Abstract
Antibiotics are regarded as a miracle in the medical field as it prevents disease caused by pathogenic bacteria. Since the discovery of penicillin, antibiotics have become the foundation for modern medical discoveries. However, bacteria soon became resistant to antibiotics, which puts a burden on the healthcare system. Methicillin-resistant Staphylococcus aureus (MRSA) has become one of the most prominent antibiotic-resistant bacteria in the world since 1961. MRSA primarily developed resistance to beta-lactamases antibiotics and can be easily spread in the healthcare system. Thus, alternatives to combat MRSA are urgently required. Antimicrobial peptides (AMPs), an innate host immune agent and silver nanoparticles (AgNPs), are gaining interest as alternative treatments against MRSA. Both agents have broad-spectrum properties which are suitable candidates for controlling MRSA. Although both agents can exhibit antimicrobial effects independently, the combination of both can be synergistic and complementary to each other to exhibit stronger antimicrobial activity. The combination of AMPs and AgNPs also reduces their own weaknesses as their own, which can be developed as a potential agent to combat antibiotic resistance especially towards MRSA. Thus, this review aims to discuss the potential of antimicrobial peptides and silver nanoparticles towards controlling MRSA pathogen growth.
1. Introduction
Antibiotics are one of the outstanding discoveries in the medical field in treating infectious diseases caused by pathogenic bacteria. Before the antibiotic discovery era, the lethality and death rate caused by pathogenic microorganisms was high until the accidental rediscovery of penicillin in 1928 by Alexander Fleming [1]. This rediscovery grants the exploration of other types of antibiotics such as sulphonamides, lipopeptides, aminoglycosides, fluoroquinolones, and many more [1,2]. Antibiotics also allow modern medical technology to exist as it aids in preventing infection in chemotherapy and various surgical wounds.
Although antibiotics give significant advantages in treating diseases caused by pathogenic bacteria, Alexander Fleming warns of the danger of uncontrolled antibiotic usage where resistance can be developed. The warning appeared to be true as Escherichia coli started to exhibit antibiotic resistance (AR) towards penicillin in 1940 [3]. Up until this day, antibiotic resistance has been a significant threat in the healthcare system as more bacteria developed resistance towards various classes of antibiotics. It is predicted that, by 2050, AR related death may reach 10 million per year [4,5].
A recent comprehensive report released in The Lancet [6] stated that 4.95 million AR associated death and 1.27 million AR attributed death were estimated from 204 countries in 2019. Highest AR related death can be found in Western Sub-Saharan Africa with estimated 27.3 AR attributed death per 100,000 and 114.8 AR associated death per 100,000. Meanwhile, the lowest death can be found in Australasia where only 6.5 AR attributed deaths per 100,000 and 28 AR associated deaths per 100,000. The same report also lists out six pathogenic bacteria that cause the most death in 2019 [6]. In order of the number of deaths, E. coli, S. aureus, K. pneumoniae, A. baumannii, and P. aeruginosa caused 929,000 AR attributed deaths and 3.57 million AR associated deaths.
Methicillin-resistant Staphylococcus aureus (MRSA) is an antibiotic-resistant type of S. aureus that is generally resistant towards beta-lactam antibiotics such as penicillin (methicillin and oxacillin) and cephalosporin [7,8,9]. Beta-lactam inhibits the bacterial growth by halting the cell wall synthesis process [10,11,12]. MRSA generally overcomes the beta-lactam effects by producing beta-lactamase and altering the binding site for cell wall synthesis [7,8,9,13]. The current clinically approved method to treat MRSA infection involves different antibiotic classes such as vancomycin and teicoplanin [14,15]. These glycopeptide antibiotics act on the bacterial cell wall similar to beta-lactam, but it utilises different target by binding to the peptidoglycan side chain, which prevents peptidoglycan crosslinking [13,14,15]. However, the newer MRSA strain started to exhibit resistance towards glycopeptide antibiotics, which makes it difficult to treat the infection [13,14]. Other types of antibiotics such as mupirocin, clindamycin, fusidic acid, and co-trimoxazole also used a second line option in treating MRSA [16]. However, these antibiotics can only be prescribed when there is no other alternative available due to the risk of resistance [16,17]. Thus, alternatives to treat MRSA without the use of different classes of antibiotics are greatly needed.
Recent scientific development showed some promising potential in inhibiting MRSA through the usage of antimicrobial peptides (AMPs) and silver nanoparticles (AgNPs). These two agents exhibit broad-spectrum antimicrobial properties, which makes them the suitable candidates to combat MRSA threat [18,19,20,21]. AMPs are naturally occurring molecules that can be found in all types of life, which are involved in innate immunity defense [20,21]. AMP mainly takes action on the bacterial membrane, and it can be simplified into two mechanisms of action: membranolytic and non-membranolytic action [21,22,23]. Membranolytic action can be defined as direct AMP action on the bacterial membrane, which greatly alters its structural integrity [23,24,25]. Meanwhile, non-membranolytic action is when AMPs were internalised into the cells without causing major damage to the membrane, but it targets the vital intracellular components [26,27,28].
AgNPs are metallic nanoparticles that have unique physicochemical properties including optical, thermal, electrical and high electrical conductivity in comparison to its bulk form due to its nano size [29,30]. Their enhanced antimicrobial properties mainly contributed with their large surface area per volume area, which allows more antibacterial contact with the pathogenic bacteria [19,31,32]. AgNPs are also steadily gaining interest due to its multiple mechanism of actions. AgNPs generally act on membranes by disrupting it through hole formation, direct adhesion and internalisation into the membrane, excessive ROS generation and alteration in signalling pathways [30,33,34,35]. Despite their excellent antimicrobial properties, AMPs and AgNPs have their own limitations, but, through the combination of both agents, a positive synergistic effect can be observed [36,37]. Thus, this review discusses about MRSA, its mechanism of resistance, advantages and limitations of AMPs and AgNPs as its own. This topical review also discusses the combination of AMPs–AgNPs in combating bacteria, particularly MRSA and S. aureus, as no other review has been reported with the combination of the two antimicrobial agents. The review process was done based on a literature search in Google Scholar with the keywords, antimicrobial peptide, antibiotic resistance, MRSA, silver nanoparticles and S. aureus, with most of it published from 2016–2022 and some of the papers published before 2016.
2. Methicillin-Resistant Staphylococcus aureus
Staphylococcus aureus is Gram-positive bacteria with round shape morphology that commonly can be found in the body as a part of its microbiota. Despite it acting commensally on the human body, it can be opportunistic bacteria since it can cause skin infections and food poisoning. Methicillin-resistant Staphylococcus aureus (MRSA) is an antibiotic-resistant strain of S. aureus that are mainly resistant to beta-lactam antibiotics. MRSA was first identified in 1961 in United Kingdom just a year after methicillin was introduced to treat S. aureus infection [38,39]. Despite methicillin no longer being used clinically, the term methicillin-resistant is still used to reflect S. aureus resistance towards commercial antibiotics such as beta-lactams antibiotics including oxacillin. According to World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC), MRSA has been a big and serious threat on the pathogenic bacteria watch list respectively [40,41]. According to recent systematic analysis in the Lancet in 2019, MRSA alone caused more than 100,000 deaths [6]. Originally, MRSA are common in the healthcare setting, and this type of MRSA is often dubbed as healthcare-associated or hospital-acquired MRSA (HA-MRSA) [42]. The infection can be spread through direct contact with an infected wound or contaminated hands. Untreated infection can cause serious bloodstream infections, surgical site infections, sepsis and pneumonia [7,43]. Other types of MRSA are community-associated (CA-MRSA) and livestock-associated MRSA (LA-MRSA) [39,43].
Beta-lactam antibiotics act on the bacterial cell wall by binding to the penicillin binding protein (PBP), which is responsible for the crosslinking of N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc) [10,11]. This crosslinking will form a cell wall that protects the bacteria from external threats. MurNAc subunits have pentapeptide chains attached to it, typically with a sequence of l-Ala-γ-d-Glu-l-lysine (or -meso-diaminopimelic acid)-d-Ala-d-Ala [11]. Beta-lactam antibiotics such as penicillin, cephalosporin, carbapenem and monobactams have a beta-lactam ring which shared similar structural homology to d-Ala-d-Ala of the pentapeptide chain [10,44]. The d-Ala-d-Ala substrate is responsible for the PBP binding site for crosslinking, and this similarity causes beta-lactam antibiotics bind to PBP, causing the crosslinking between the glycan stands to be halted [11]. The binding between beta-lactam and PBP causes the build-up of peptidoglycan precursors which trigger autolytic digestion of old peptidoglycan by hydrolase [10]. Without the production of new peptidoglycan, the structural integrity of the cell wall is significantly disrupted and led to cell damage due to high internal osmotic pressure [11,12].
MRSA overcomes this detrimental effect by producing beta-lactamase, an enzyme to break down the antibacterial effect of beta-lactam antibiotics and production of the mecA gene, which changes the penicillin-binding protein (PBP) confirmation. Beta-lactamase is an enzyme produced by bacteria to counteract the effects of beta-lactam antibiotics. This enzyme hydrolyses beta-lactam in the periplasmic space, thus deactivating it before PBP interaction [4]. Beta-lactamase production in staphylococci is controlled by the repressor BlaI and the sensor protein BlaR1 (Figure 1a) [44]. The genes encoding beta-lactamase, the blaZ-blaR1-blaI genes, are repressed by BlaI is from transcribing beta-lactamase when beta-lactam is absent [10,12]. Once beta-lactam is presented, the transmembrane sensor, BlaR1, covalently binds to it and irreversibly acylated at its active site serine. This will activate the intracellular zinc metalloprotease domain of BlaR1 and cause BlaI that are bound to blaI-blaRI operator to proteolytically cleave and dissociate from its binding site [12]. The dissociation allows blaZ to be upregulated and transcribed beta-lactamase enzyme. The produced beta-lactamase enzyme later hydrolyses beta-lactam antibiotic by hindering it from binding with PBP [10,12]. Thus, the peptidoglycan synthesis of the bacteria can be initiated as usual.
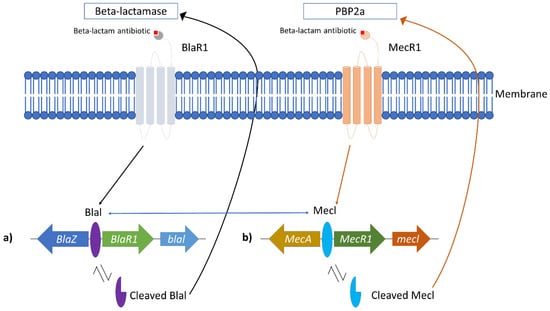
Figure 1.
Correlation between Blal and MecA role in MRSA resistance. (a) bla operon responsible for beta-lactamase production and (b) mec operon responsible for the alteration of normal PBP to PBP2a. The blue arrows indicate that bla and mec operon shared similarities, which allows the repressor (Blal and Mecl) to bind to each operon.
In MRSA, the PBP responsible for the peptidoglycan cross-linking is altered to novel penicillin-binding protein 2a (PBP2a), which has a lower binding affinity to beta-lactam antibiotics [39]. The resistance arose from the mecA gene located in the staphylococcal cassette chromosome mec (SCCmec), and this resistance gene can be passed to other populations through horizontal gene transfer [12]. Upon acquiring the mecA gene, it will be localized in the S. aureus chromosome. The production of PBP2a is controlled by MecI repressor and transmembrane MecR1 sensor protein (Figure 1b) [10]. In the absence of beta-lactam antibiotics, MecI represses mecA gene expression by binding to the promoter region of mec operon [10,39]. In the presence of beta-lactam antibiotics, the antibiotic binds to the MecR1 sensor protein. It triggers autolytic activation of the metalloproteinase domain in the cytoplasm part of MecR1, causing signal transduction to be activated [12]. The latter caused the MecI repressor to be proteolytically cleaved from its binding site, and this allows the expression mecA to produce PBP2a [10]. The PBP2a production allows the peptidoglycan wall synthesis to continue without the interaction of beta-lactam antibiotics due to its low binding affinity to the antibiotic [7,39]. Interestingly, the mec operon shared a similar structure and function with the bla operon, which produces beta-lactamase [7,12]. This similarity allows the Blal repressor to bind to the mec operon to repress mecA transcription (Figure 1) [10].
3. Antimicrobial Peptides (AMPs)
Antimicrobial peptides (AMPs) are naturally occurring host defense mechanisms against infections. AMPs can be found in all living organisms such as plants, microorganisms and animals [20,23]. Typical AMPs consist of 5–50 amino acid chains and have amphipathic or cationic structure. Despite AMPs being naturally occurring, synthetic AMPs have been developed by the scientist to overcome the naturally occurring AMP limitations [21]. While naturally occurring AMPs are susceptible to proteolytic degradation, synthetic AMPs have a longer half-life, and it is designed to improve their antimicrobial properties. AMPs then can be divided into four main groups based on its secondary structure including amphipathic alpha-helices, beta-sheets, a combination of both alpha and beta structure (mixed) and extended structure (without alpha and beta structure) (Figure 2) [21].
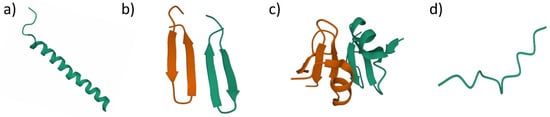
Figure 2.
Antimicrobial peptides structural classification based on their secondary structure. (a) alpha-helices AMPs, human LL-37 (PDB ID: 2K60); (b) beta-sheets AMPs, protegrin-1 (PDB ID: 1ZY6); (c) mixed structure AMPs, human beta-defensin-2 (PDB ID: 1FD4); (d) extended structure AMPs, indolicidin (PDB ID: 5ZVN).
Alpha helices’ AMPs generally contain two amino acids that are adjacent to each other with the distance of 1.5 Å (0.15 nm) [23]. The most studied AMPs in this group is LL-37 (amino acid sequence: LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES), AMPs that can be found in the human body that act as the host defense towards bacterial infections [21,23,45,46]. Beta-sheets’ AMPs have at least two beta strands with disulfide bonds. Protegrin-1 (amino acid sequence: RGGRLCYCRRRFCVCVGR-NH2), which is isolated from pigs, is one of the examples in this group which exhibits antimicrobial activity against fungi, bacteria and some enveloped viruses [23,26]. Mixed structure usually consists of a combination of alpha-helix and beta-sheet that are packed against each other. Human-beta-defensin-2 is one of the well-studied AMPs in this group [23,28]. The extended structure is a unique group of AMPs that consists of two or more tryptophan, arginine, histidine and proline structure in single molecules. Cattle neutrophil isolated AMPs, indolicidin (amino acid sequence: ILPWKWPWWPWRR-NH2), are in this group [23,45].
In terms of AMP mechanism of actions, it can be divided into two main categories, membrane disruptive and non-membrane disruptive AMPs [22,26,28]. Membrane disruptive AMP can be further divided into the toroidal-pore model, barrel-stave model and carpet model (Figure 3) [26,27]. The toroidal-pore model is where AMPs form pores in the membrane (1–2 nm diameter) vertically [27]. This will also cause the phospholipid head to bend due to the insertion of AMPs [28]. In the barrel-stave model, AMPs bind to the cell membrane and aggregate before penetrating the membrane [26]. During this process, hydrophobic regions of AMPs are inserted into the phospholipid membrane, while the hydrophilic regions of AMPs are facing the outer part of the membrane pore [27]. This will cause uncontrolled cellular movement for the cell, which will lead to cell death. The carpet-like model destroys the membrane in a detergent-like manner [22]. AMPs are first arranged onto the cell membrane by their hydrophobic part facing the phospholipid bilayer, which alters its surface tension. The altered surface tension later causes micelles formation as the results of peptide accumulation and destroys the membrane [22,27].
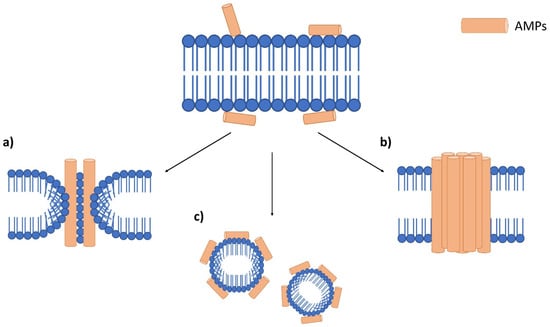
Figure 3.
Antimicrobial peptide mechanism on the bacterial membrane. Accumulation of AMPs on the bacterial membrane surface, which leads to three main membranolytic mechanisms. (a) toroidal pore model which forms pores on the membrane, (b) barrel-stove model which AMPs aggregate before entering the membrane and (c) a carpet-like model which promotes the formation of micelles.
A non-membrane disruptive mechanism is rarely studied in AMP research, but recent advancement showed that AMPs are internalised into cells and interacts with vital intracellular targets and even inhibits cell wall biosynthesis [28]. This includes inhibition of protein and nucleic acid synthesis, cell division and protease activity [23,26,28]. AMPs inhibit protein synthesis by directly interacting with the transcription and translation process. PR-39 AMPs isolated from a pig’s small intestine can inhibit protein synthesis, which causes proteins degradation that are required for DNA synthesis [28]. Indolicilin induces degradation of nucleic acid by binding to the double stranded DNA, which causes the DNA synthesis to be halted [23,26]. Teixobactin AMPs bind to lipid II and lipid III (precursors of cell wall), which later inhibits the cell wall synthesis process [28]. Based on the promising antimicrobial action of AMPs, Table 1 showed some examples of AMPs that are effective towards MRSA and wild type S. aureus.

Table 1.
Examples of natural antimicrobial peptides that are effective towards Staphylococcus aureus (methicillin-susceptible and MRSA).
As the innate immune system, AMPs have broad spectrum antimicrobial properties which are said to be effective towards pathogenic microorganisms [21,23,45]. These antimicrobial properties are greatly enhanced as AMPs can be found abundantly at the site of the infection, which makes it more time efficient since it can react faster to combat the infection [23,28]. Resistance towards AMPs is also said to be low, which makes it one of the suitable candidates to combat MRSA [28]. Besides that, AMPs also have good water solubility and thermal stability [23,45]. However, AMPs do possess weakness as naturally occurring AMPs are susceptible towards proteolytic degradation, which limits their potential [23,28]. In addition, AMP production and purification can be costly sometimes. Despite their broad antimicrobial spectrum, it can be a challenge to be used medically as some AMPs might induce hypersensitivity after application and might cause immunogenicity and toxicity when it is administered in humans [21,23,45].
4. Silver Nanoparticles (AgNPs)
Silver nanoparticles are the product of nanotechnology which are particles of silver that are ranging in size from 1 to 100 nm [38,39]. Their nanosized greatly enhances its broad-spectrum antibacterial properties as it has larger surface area per volume ratio [29,33,35,51]. Due to their unique properties in terms of optical, electrical, magnetic and antibacterial, AgNPs have various applications, which include medical appliances, optical sensors, cosmetics, drug delivery, textiles, keyboards, wound dressings and food packaging [35,51,52,53]. AgNPs are gaining popularity due to their multiple mechanisms of action on bacteria (Figure 4), which include direct adhesion of AgNPs on the bacterial surface and altering the membrane structural integrity [32,54]. Next, AgNPs penetrate inside the bacterial cell and interact with its intracellular components, damaging it until it cannot perform vital cellular processes [32,54]. AgNPs are also able to induce reaction oxygen species and free radical generation, thus causing irreversible oxidative damage to the bacteria [55,56]. Alteration of vital signaling transduction, which is necessary for the bacterial life cycle, is also one of the mechanisms exhibited by AgNPs [52,57].
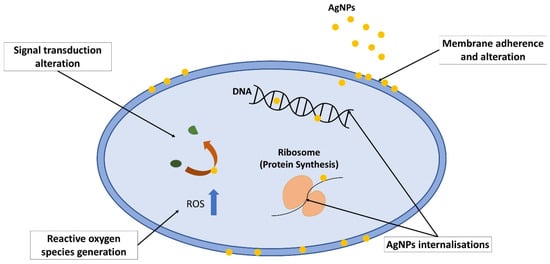
Figure 4.
Mechanism of action exhibited by silver nanoparticles on bacteria, which includes membrane adherence and alteration, AgNP internalisations, which later induces cellular damage, reactive oxygen species generation, which causes oxidative stress, and signal transduction alteration.
When AgNPs is in contact with the outer membrane of the bacteria, it adheres to it due to the difference in electrostatic charge [58]. This electrostatic force is driven by the positively charged AgNPs and negatively charged bacterial cell membrane [34,59]. The negative charge on the membrane is contributed by the presence of the amino, carboxyl and phosphate group [60,61]. This metal depletion on the membrane causes pit formation on the membrane as first visualized using transmission electron microscopy by Sondi and Salopek-Sondi [62]. The phenomenon causes the membrane to fail to regulate vital cellular content movements and may lead to cell death.
After AgNPs attached on the membrane, its permeability and structural integrity are greatly altered, and this causes some portion of AgNPs to infuse into the cell. This statement is further strengthened by several studies which revealed that AgNPs penetrate the cells through transmission electron microscope analysis on bacteria [19,62,63]. In addition, in the presence of oxygen and proton, AgNPs dissociate to Ag+ ions, which also facilitates the infusion [64]. When AgNPs are internalised into the cell, it interacts with cellular molecules and structures such as protein, DNA and lipids. For instance, AgNPs interrupt protein synthesis by interacting with ribosome by denaturing it, which halts the translation process [65,66]. AgNPs also interacts with DNA molecules, which may cause denaturation and shearing of DNA and also cell division interruption [32,67]. The interaction causes the bacteria to lose the ability to undergo division, reproduction and eventually cell death [54,68].
Reactive oxygen species or ROS are also the culprits for the bacterial growth inhibition. AgNPs generate a high level of ROS, which induces oxidative stress in the cell [55,56]. Oxidative stress causes a vital cellular component breakdown, such as protein, RNA and DNA, which led to the alteration of membrane permeability and increased cellular component leakage from the cell [52,69]. This will cause irreversible oxidative damage to the bacteria and cell death [33,65].
Tyrosine phosphorylation is important in activation of various proteins such as RNA polymerase sigma factor that are essential in the bacterial transcription process [34,57]. Increased dephosphorylation of tyrosine profile might inhibit vital processes in bacteria such as polysaccharide biosynthesis of bacterial capsule in bacteria [33,34]. AgNPs can alter tyrosine phosphorylation, which led to failed regulation of the cellular process and homeostasis, which later destroys the cells [52,57]. Since AgNPs managed to exhibit multiple mechanisms of action on bacteria, it is said to be effective to combat MRSA. Table 2 showed some AgNPs action on inhibiting the growth of MRSA.

Table 2.
Antibacterial effect of AgNPs on MRSA.
Despite AgNPs possessing broad spectrum antibacterial activity on MRSA, it tends to aggregate and reduces its antibacterial properties [36,77]. Other than that, some AgNPs can be toxic in vitro and in vivo when it is administered as its own without a capping agent, which limits its toxicity effect towards cells [78]. Oxidation of AgNPs also contributes to its weakness to be developed as a promising antibacterial agent [78,79,80].
5. AMP and AgNPs Combination on MRSA or MSSA
Despite AMPs and AgNPs having their own weaknesses on their own, the combination of these two, or sometimes with the addition of polymer, enhances its antibacterial properties while greatly reducing their toxicity effects. Synergistic effect in terms of stronger antibacterial activity of these two agents can also be observed once they are administered together.
A study by Jin et al. utlises AMPs, Tet-213 and AgNPs that are loaded onto porous silicon microparticles [36]. Tet-213 is a 10 amino acid peptide (sequence: KRWWKWWRRC) that possesses broad spectrum activity due to the presence of thiol group and, with the combination of AgNPs, the antimicrobial effect increases drastically. The presence of porous silicon microparticles (PSiMPs) acts as a carrier for effective delivery of the antimicrobial agent to the infected site [36,81]. PSiMPs was chosen due to its tunable pore size, biocompatibility and decompatibility. However, PSiMPs only dissociate in an alkaline condition as it is normally acidic during the early stage of infection [82]. Despite the carrier only being able to dissociate in alkaline conditions, the presence of ROS also allows PSiMPs be to be dissociated easily. When ROS is high during the wound infection, it allows the carrier to be disintegrated and releases silver ions from AgNPs together with Tet-213. The acidic condition also allows a gradual release of AgNPs-AMPs, which allows more effective and stable antimicrobial action. In this study, for the combination of these agents, the MIC value was greatly reduced to 2 mg/mL in comparison to AgNPs-PsiMPs (2.5 mg/mL) and AMPs-PsiMPs (>5 mg/mL) on S. aureus [36]. In-vitro testing on mouse fibroblast (NIH3T3) cells and human immortal keratinocyte (HaCaT) showed low toxicity effects as this complex does not affect the cells’ proliferation. This AgNPs-AMPs-PSiMPs combination also exhibits low toxicity and faster wound healing on rats infected with S. aureus [36]. The faster wound healing contributed with the release of silicon ions in the complex, with the help of AgNPs and AMPs to reduce the bacterial infection in the wound. Note that silicon ions promote wound healing by activating the epidermal growth factor receptor (EGFR), epidermal growth factor (EGF) and extracellular signal-related kinase (ERK) signaling pathway [36,83,84].
A star conjugated PCL-b-AMPs nanocomposite was also used in stabilising AgNPs and enhancing antimicrobial activity of it with the help of AMPs [77]. Star conjugated PCL-b-AMPs consist of polycaprolactone (PCL) and polypeptide (Phe8-stat-Lys32), which are later loaded with AgNPs. This complex is relatively stable at room temperature for three months with any sign of aggregations. In this case, PCL-b-AMPs penetrate the negatively charged membrane since this complex is positively charged. This penetration allows AgNPs to be released in the cytoplasm and the deactivating of vital cellular components. This complex managed to exhibit enhanced inhibition on S. aureus (27.6 mm) when compared to the combination of PCL-b-AMPs (19.1 mm) and AgNPs (12.7 mm) alone. A low MIC value (4 µg/mL) is also observed when PCL-b-AMPs with AgNPs is tested on MRSA [77]. This suggests that a synergistic effect of AMPs and AgNPs allows higher inhibition on the bacterial growth. A damaged membrane was also observed on MRSA, which later led to cell death [77,85]. This complex also showed no sign of resistance even after 21 passage exposure with a sub-lethal MIC value of the complex when tested on the wild type S. aureus [77]. It also showed low cytotoxicity towards normal mouse fibroblast cells (L929) as it managed to retain up to 80% of cell viability after 48 h. The PCL-b-AMPs managed to reduce AgNPs toxicity by only releasing it to the target site besides from their biocompatibility.
Polymersomes, which are polymeric biocompatible vesicle, were also used for an effective synergistic antimicrobial effect of AMPs and AgNPs [85]. PR-39 peptide was utilised in the polymeric compound as it is effective towards inhibiting bacterial growth. Originally, porcine PR-39 peptide could not translocate across the bacterial membrane as MRSA produces protease which degrades the AMPs. For the addition of polymersomes and AgNPs, the MRSA growth was totally eradicated under 23 h [85]. Polymersomes and AgNPs allow the complex to translocate the cells and release the antimicrobial agent to inhibit the bacterial growth. From the scanning electron microscopy, apparent damage on MRSA membrane can be observed, which led to cell death [27,85]. A low toxicity level toward CCL-110 human dermal fibroblast (HDF) cell lines can be observed since the coating reduces the toxicity effects of AgNPs and stabilises AMPs [77,85].
A combination of protegrin-1 AMPs and gelatinized coated AgNPs also greatly enhances its antimicrobial properties as it exhibits low MIC value (6 µg/mL) in comparison to AgNPs (48 µg/mL) and AMPs (8.5 µg/mL) treatment alone [79]. It is said that this complex limits MRSA growth by membrane permeabilisation (possibly through the toroidal pores model) [28,79]. The same study also combines AgNPs with another type of AMPs, Indolicidin [79]. This combination also exhibits excellent antimicrobial properties as its MIC value to inhibit MRSA is 12 µg/mL. The MIC value for indolicidin alone on MRSA 40 µg/mL is relatively high in comparison to the AgNPs-Indolicidin complex. This complex acted on MRSA by self-translocating into the cells by forming an apparent pore on the membrane and interacting with nucleic acid, which halts the DNA synthesis [22,27]. Low haemolytic activity can be observed when the complex was tested with human erythrocytes. However, more optimisations are required as they showed a cytotoxicity effect towards cancerous and normal cell lines, which grants in vivo assessment to elucidate the actual toxicity.
A novel composite of AgNPs and designed AMPs P-13 (amino acid sequence: KRWWKWWRRCECG) were tested against S. aureus (ATCC 25923) [86]. Based on the MIC values, this composite manages to inhibit bacteria effectively at lower concentration (7.8 ± 0.05 µg/mL) compared to AgNPs and AMPs alone with 7.8 ± 0.05 µg/mL and >500 ± 0.04 µg/mL, respectively. Interestingly, with the addition of P-13 to AgNPs, a drastic toxicity reduction can be observed on mouse fibroblast cells (NIH-3T3) [86]. This addition allows AMPs to stabilise AgNPs and reduce its cytotoxicity effect in comparison to AgNPs alone. It is proposed that this complex inhibits bacteria growth by adhesion to the bacteria through electrostatic force and was internalised into the cell reacting with vital cellular components. This causes cellular leakage out of the cell, which led to cell death [27,86].
Another study by Li et al. developed multifunctional peptide (MFP)-coated silver nanoparticles as an alternative to combat antibiotic resistance [78]. In this study, AMPs tachyplesin-1 and target peptide N-ac-PGP-PEG were combined to adsorb AgNPs through electrostatic interaction. This complex was proven to be effective at inhibiting S. aureus and MRSA growth with MIC values of 8 µg/mL and 16 µg/mL, respectively [78]. Despite the MIC for vancomycin, an antibiotic control in this experiment is much lower than the complex (2 µg/mL); this complex was proven to be a promising agent to inhibit the bacterial growth with future optimisations.
The AMP@PDA@AgNPs nanocomposite was created through polymerisation to inhibit biofilm formation by S. aureus [80]. PDA was added as a delivery agent, which allows more effective AMPs and AgNPs delivery to the target site. This allows more effective internalisation into the cell to exhibit its antimicrobial activity. This nanocomposite showed no cytotoxicity effect even at a high concentration (400 µg/mL) when tested on human embryonic kidney (HEK293T) cells. To inhibit S. aureus growth, only a concentration of 25 μg/mL was required, which is much lower than the concentration used in the cytotoxicity assessments. This complex also managed to reduce biofilm formed by the bacteria by reducing the expression of biofilm forming genes (las I and rh II, fim H) [80]. Table 3 showed other combinations of AMPs with AgNPs that are able to inhibit S. aureus or MRSA growth effectively.

Table 3.
Combinations of AMPs and AgNPs with addition of polymer for inhibiting S. aureus or MRSA growth.
In general, AMPs and AgNPs can be combined together to exhibit synergistic antimicrobial effect or carrier/polymer can be added to the complex to allow more effective AMPs and AgNPs for the target site without exhibiting a high toxicity effect (Figure 5).
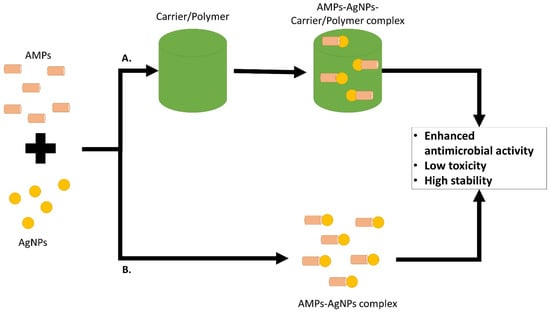
Figure 5.
Combination of AMPs and AgNPs or together with the addition of carrier/polymer for a more effective delivery system to the target site, enhanced antimicrobial activity and lower toxicity effect in comparison to the agents on their own.
6. Conclusions and Future Perspectives
Despite as antibiotic resistance threat that keeps on increasing year by year, scientists are never giving up on finding alternatives to curb the spreading of antibiotic resistance. The emergence of antimicrobial peptides with multiple membranolytic and non-membranolytic mechanism cast light on the antibiotic resistance research. A lower rate of microbial resistance towards AMPs also allows it to be studied intensively in combating MRSA, a worldwide pathogenic threat. The introduction of silver nanoparticles in this modern era also allows its utilisation in combating antibiotic-resistant bacteria including MRSA. Multiple mechanisms, such as direct adherence and internalization of AgNPs, allow it to exhibit an antimicrobial effect effectively by inducing ROS and alteration of signal transduction. Nevertheless, AMPs and AgNPs each possess their own weaknesses, which include toxicity and instability. A combination of these two agents somehow overcomes these weaknesses by stabilising these agents to the target site. A synergistic effect can also be observed once these two agents are combined to inhibit bacterial growth. A lower toxicity effect in vitro and in vivo can also be observed. Although this research is still the beginning, future optimisations can be done, especially in terms of enhanced complex stability, lower dosage required in inhibiting bacteria and lower toxicity, in vitro and in vivo need to be done. Clinical research also needs to be done before the co-application of AMPs and AgNPs can be truly used in combating antibiotic resistance, especially towards MRSA in humans.
Author Contributions
Conceptualization, M.A.A.M. and W.I.W.I.; writing—original draft preparation, M.A.A.M.; writing—review and editing, M.A.A.M., N.A.H., M.M. and W.I.W.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable.
Acknowledgments
Special thanks to Research Management Centre and Faculty of Science and Marine Environment, Universiti Malaysia Terengganu for supporting this project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Rigol, S. A Brief History of Antibiotics and Select Advances in Their Synthesis. J. Antibiot. 2018, 71, 153–184. [Google Scholar] [CrossRef]
- Abraham, E.P.; Chain, E. An Enzyme from Bacteria Able to Destroy Penicillin. Nature 1940, 146, 837. [Google Scholar] [CrossRef]
- Dixit, A.; Kumar, N.; Kumar, S.; Trigun, V. Antimicrobial Resistance: Progress in the Decade since Emergence of New Delhi Metallo-β-Lactamase in India. Indian J. Community Med. Off. Publ. Indian Assoc. Prev. Soc. Med. 2019, 44, 4–8. [Google Scholar] [CrossRef]
- Lv, J.; Deng, S.; Zhang, L. A Review of Artificial Intelligence Applications for Antimicrobial Resistance. Biosaf. Health 2021, 3, 22–31. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Kirmusaolu, S. MRSA and MSSA: The Mechanism of Methicillin Resistance and the Influence of Methicillin Resistance on Biofilm Phenotype of Staphylococcus Aureus. In The Rise of Virulence and Antibiotic Resistance in Staphylococcus aureus; Enany, S., Crotty Alexander, L.E., Eds.; InTech: Hong Kong, China, 2017; ISBN 978-953-51-2983-7. [Google Scholar]
- Vestergaard, M.; Frees, D.; Ingmer, H. Antibiotic Resistance and the MRSA Problem. Microbiol. Spectr. 2019, 7, 18. [Google Scholar] [CrossRef]
- Nandhini, P.; Kumar, P.; Mickymaray, S.; Alothaim, A.S.; Somasundaram, J.; Rajan, M. Recent Developments in Methicillin-Resistant Staphylococcus Aureus (MRSA) Treatment: A Review. Antibiotics 2022, 11, 606. [Google Scholar] [CrossRef]
- King, D.T.; Sobhanifar, S.; Strynadka, N.C.J. The Mechanisms of Resistance to β-Lactam Antibiotics. In Handbook of Antimicrobial Resistance; Berghuis, A., Matlashewski, G., Wainberg, M.A., Sheppard, D., Eds.; Springer: New York, NY, USA, 2017; pp. 177–201. ISBN 978-1-4939-0693-2. [Google Scholar]
- Pandey, N.; Cascella, M. Beta Lactam Antibiotics. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Kırmusaoğlu, S.; Gareayaghi, N.; Kocazeybek, S.B. Introductory Chapter: The Action Mechanisms of Antibiotics and Antibiotic Resistance. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; Kırmusaoğlu, S., Ed.; IntechOpen: Hong Kong, China, 2019; ISBN 978-1-78985-789-4. [Google Scholar]
- Cameron, D.R.; Jiang, J.-H.; Kostoulias, X.; Foxwell, D.J.; Peleg, A.Y. Vancomycin Susceptibility in Methicillin-Resistant Staphylococcus Aureus Is Mediated by YycHI Activation of the WalRK Essential Two-Component Regulatory System. Sci. Rep. 2016, 6, 30823. [Google Scholar] [CrossRef]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef]
- Stogios, P.J.; Savchenko, A. Molecular Mechanisms of Vancomycin Resistance. Protein Sci. Publ. Protein Soc. 2020, 29, 654–669. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.M.; Goodman, A.L.; Horner, C.; Jenkins, A.; Brown, E.M. Treatment of Methicillin-Resistant Staphylococcus Aureus (MRSA): Updated Guidelines from the UK. JAC-Antimicrob. Resist. 2021, 3, dlaa114. [Google Scholar] [CrossRef] [PubMed]
- Montravers, P.; Eckmann, C. Cotrimoxazole and Clindamycin in Skin and Soft Tissue Infections. Curr. Opin. Infect. Dis. 2021, 34, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Ciandrini, E.; Morroni, G.; Arzeni, D.; Kamysz, W.; Neubauer, D.; Kamysz, E.; Cirioni, O.; Brescini, L.; Baffone, W.; Campana, R. Antimicrobial Activity of Different Antimicrobial Peptides (AMPs) against Clinical Methicillin-Resistant Staphylococcus aureus (MRSA). Curr. Top. Med. Chem. 2018, 18, 2116–2126. [Google Scholar] [CrossRef]
- Ansari, M.A.; Alzohairy, M.A. One-Pot Facile Green Synthesis of Silver Nanoparticles Using Seed Extract of Phoenix Dactylifera and Their Bactericidal Potential against MRSA. Evid. Based Complement. Altern. Med. 2018, 2018, 1860280. [Google Scholar] [CrossRef]
- Baharin, N.H.Z.; Mokhtar, N.F.K.; Desa, M.N.M.; Gopalsamy, B.; Zaki, N.N.M.; Yuswan, M.H.; Muthanna, A.; Dzaraly, N.D.; Abbasiliasi, S.; Hashim, A.M.; et al. The Characteristics and Roles of Antimicrobial Peptides as Potential Treatment for Antibiotic-Resistant Pathogens: A Review. PeerJ 2021, 9, e12193. [Google Scholar] [CrossRef]
- Patrulea, V.; Borchard, G.; Jordan, O. An Update on Antimicrobial Peptides (AMPs) and Their Delivery Strategies for Wound Infections. Pharmaceutics 2020, 12, 840. [Google Scholar] [CrossRef]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial Peptides as Therapeutic Agents: Opportunities and Challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef]
- Frimodt-Møller, J.; Campion, C.; Nielsen, P.E.; Løbner-Olesen, A. Translocation of Non-Lytic Antimicrobial Peptides and Bacteria Penetrating Peptides across the Inner Membrane of the Bacterial Envelope. Curr. Genet. 2022, 68, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef] [PubMed]
- Seyfi, R.; Kahaki, F.A.; Ebrahimi, T.; Montazersaheb, S.; Eyvazi, S.; Babaeipour, V.; Tarhriz, V. Antimicrobial Peptides (AMPs): Roles, Functions and Mechanism of Action. Int. J. Pept. Res. Ther. 2020, 26, 1451–1463. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial Peptides: Mechanism of Action, Activity and Clinical Potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Rosman, N.S.R.; Masimen, M.A.A.; Harun, N.A.; Idris, I.; Ismail, W.I.W. Biogenic Silver Nanoparticles (AgNPs) from Marphysa Moribidii Extract: Optimization of Synthesis Parameters. Int. J. Technol. 2021, 12, 635. [Google Scholar] [CrossRef]
- Huq, M.A.; Ashrafudoulla, M.; Rahman, M.M.; Balusamy, S.R.; Akter, S. Green Synthesis and Potential Antibacterial Applications of Bioactive Silver Nanoparticles: A Review. Polymers 2022, 14, 742. [Google Scholar] [CrossRef]
- Rosman, N.S.R.; Harun, N.A.; Idris, I.; Ismail, W.I.W. Eco-Friendly Silver Nanoparticles (AgNPs) Fabricated by Green Synthesis Using the Crude Extract of Marine Polychaete, Marphysa Moribidii: Biosynthesis, Characterisation, and Antibacterial Applications. Heliyon 2020, 6, e05462. [Google Scholar] [CrossRef]
- Lee, S.; Jun, B.-H. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef]
- Mikhailova, E.O. Silver Nanoparticles: Mechanism of Action and Probable Bio-Application. J. Funct. Biomater. 2020, 11, 84. [Google Scholar] [CrossRef]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic Basis of Antimicrobial Actions of Silver Nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef]
- Salleh, A.; Naomi, R.; Utami, N.D.; Mohammad, A.W.; Mahmoudi, E.; Mustafa, N.; Fauzi, M.B. The Potential of Silver Nanoparticles for Antiviral and Antibacterial Applications: A Mechanism of Action. Nanomaterials 2020, 10, 1566. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yang, Y.; Duan, W.; Qu, X.; Wu, J. Synergistic and On-Demand Release of Ag-AMPs Loaded on Porous Silicon Nanocarriers for Antibacteria and Wound Healing. ACS Appl. Mater. Interfaces 2021, 13, 16127–16141. [Google Scholar] [CrossRef] [PubMed]
- Salouti, M.; Mirzaei, F.; Shapouri, R.; Ahangari, A. Synergistic Antibacterial Activity of Plant Peptide MBP-1 and Silver Nanoparticles Combination on Healing of Infected Wound Due to Staphylococcus Aureus. Jundishapur J. Microbiol. 2016, 9, e27997. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Robinson, D.A.; Randle, G.; Feil, E.J.; Grundmann, H.; Spratt, B.G. The Evolutionary History of Methicillin-Resistant Staphylococcus Aureus (MRSA). Proc. Natl. Acad. Sci. USA 2002, 99, 7687–7692. [Google Scholar] [CrossRef]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organisation: Geneva, Switzerland, 2017. [Google Scholar]
- Centers for Disease Control and Prevention (U.S.). Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention (U.S.): Atlanta, GS, USA, 2019.
- Hibbitts, A.; O’Leary, C. Emerging Nanomedicine Therapies to Counter the Rise of Methicillin-Resistant Staphylococcus aureus. Materials 2018, 11, 321. [Google Scholar] [CrossRef]
- Siddiqui, A.H.; Koirala, J. Methicillin Resistant Staphylococcus aureus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and Resistance Mechanisms of Antibiotics: A Guide for Clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The Antimicrobial Peptides and Their Potential Clinical Applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Kang, J.; Dietz, M.J.; Li, B. Antimicrobial Peptide LL-37 Is Bactericidal against Staphylococcus Aureus Biofilms. PLoS ONE 2019, 14, e0216676. [Google Scholar] [CrossRef]
- Garbacz, K.; Kamysz, W.; Piechowicz, L. Activity of Antimicrobial Peptides, Alone or Combined with Conventional Antibiotics, against Staphylococcus aureus Isolated from the Airways of Cystic Fibrosis Patients. Virulence 2017, 8, 94–100. [Google Scholar] [CrossRef]
- Rodrigues de Almeida, N.; Catazaro, J.; Krishnaiah, M.; Singh Chhonker, Y.; Murry, D.J.; Powers, R.; Conda-Sheridan, M. Understanding Interactions of Citropin 1.1 Analogues with Model Membranes and Their Influence on Biological Activity. Peptides 2019, 119, 170119. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, O.; Cirioni, O.; Goteri, G.; Lucarini, G.; Kamysz, E.; Kamysz, W.; Orlando, F.; Rizzetto, G.; Molinelli, E.; Morroni, G.; et al. Efficacy of Cathelicidin LL-37 in an MRSA Wound Infection Mouse Model. Antibiotics 2021, 10, 1210. [Google Scholar] [CrossRef] [PubMed]
- Demirci, M.; Yigin, A.; Demir, C. Efficacy of Antimicrobial Peptide LL-37 against Biofilm Forming Staphylococcus Aureus Strains Obtained from Chronic Wound Infections. Microb. Pathog. 2022, 162, 105368. [Google Scholar] [CrossRef] [PubMed]
- Bedlovičová, Z.; Salayová, A. Green-Synthesized Silver Nanoparticles and Their Potential for Antibacterial Applications. In Bacterial Pathogenesis and Antibacterial Control; Kırmusaoğlu, S., Ed.; InTech: Hong Kong, China, 2018; ISBN 978-1-78923-160-1. [Google Scholar]
- Vigneswari, S.; Amelia, T.S.M.; Hazwan, M.H.; Mouriya, G.K.; Bhubalan, K.; Amirul, A.-A.A.; Ramakrishna, S. Transformation of Biowaste for Medical Applications: Incorporation of Biologically Derived Silver Nanoparticles as Antimicrobial Coating. Antibiotics 2021, 10, 229. [Google Scholar] [CrossRef]
- de Alwis Weerasekera, H.; Griffith, M.; Alarcon, E.I. Biomedical Uses of Silver Nanoparticles: From Roman Wine Cups to Biomedical Devices. In Silver Nanoparticle Applications; Engineering Materials; Springer: Cham, Switzerland, 2015; pp. 93–125. ISBN 978-3-319-11261-9. [Google Scholar]
- Prabhu, S.; Poulose, E.K. Silver Nanoparticles: Mechanism of Antimicrobial Action, Synthesis, Medical Applications, and Toxicity Effects. Int. Nano Lett. 2012, 2, 32–42. [Google Scholar] [CrossRef]
- Durán, N.; Durán, M.; de Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver Nanoparticles: A New View on Mechanistic Aspects on Antimicrobial Activity. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 789–799. [Google Scholar] [CrossRef]
- Yan, X.; He, B.; Liu, L.; Qu, G.; Shi, J.; Hu, L.; Jiang, G. Antibacterial Mechanism of Silver Nanoparticles in Pseudomonas aeruginosa: Proteomics Approach. Metallomics 2018, 10, 557–564. [Google Scholar] [CrossRef]
- Shrivastava, S.; Bera, T.; Roy, A.; Singh, G.; Ramachandrarao, P.; Dash, D. Characterization of Enhanced Antibacterial Effects of Novel Silver Nanoparticles. Nanotechnology 2007, 18, 225103–225112. [Google Scholar] [CrossRef]
- López-Heras, M.; Theodorou, I.G.; Leo, B.F.; Ryan, M.P.; Porter, A.E. Towards Understanding the Antibacterial Activity of Ag Nanoparticles: Electron Microscopy in the Analysis of the Materials-Biology Interface in the Lung. Environ. Sci. Nano 2015, 2, 312–326. [Google Scholar] [CrossRef]
- Abbaszadegan, A.; Ghahramani, Y.; Gholami, A.; Hemmateenejad, B.; Dorostkar, S.; Nabavizadeh, M.; Sharghi, H. The Effect of Charge at the Surface of Silver Nanoparticles on Antimicrobial Activity against Gram-Positive and Gram-Negative Bacteria: A Preliminary Study. J. Nanomater. 2015, 2015, 720654. [Google Scholar] [CrossRef]
- Auer, G.K.; Weibel, D.B. Bacterial Cell Mechanics. Biochemistry 2017, 56, 3710–3724. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Sondi, I.; Salopek-Sondi, B. Silver Nanoparticles as Antimicrobial Agent: A Case Study on E. coli as a Model for Gram-Negative Bacteria. J. Colloid Interface Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Salvioni, L.; Galbiati, E.; Collico, V.; Alessio, G.; Avvakumova, S.; Corsi, F.; Tortora, P.; Prosperi, D.; Colombo, M. Negatively Charged Silver Nanoparticles with Potent Antibacterial Activity and Reduced Toxicity for Pharmaceutical Preparations. Int. J. Nanomed. 2017, 12, 2517–2530. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Sato, M.; Sato, Y.; Ando, N.; Takayama, T.; Fujita, M.; Ishihara, M. Synthesis and Application of Silver Nanoparticles (AgNPs) for the Prevention of Infection in Healthcare Workers. Int. J. Mol. Sci. 2019, 20, 3620. [Google Scholar] [CrossRef]
- Qing, Y.; Cheng, L.; Li, R.; Liu, G.; Zhang, Y.; Tang, X.; Wang, J.; Liu, H.; Qin, Y. Potential Antibacterial Mechanism of Silver Nanoparticles and the Optimization of Orthopedic Implants by Advanced Modification Technologies. Int. J. Nanomed. 2018, 13, 3311–3327. [Google Scholar] [CrossRef]
- Rai, M.K.; Deshmukh, S.D.; Ingle, A.P.; Gade, A.K. Silver Nanoparticles: The Powerful Nanoweapon against Multidrug-Resistant Bacteria: Activity of Silver Nanoparticles against MDR Bacteria. J. Appl. Microbiol. 2012, 112, 841–852. [Google Scholar] [CrossRef]
- Kumar, N.; Das, S.; Jyoti, A.; Kaushik, S. Synergistic Effect of Silver Nanoparticles with Doxycycline against Klebsiella pneumonia. Int. J. Pharm. Pharm. Sci. 2016, 8, 183–186. [Google Scholar]
- Liao, C.; Li, Y.; Tjong, S. Bactericidal and Cytotoxic Properties of Silver Nanoparticles. Int. J. Mol. Sci. 2019, 20, 449. [Google Scholar] [CrossRef]
- Xu, H.; Qu, F.; Xu, H.; Lai, W.; Andrew Wang, Y.; Aguilar, Z.P.; Wei, H. Role of Reactive Oxygen Species in the Antibacterial Mechanism of Silver Nanoparticles on Escherichia coli O157:H7. Biomet. Int. J. Role Met. Ions Biol. Biochem. Med. 2012, 25, 45–53. [Google Scholar] [CrossRef]
- Salah, R.; Karmy, M.; Abdelraouf, A.; Kotb, S. Evaluation of the Bactericidal Effect of Silver Nanoparticles against Methicillin Resistant Staphylococcus Aureus (MRSA) and Methicillin Sensitive Staphylococcus Aureus (MSSA) Strains Isolated from Mastitic Milk of Small Ruminants and Their Surrounding Environment in Aswan. J. Vet. Med. Res. 2021, 27, 143–151. [Google Scholar] [CrossRef]
- Ansari, M.; Khan, H.; Khan, A.; Cameotra, S.; Alzohairy, M. Anti-Biofilm Efficacy of Silver Nanoparticles against MRSA and MRSE Isolated from Wounds in a Tertiary Care Hospital. Indian J. Med. Microbiol. 2015, 33, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Sedrizadeh-Bami, S.; Kariminik, A.; Ranjbar, M. Synthesis and Characterization of Silver Nanoparticles with Ultrasound-Assisted Reverse Micelles Method and Their Antibacterial Effects on Methicillin-Resistant Staphylococcus Aureus Isolates. Avicenna J. Clin. Microbiol. Infect. 2020, 7, 99–103. [Google Scholar] [CrossRef]
- Paredes, D.; Ortiz, C.; Torres, R. Synthesis, Characterization, and Evaluation of Antibacterial Effect of Ag Nanoparticles against Escherichia coli O157:H7 and Methicillin-Resistant Staphylococcus aureus (MRSA). Int. J. Nanomed. 2014, 9, 1717–1729. [Google Scholar] [CrossRef]
- Wady, A.F.; Machado, A.L.; Foggi, C.C.; Zamperini, C.A.; Zucolotto, V.; Moffa, E.B.; Vergani, C.E. Effect of a Silver Nanoparticles Solution on Staphylococcus aureus and Candida spp. J. Nanomater. 2014, 2014, 545279. [Google Scholar] [CrossRef]
- Das, B.; Dash, S.K.; Mandal, D.; Ghosh, T.; Chattopadhyay, S.; Tripathy, S.; Das, S.; Dey, S.K.; Das, D.; Roy, S. Green Synthesized Silver Nanoparticles Destroy Multidrug Resistant Bacteria via Reactive Oxygen Species Mediated Membrane Damage. Arab. J. Chem. 2017, 10, 862–876. [Google Scholar] [CrossRef]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Khalil, M.I.; Al-Zaban, M.I. Novel Biogenic Silver Nanoparticle-Induced Reactive Oxygen Species Inhibit the Biofilm Formation and Virulence Activities of Methicillin-Resistant Staphylococcus Aureus (MRSA) Strain. Front. Bioeng. Biotechnol. 2020, 8, 433. [Google Scholar] [CrossRef]
- Zhen, J.-B.; Kang, P.-W.; Zhao, M.-H.; Yang, K.-W. Silver Nanoparticle Conjugated Star PCL- b -AMPs Copolymer as Nanocomposite Exhibits Efficient Antibacterial Properties. Bioconjug. Chem. 2020, 31, 51–63. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Sun, P.; Zhang, N.; Zhao, Y.; Qin, S.; Zhao, Y. Antimicrobial Peptide-Modified Silver Nanoparticles for Enhancing the Antibacterial Efficacy. RSC Adv. 2020, 10, 38746–38754. [Google Scholar] [CrossRef]
- Zharkova, M.S.; Golubeva, O.Y.; Orlov, D.S.; Vladimirova, E.V.; Dmitriev, A.V.; Tossi, A.; Shamova, O.V. Silver Nanoparticles Functionalized with Antimicrobial Polypeptides: Benefits and Possible Pitfalls of a Novel Anti-Infective Tool. Front. Microbiol. 2021, 12, 750556. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Wang, H.; Zhu, M.; Feng, W.; Liang, G. Enhanced Antibacterial and Anti-Biofilm Activities of Antimicrobial Peptides Modified Silver Nanoparticles. Int. J. Nanomed. 2021, 16, 4831–4846. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Duan, W.; Wo, F.; Wu, J. Two-Dimensional Fluorescent Strategy Based on Porous Silicon Quantum Dots for Metal-Ion Detection and Recognition. ACS Appl. Nano Mater. 2019, 2, 6110–6115. [Google Scholar] [CrossRef]
- Chen, X.; Wo, F.; Jin, Y.; Tan, J.; Lai, Y.; Wu, J. Drug-Porous Silicon Dual Luminescent System for Monitoring and Inhibition of Wound Infection. ACS Nano 2017, 11, 7938–7949. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Tang, H.; Bian, X.; Ma, K.; Chang, J.; Fu, X.; Zhang, C. Calcium Silicate Accelerates Cutaneous Wound Healing with Enhanced Re-Epithelialization through EGF/EGFR/ERK-Mediated Promotion of Epidermal Stem Cell Functions. Burns Trauma 2021, 9, tkab029. [Google Scholar] [CrossRef] [PubMed]
- Awad, K.; Ahuja, N.; Fiedler, M.; Peper, S.; Wang, Z.; Aswath, P.; Brotto, M.; Varanasi, V. Ionic Silicon Protects Oxidative Damage and Promotes Skeletal Muscle Cell Regeneration. Int. J. Mol. Sci. 2021, 22, 497. [Google Scholar] [CrossRef]
- Bassous, N.J.; Webster, T.J. The Binary Effect on Methicillin-Resistant Staphylococcus aureus of Polymeric Nanovesicles Appended by Proline-Rich Amino Acid Sequences and Inorganic Nanoparticles. Small 2019, 15, 1804247. [Google Scholar] [CrossRef]
- Gao, J.; Na, H.; Zhong, R.; Yuan, M.; Guo, J.; Zhao, L.; Wang, Y.; Wang, L.; Zhang, F. One Step Synthesis of Antimicrobial Peptide Protected Silver Nanoparticles: The Core-Shell Mutual Enhancement of Antibacterial Activity. Colloids Surf. B Biointerfaces 2020, 186, 110704. [Google Scholar] [CrossRef]
- Zhao, X.; Kuipers, O.P. Synthesis of Silver-Nisin Nanoparticles with Low Cytotoxicity as Antimicrobials against Biofilm-Forming Pathogens. Colloids Surf. B Biointerfaces 2021, 206, 111965. [Google Scholar] [CrossRef]
- Zheng, K.; Setyawati, M.I.; Lim, T.-P.; Leong, D.T.; Xie, J. Antimicrobial Cluster Bombs: Silver Nanoclusters Packed with Daptomycin. ACS Nano 2016, 10, 7934–7942. [Google Scholar] [CrossRef]
- Ye, Z.; Sang, T.; Li, K.; Fischer, N.G.; Mutreja, I.; Echeverría, C.; Kumar, D.; Tang, Z.; Aparicio, C. Hybrid Nanocoatings of Self-Assembled Organic-Inorganic Amphiphiles for Prevention of Implant Infections. Acta Biomater. 2022, 140, 338–349. [Google Scholar] [CrossRef]
- Golubeva, O.Y.; Shamova, O.V.; Orlov, D.S.; Pazina, T.Y.; Boldina, A.S.; Drozdova, I.A.; Kokryakov, V.N. Synthesis and Study of Antimicrobial Activity of Bioconjugates of Silver Nanoparticles and Endogenous Antibiotics. Glass Phys. Chem. 2011, 37, 78–84. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).