheppank, C.; et al. Detecting Early Markers of Ventilator-Associated Pneumonia by Analysis of Exhaled Gas. Crit. Care Med. 2019, 47, e234–e240. [Google Scholar] [CrossRef] [PubMed]
- Reupke, V.; Walliser, K.; Perl, T.; Kimmina, S.; Schraepler, A.; Quintel, M.; Kunze-Szikszay, N. Total intravenous anaesthesia using propofol and sufentanil allows controlled long-term ventilation in rabbits without neuromuscular blocking agents. Lab. Anim. 2017, 51, 284–291. [Google Scholar] [CrossRef]
- van Amsterdam, J.G.; van den Berg, C.; Zuidema, J.; te Biesebeek, J.D.; Rokos, H. Effect of septicaemia on the plasma levels of biopterin and nitric oxide metabolites in rats and rabbits. Biochem. Pharmacol. 1996, 52, 1447–1451. [Google Scholar] [CrossRef]

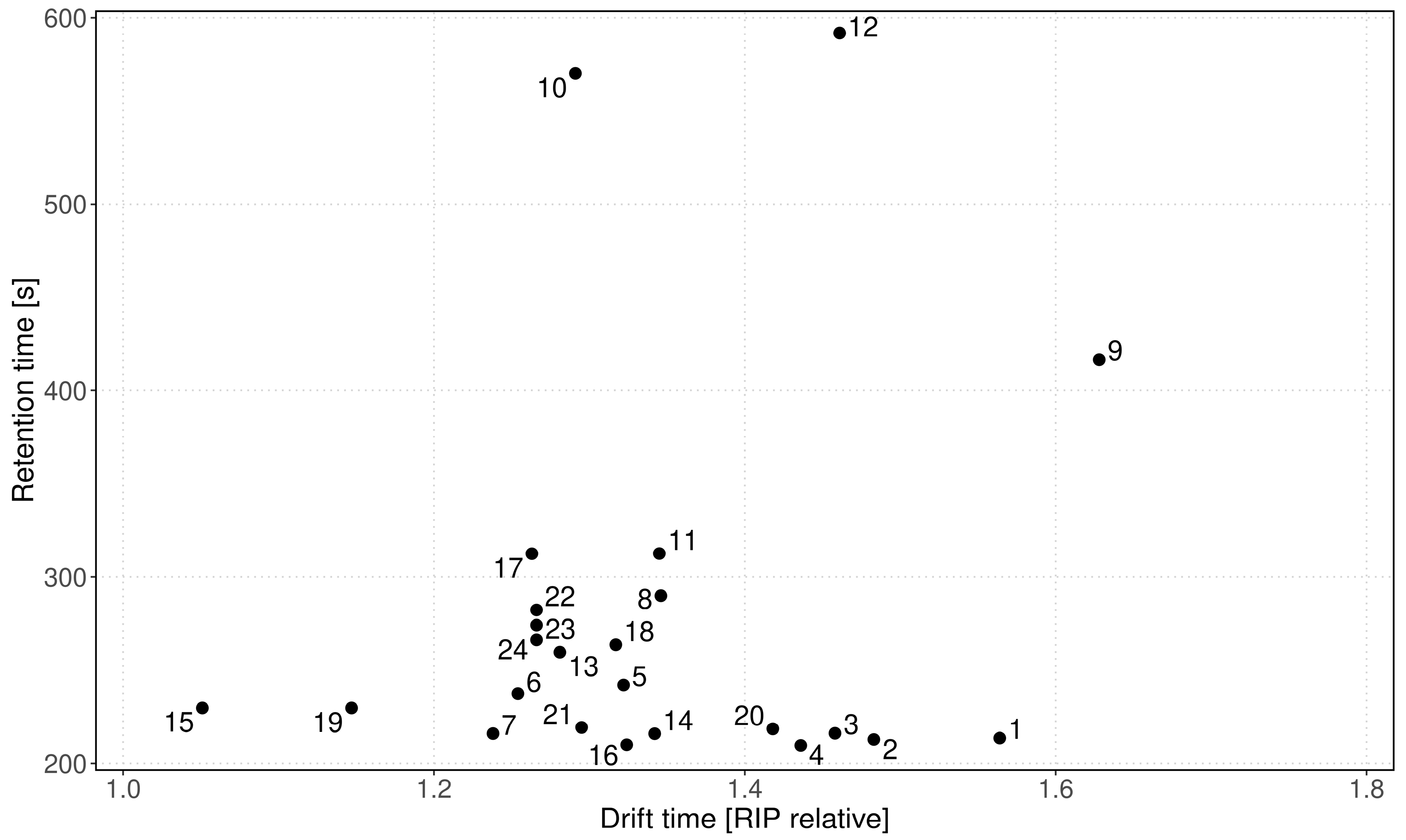

| Peak | Dt [RIP Rel] | 1/K0 [Vs/cm−2] | Rt [s] |
|---|---|---|---|
| P_1 | 1.564 | 0.779 | 213.49 |
| P_2 | 1.483 | 0.738 | 212.75 |
| P_3 | 1.458 | 0.726 | 216.07 |
| P_4 | 1.436 | 0.715 | 209.53 |
| P_5 | 1.322 | 0.658 | 241.95 |
| P_6 | 1.254 | 0.625 | 237.36 |
| P_7 | 1.238 | 0.616 | 215.91 |
| P_8 | 1.346 | 0.670 | 289.96 |
| P_9 | 1.628 | 0.811 | 416.53 |
| P_10 | 1.291 | 0.642 | 570.18 |
| P_11 | 1.345 | 0.670 | 312.51 |
| P_12 | 1.461 | 0.727 | 591.79 |
| P_13 | 1.281 | 0.638 | 259.62 |
| P_14 | 1.342 | 0.668 | 215.85 |
| P_15 | 1.051 | 0.523 | 229.73 |
| P_16 | 1.324 | 0.659 | 209.89 |
| P_17 | 1.263 | 0.629 | 312.43 |
| P_18 | 1.317 | 0.656 | 263.64 |
| P_19 | 1.147 | 0.571 | 229.73 |
| P_20 | 1.418 | 0.706 | 218.46 |
| P_21 | 1.295 | 0.645 | 219.22 |
| P_22 | 1.266 | 0.630 | 282.26 |
| P_23 | 1.266 | 0.630 | 274.07 |
| P_24 | 1.266 | 0.630 | 266.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Euler, M.; Perl, T.; Eickel, I.; Dudakova, A.; Maguilla Rosado, E.; Drees, C.; Vautz, W.; Wieditz, J.; Meissner, K.; Kunze-Szikszay, N. Blood Culture Headspace Gas Analysis Enables Early Detection of Escherichia coli Bacteremia in an Animal Model of Sepsis. Antibiotics 2022, 11, 992. https://doi.org/10.3390/antibiotics11080992
Euler M, Perl T, Eickel I, Dudakova A, Maguilla Rosado E, Drees C, Vautz W, Wieditz J, Meissner K, Kunze-Szikszay N. Blood Culture Headspace Gas Analysis Enables Early Detection of Escherichia coli Bacteremia in an Animal Model of Sepsis. Antibiotics. 2022; 11(8):992. https://doi.org/10.3390/antibiotics11080992
Chicago/Turabian StyleEuler, Maximilian, Thorsten Perl, Isabell Eickel, Anna Dudakova, Esther Maguilla Rosado, Carolin Drees, Wolfgang Vautz, Johannes Wieditz, Konrad Meissner, and Nils Kunze-Szikszay. 2022. "Blood Culture Headspace Gas Analysis Enables Early Detection of Escherichia coli Bacteremia in an Animal Model of Sepsis" Antibiotics 11, no. 8: 992. https://doi.org/10.3390/antibiotics11080992
APA StyleEuler, M., Perl, T., Eickel, I., Dudakova, A., Maguilla Rosado, E., Drees, C., Vautz, W., Wieditz, J., Meissner, K., & Kunze-Szikszay, N. (2022). Blood Culture Headspace Gas Analysis Enables Early Detection of Escherichia coli Bacteremia in an Animal Model of Sepsis. Antibiotics, 11(8), 992. https://doi.org/10.3390/antibiotics11080992






