Antimicrobial Resistance Development Pathways in Surface Waters and Public Health Implications
Abstract
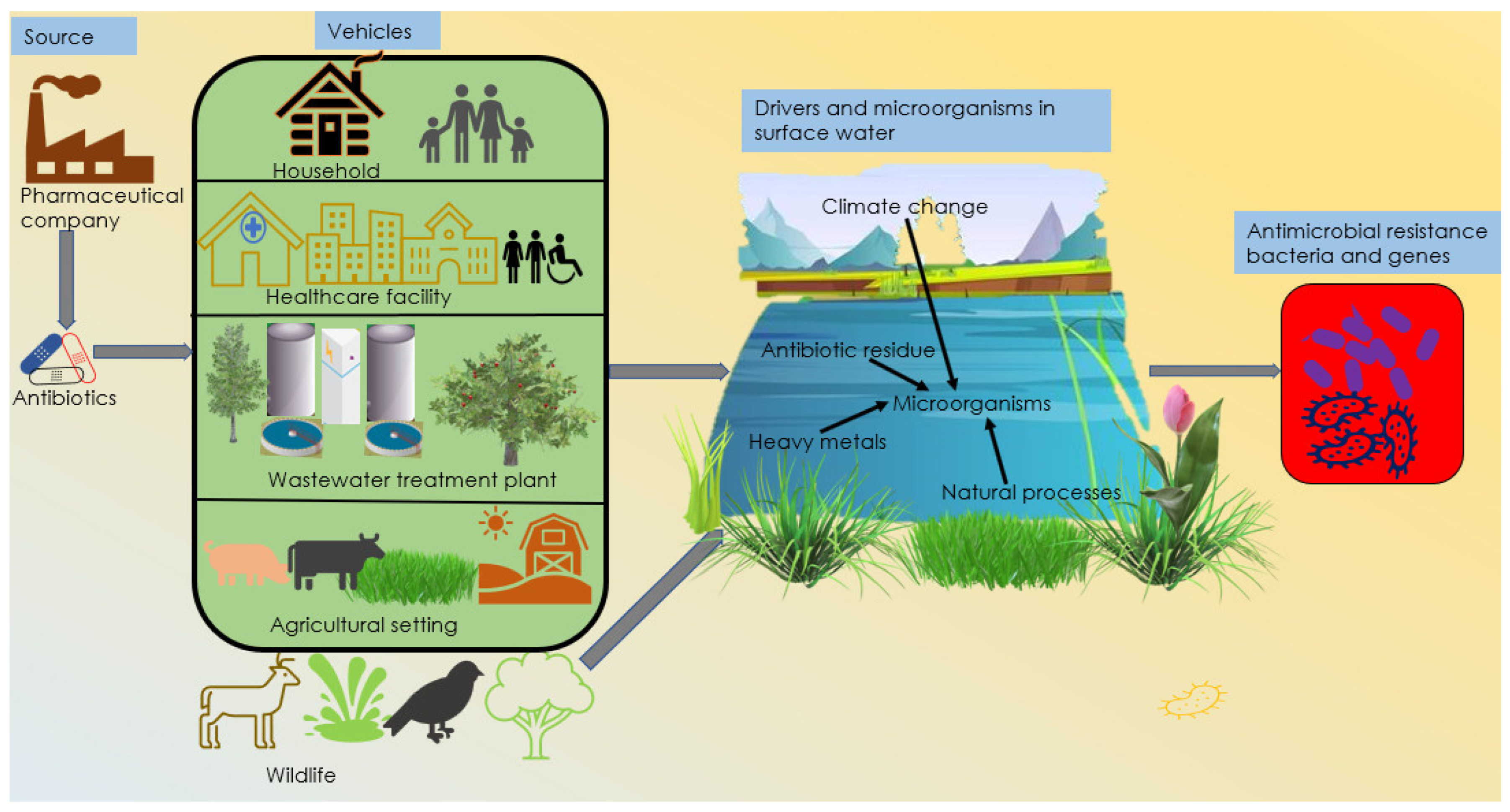
:1. Introduction
2. Primary Source of Antibiotics
3. Vehicles/Pathways for Antimicrobial Resistance
3.1. Healthcare Facilities
3.2. Wastewater
3.2.1. Microorganisms in Wastewater
3.2.2. Controversy over Wastewater Treatment Process
3.3. Agricultural Settings
3.4. Food
| Source | Exposure Route | Risk Group | Resistant Bacteria/Gene | Reference |
|---|---|---|---|---|
| Maize | Ingestion Direct contact | Poultry workers Market workers | E. coli | [68] |
| Chicken | Direct contact Ingestion | Farmworkers, slaughterhouse workers, veterinarians | Methicillin-resistant S. aureus (LA-MRSA). E. coli | [68,110] [69] |
| Vegetables, fruits, fish, and dairy products | Ingestion Direct contact | Long term storage consumers Farmers | Sitotroga cerealla Salmonella Campylobacter | [68,87,111,112] |
| Beef | Direct contact with livestock Fecal oral route | Agricultural workers Meat consumer | S. typhimurium DT104 | [113] |
| Chicken, beef, pork | Ingestion Skin contact | General population | Salmonella enterica | [114] |
| Chicken, beef, fish | Direct contact | Veterinarians, Farm workers, | E. coli methicillin-resistant S. aureus | [115] |
| Chicken, turkey, bovine, porcine meat | Ingestion Direct contact | Food handler Health workers | E. coli, S. typhimurium Klebsiella pneumoniae | [115,116] |
| Beef, chicken, pork, lamb, duck, egg, milk, vegetables, seafood | Food contact surface Ingestion Direct contact | Farm workers Food service worker | Leuconostoc pseudomesenteroides, Lactobacillus pentosus, Salmonella enteritidis | [117,118,119] |
| Milk | Direct contact Ingestion | Poultry workers Market workers | E. coli, S. aureus | [68,112] |
3.5. Wildlife Populations
4. Drivers of Antimicrobial Resistance
4.1. Natural Processes
4.2. Heavy Metals
4.3. Climate Change
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murugaiyan, J.; Anand Kumar, P.; Rao, G.S.; Iskandar, K.; Hawser, S.; Hays, J.P.; Mohsen, Y.; Adukkadukkam, S.; Awuah, W.A.; Jose, R.A.M.; et al. Progress in Alternative Strategies to Combat Antimicrobial Resistance: Focus on Antibiotics. Antibiotics 2022, 11, 200. [Google Scholar] [CrossRef]
- Rogers, J.V.; Hall, V.L.; McOsker, C.C. Crumbling the Castle: Targeting DNABII Proteins for Collapsing Bacterial Biofilms as a Therapeutic Approach to Treat Disease and Combat Antimicrobial Resistance. Antibiotics 2022, 11, 104. [Google Scholar] [CrossRef]
- Center for Disease Control. Antibiotic Use in the United States, 2018: Progress and Opportunities; United States Department of Health and Human Services: Washington, DC, USA, 2018; pp. 1–37. [Google Scholar]
- Ge, B.; Domesle, K.J.; Yang, Q.; Young, S.R.; Rice-Trujillo, C.L.; Bodeis Jones, S.M.; Gaines, S.A.; Keller, M.W.; Li, X.; Piñeiro, S.A.; et al. Effects of Low Concentrations of Erythromycin, Penicillin, and Virginiamycin on Bacterial Resistance Development in Vitro. Sci. Rep. 2017, 7, 11017. [Google Scholar] [CrossRef]
- Frost, I.; Laxminarayan, R.; McKenna, N.; Chai, S.; Joshi, J. Antimicrobial Resistance and Primary Health Care; World Health Organization: Geneva, Switzerland, 2018; pp. 3–6. [Google Scholar]
- Albrich, W.C.; Monnet, D.L.; Harbarth, S. Antibiotic Selection Pressure and Resistance in Streptococcus Pneumoniae and Streptococcus Pyogenes. Emerg. Infect. Dis. 2004, 10, 514–517. [Google Scholar] [CrossRef]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global Contributors to Antibiotic Resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef]
- Hiller, C.X.; Hübner, U.; Fajnorova, S.; Schwartz, T.; Drewes, J.E. Antibiotic Microbial Resistance (AMR) Removal Efficiencies by Conventional and Advanced Wastewater Treatment Processes: A Review. Sci. Total Environ. 2019, 685, 596–608. [Google Scholar] [CrossRef]
- Fletcher, S. Understanding the Contribution of Environmental Factors in the Spread of Antimicrobial Resistance. Environ. Health Prev. Med. 2015, 20, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-Resistant Bacteria and Antimicrobial Residues in Wastewater and Process Water from German Pig Slaughterhouses and Their Receiving Municipal Wastewater Treatment Plants. Sci. Total Environ. 2020, 727, 138788. [Google Scholar] [CrossRef]
- Danner, M.C.; Robertson, A.; Behrends, V.; Reiss, J. Antibiotic Pollution in Surface Fresh Waters: Occurrence and Effects. Sci. Total Environ. 2019, 664, 793–804. [Google Scholar] [CrossRef]
- Petrovich, M.L.; Zilberman, A.; Kaplan, A.; Eliraz, G.R.; Wang, Y.; Langenfeld, K.; Duhaime, M.; Wigginton, K.; Poretsky, R.; Avisar, D.; et al. Microbial and Viral Communities and Their Antibiotic Resistance Genes Throughout a Hospital Wastewater Treatment System. Front. Microbiol. 2020, 11, 153. [Google Scholar] [CrossRef] [Green Version]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial Use in Aquaculture Re-Examined: Its Relevance to Antimicrobial Resistance and to Animal and Human Health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef]
- Shallcross, L.J.; Howard, S.J.; Fowler, T.; Davies, S.C. Tackling the Threat of Antimicrobial Resistance: From Policy to Sustainable Action. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140082. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, J.; Fong, K.; Nadya, S.; Allen, K.; Laing, C.; Ziebell, K.; Topp, E.; Carroll, L.M.; Wiedmann, M.; et al. Antibiotic Resistance in Shiga Toxigenic Escherichia Coli Isolates from Surface Waters and Sediments in a Mixed Use Urban Agricultural Landscape. Antibiotics 2021, 10, 237. [Google Scholar] [CrossRef]
- Bartley, P.S.; Domitrovic, T.N.; Moretto, V.T.; Santos, C.S.; Ponce-Terashima, R.; Reis, M.G.; Barbosa, L.M.; Blanton, R.E.; Bonomo, R.A.; Perez, F. Antibiotic Resistance in Enterobacteriaceae from Surface Waters in Urban Brazil Highlights the Risks of Poor Sanitation. Am. J. Trop. Med. Hyg. 2019, 100, 1369–1377. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Rivera, J.I.; Coradin, M.; Toranzos, G.A. Antibiotic-Resistance and Virulence Genes in Enterococcus Isolated from Tropical Recreational Waters. J. Water Health 2013, 11, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Silva, V.; Ferreira, E.; Manageiro, V.; Reis, L.; Tejedor-Junco, M.T.; Sampaio, A.; Capelo, J.L.; Caniça, M.; Igrejas, G.; Poeta, P. Distribution and Clonal Diversity of Staphylococcus Aureus and Other Staphylococci in Surface Waters: Detection of ST425-T742 and ST130-T843 MecC-Positive MRSA Strains. Antibiotics 2021, 10, 1416. [Google Scholar] [CrossRef]
- Hay, S.I.; Rao, P.C.; Dolecek, C.; Day, N.P.J.; Stergachis, A.; Lopez, A.D.; Murray, C.J.L. Measuring and Mapping the Global Burden of Antimicrobial Resistance. BMC Med. 2018, 16, 78. [Google Scholar] [CrossRef] [Green Version]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental Antimicrobial Resistance and Its Drivers: A Potential Threat to Public Health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Raffatellu, M. Learning from Bacterial Competition in the Host to Develop Antimicrobials. Nat. Med. 2018, 24, 1097–1103. [Google Scholar] [CrossRef]
- McInnes, R.S.; uz-Zaman, M.H.; Alam, I.T.; Ho, S.F.S.; Moran, R.A.; Clemens, J.D.; Islam, M.S.; van Schaik, W. Metagenome-Wide Analysis of Rural and Urban Surface Waters and Sediments in Bangladesh Identifies Human Waste as a Driver of Antibiotic Resistance. mSystems 2021, 6, e0013721. [Google Scholar] [CrossRef]
- Zhang, C.M.; Liang, J.; Liu, W.Y. Comparative Study on the Bacterial Diversity and Antibiotic Resistance Genes of Urban Landscape Waters Replenished by Reclaimed Water and Surface Water in Xi’an, China. Environ. Sci. Pollut. Res. 2021, 28, 41396–41406. [Google Scholar] [CrossRef]
- Zhang, S.H.; Lv, X.; Han, B.; Gu, X.; Wang, P.F.; Wang, C.; He, Z. Prevalence of Antibiotic Resistance Genes in Antibiotic-Resistant Escherichia Coli Isolates in Surface Water of Taihu Lake Basin, China. Environ. Sci. Pollut. Res. 2015, 22, 11412–11421. [Google Scholar] [CrossRef]
- Liang, X.; Guan, F.; Chen, B.; Luo, P.; Guo, C.; Wu, G.; Ye, Y.; Zhou, Q.; Fang, H. Spatial and Seasonal Variations of Antibiotic Resistance Genes and Antibiotics in the Surface Waters of Poyang Lake in China. Ecotoxicol. Environ. Saf. 2020, 196, 110543. [Google Scholar] [CrossRef]
- Zhao, B.; Xu, J.; Zhang, G.; Lu, S.; Liu, X.; Li, L.; Li, M. Occurrence of Antibiotics and Antibiotic Resistance Genes in the Fuxian Lake and Antibiotic Source Analysis Based on Principal Component Analysis-Multiple Linear Regression Model. Chemosphere 2021, 262, 127741. [Google Scholar] [CrossRef]
- Zhang, S.; Pang, S.; Wang, P.F.; Wang, C.; Han, N.; Liu, B.; Han, B.; Li, Y.; Anim-Larbi, K. Antibiotic Concentration and Antibiotic-Resistant Bacteria in Two Shallow Urban Lakes after Stormwater Event. Environ. Sci. Pollut. Res. 2016, 23, 9984–9992. [Google Scholar] [CrossRef]
- Wang, Z.; Han, M.; Li, E.; Liu, X.; Wei, H.; Yang, C.; Lu, S.; Ning, K. Distribution of Antibiotic Resistance Genes in an Agriculturally Disturbed Lake in China: Their Links with Microbial Communities, Antibiotics, and Water Quality. J. Hazard. Mater. 2020, 393, 122426. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, C.; Cao, X.; Lin, H.; Wang, J. Antibiotic Resistance Genes in Surface Water of Eutrophic Urban Lakes Are Related to Heavy Metals, Antibiotics, Lake Morphology and Anthropic Impact. Ecotoxicology 2017, 26, 831–840. [Google Scholar] [CrossRef]
- Wang, C.; Gu, X.; Zhang, S.; Wang, P.; Guo, C.; Gu, J.; Hou, J. Characterization of Antibiotic-Resistance Genes in Antibiotic Resistance Escherichia Coli Isolates from a Lake. Arch. Environ. Contam. Toxicol. 2013, 65, 635–641. [Google Scholar] [CrossRef]
- Moussa, J.; Abboud, E.; Tokajian, S. The Dissemination of Antimicrobial Resistance Determinants in Surface Water Sources in Lebanon. FEMS Microbiol. Ecol. 2021, 97, fiab113. [Google Scholar] [CrossRef]
- Reichert, G.; Hilgert, S.; Alexander, J.; Rodrigues de Azevedo, J.C.; Morck, T.; Fuchs, S.; Schwartz, T. Determination of Antibiotic Resistance Genes in a WWTP-Impacted River in Surface Water, Sediment, and Biofilm: Influence of Seasonality and Water Quality. Sci. Total Environ. 2021, 768, 144526. [Google Scholar] [CrossRef]
- Cheng, J.; Tang, X.; Liu, C. Occurrence and Distribution of Antibiotic Resistance Genes in Various Rural Environmental Media. Environ. Sci. Pollut. Res. 2020, 27, 29191–29203. [Google Scholar] [CrossRef]
- Wu, D.L.; Zhang, M.; He, L.X.; Zou, H.Y.; Liu, Y.S.; Li, B.B.; Yang, Y.Y.; Liu, C.; He, L.Y.; Ying, G.G. Contamination Profile of Antibiotic Resistance Genes in Ground Water in Comparison with Surface Water. Sci. Total Environ. 2020, 715, 136975. [Google Scholar] [CrossRef]
- Stange, C.; Yin, D.; Xu, T.; Guo, X.; Schäfer, C.; Tiehm, A. Distribution of Clinically Relevant Antibiotic Resistance Genes in Lake Tai, China. Sci. Total Environ. 2019, 655, 337–346. [Google Scholar] [CrossRef]
- Jiang, H.; Zhou, R.; Yang, Y.; Chen, B.; Cheng, Z.; Zhang, M.; Li, J.; Zhang, G.; Zou, S. Characterizing the Antibiotic Resistance Genes in a River Catchment: Influence of Anthropogenic Activities. J. Environ. Sci. 2018, 69, 125–132. [Google Scholar] [CrossRef]
- Jia, J.; Gomes-Silva, G.; Plath, M.; Pereira, B.B.; UeiraVieira, C.; Wang, Z. Shifts in Bacterial Communities and Antibiotic Resistance Genes in Surface Water and Gut Microbiota of Guppies (Poecilia Reticulata) in the Upper Rio Uberabinha, Brazil. Ecotoxicol. Environ. Saf. 2021, 211, 111955. [Google Scholar] [CrossRef]
- Arsand, J.B.; Hoff, R.B.; Jank, L.; Bussamara, R.; Dallegrave, A.; Bento, F.M.; Kmetzsch, L.; Falção, D.A.; do Carmo Ruaro Peralba, M.; de Araujo Gomes, A.; et al. Presence of Antibiotic Resistance Genes and Its Association with Antibiotic Occurrence in Dilúvio River in Southern Brazil. Sci. Total Environ. 2020, 738, 139781. [Google Scholar] [CrossRef]
- Garner, E.; Benitez, R.; von Wagoner, E.; Sawyer, R.; Schaberg, E.; Hession, W.C.; Krometis, L.A.H.; Badgley, B.D.; Pruden, A. Stormwater Loadings of Antibiotic Resistance Genes in an Urban Stream. Water Res. 2017, 123, 144–152. [Google Scholar] [CrossRef]
- Liyanage, G.Y.; Illango, A.; Manage, P.M. Prevalence and Quantitative Analysis of Antibiotic Resistance Genes (ARGs) in Surface and Groundwater in Meandering Part of the Kelani River Basin in Sri Lanka. Water. Air. Soil Pollut. 2021, 232, 351. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, L.; Chen, J.; Fan, X.; Xie, S.; Huang, J.; Yu, G. Antibiotic Resistance Genes and Mobile Genetic Elements in a Rural River in Southeast China: Occurrence, Seasonal Variation and Association with the Antibiotics. Sci. Total Environ. 2021, 778, 146131. [Google Scholar] [CrossRef]
- Tang, Y.; Liang, Z.; Li, G.; Zhao, H.; An, T. Metagenomic Profiles and Health Risks of Pathogens and Antibiotic Resistance Genes in Various Industrial Wastewaters and the Associated Receiving Surface Water. Chemosphere 2021, 283, 131224. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Chowdhary, P.; Singh, A.; Chaurasia, D.; Pandey, A.; Chandra, R.; Gupta, P. Dissemination of Antibiotic Resistance Genes, Mobile Genetic Elements, and Efflux Genes in Anthropogenically Impacted Riverine Environments. Chemosphere 2021, 273, 129693. [Google Scholar] [CrossRef]
- Stange, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Antibiotic Resistance and Virulence Genes in Coliform Water Isolates. Int. J. Hyg. Environ. Health 2016, 219, 823–831. [Google Scholar] [CrossRef] [Green Version]
- Guruge, K.S.; Tamamura, Y.A.; Goswami, P.; Tanoue, R.; Jinadasa, K.B.S.N.; Nomiyama, K.; Ohura, T.; Kunisue, T.; Tanabe, S.; Akiba, M. The Association between Antimicrobials and the Antimicrobial-Resistant Phenotypes and Resistance Genes of Escherichia Coli Isolated from Hospital Wastewaters and Adjacent Surface Waters in Sri Lanka. Chemosphere 2021, 279, 130591. [Google Scholar] [CrossRef]
- Molale, L.G.; Bezuidenhout, C.C. Antibiotic Resistance, Efflux Pump Genes and Virulence Determinants in Enterococcus Spp. from Surface Water Systems. Environ. Sci. Pollut. Res. 2016, 23, 21501–21510. [Google Scholar] [CrossRef]
- Kaushik, M.; Khare, N.; Kumar, S.; Gulati, P. High Prevalence of Antibiotic Resistance and Integrons in Escherichia Coli Isolated from Urban River Water, India. Microb. Drug Resist. 2019, 25, 359–370. [Google Scholar] [CrossRef]
- Azevedo, J.S.N.; Araújo, S.; Oliveira, C.S.; Correia, A.; Henriques, I. Analysis of Antibiotic Resistance in Bacteria Isolated from the Surface Microlayer and Underlying Water of an Estuarine Environment. Microb. Drug Resist. 2013, 19, 64–71. [Google Scholar] [CrossRef]
- Veljović, K.; Popović, N.; Vidojević, A.T.; Tolinački, M.; Mihajlović, S.; Jovčić, B.; Kojić, M. Environmental Waters as a Source of Antibiotic-Resistant Enterococcus Species in Belgrade, Serbia. Environ. Monit. Assess. 2015, 187, 599. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Schwengers, O.; Schmiedel, J.; Baars, C.; Lambrecht, O.; Heß, S.; Berendonk, T.U.; Falgenhauer, J.; Chakraborty, T.; Imirzalioglu, C. Multidrug-Resistant and Clinically Relevant Gram-Negative Bacteria Are Present in German Surface Waters. Front. Microbiol. 2019, 10, 2779. [Google Scholar] [CrossRef]
- Stoll, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Prevalence of Clinically Relevant Antibiotic Resistance Genes in Surface Water Samples Collected from Germany and Australia. Environ. Sci. Technol. 2012, 46, 9716–9726. [Google Scholar] [CrossRef]
- Kotwani, A.; Joshi, J.; Kaloni, D. Pharmaceutical Effluent: A Critical Link in the Interconnected Ecosystem Promoting Antimicrobial Resistance. Environ. Sci. Pollut. Res. 2021, 28, 32111–32124. [Google Scholar] [CrossRef]
- Larsson, D.G.J. Pollution from Drug Manufacturing: Review and Perspectives. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130571. [Google Scholar] [CrossRef] [Green Version]
- Bengtsson-Palme, J.; Larsson, D.G.J. Concentrations of Antibiotics Predicted to Select for Resistant Bacteria: Proposed Limits for Environmental Regulation. Environ. Int. 2016, 86, 140–149. [Google Scholar] [CrossRef] [Green Version]
- Arya, G.; Tadayon, S.; Sadighian, J.; Jones, J.; de Mutsert, K.; Huff, T.B.; Foster, G. Pharmaceutical Chemicals, Steroids and Xenoestrogens in Water, Sediments and Fish from the Tidal Freshwater Potomac River (Virginia, USA). J. Environ. Sci. Health Part A 2017, 52, 686–696. [Google Scholar] [CrossRef]
- Burke, V.; Richter, D.; Greskowiak, J.; Mehrtens, A.; Schulz, L.; Massmann, G. Occurrence of Antibiotics in Surface and Groundwater of a Drinking Water Catchment Area in Germany. Water Environ. Res. 2016, 88, 652–659. [Google Scholar] [CrossRef]
- Dodgen, L.K.; Kelly, W.R.; Panno, S.V.; Taylor, S.J.; Armstrong, D.L.; Wiles, K.N.; Zhang, Y.; Zheng, W. Characterizing Pharmaceutical, Personal Care Product, and Hormone Contamination in a Karst Aquifer of Southwestern Illinois, USA, Using Water Quality and Stream Flow Parameters. Sci. Total Environ. 2017, 578, 281–289. [Google Scholar] [CrossRef]
- Jurado, A.; Walther, M.; Díaz-Cruz, M.S. Occurrence, Fate and Environmental Risk Assessment of the Organic Microcontaminants Included in the Watch Lists Set by EU Decisions 2015/495 and 2018/840 in the Groundwater of Spain. Sci. Total Environ. 2019, 663, 285–296. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance & Patient Safety Portal. Available online: https://arpsp.cdc.gov/about?tab=antibiotic-resistance (accessed on 24 April 2022).
- Andersson, D.I.; Hughes, D. Microbiological Effects of Sublethal Levels of Antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Rozman, U.; Duh, D.; Cimerman, M.; Turk, S.Š. Hospital Wastewater Effluent: Hot Spot for Antibiotic Resistant Bacteria. J. Water Sanit. Hyg. Dev. 2020, 10, 171–178. [Google Scholar] [CrossRef]
- Velpandian, T.; Halder, N.; Nath, M.; Das, U.; Moksha, L.; Gowtham, L.; Batta, S.P. Un-Segregated Waste Disposal: An Alarming Threat of Antimicrobials in Surface and Ground Water Sources in Delhi. Environ. Sci. Pollut. Res. 2018, 25, 29518–29528. [Google Scholar] [CrossRef]
- Han, S.; Li, X.; Huang, H.; Wang, T.; Wang, Z.; Fu, X.; Zhou, Z.; Du, P.; Li, X. Simultaneous Determination of Seven Antibiotics and Five of Their Metabolites in Municipal Wastewater and Evaluation of Their Stability under Laboratory Conditions. Int. J. Environ. Res. Public Health 2021, 18, 10640. [Google Scholar] [CrossRef]
- Cars, O.; Nordberg, P. Antibiotic Resistance—The Faceless Threat. Int. J. Risk Saf. Med. 2005, 17, 103–110. [Google Scholar]
- Morgan, D.J.; Okeke, I.N.; Laxminarayan, R.; Perencevich, E.N.; Weisenberg, S. Non-Prescription Antimicrobial Use Worldwide: A Systematic Review. Lancet Infect. Dis. 2011, 11, 692–701. [Google Scholar] [CrossRef] [Green Version]
- Esimone, C.O.; Nworu, C.S.; Udeogaranya, O.P. Utilization of Antimicrobial Agents with and without Prescription by Out-Patients in Selected Pharmacies in South-Eastern Nigeria. Pharm. World Sci. 2007, 29, 655–660. [Google Scholar] [CrossRef]
- World Health Organization. Community-Based Surveillance of Antimicrobial Use and Resistance in Resource-Constrained Settings Report on Five Pilot Projects; World Health Organization: Geneva, Switzerland, 2009; 110p. [Google Scholar]
- Alam, M.U.; Rahman, M.; Abdullah-Al-Masud; Islam, M.A.; Asaduzzaman, M.; Sarker, S.; Rousham, E.; Unicomb, L. Human Exposure to Antimicrobial Resistance from Poultry Production: Assessing Hygiene and Waste-Disposal Practices in Bangladesh. Int. J. Hyg. Environ. Health 2019, 222, 1068–1076. [Google Scholar] [CrossRef]
- Homeier-Bachmann, T.; Heiden, S.E.; Lübcke, P.K.; Bachmann, L.; Bohnert, J.A.; Zimmermann, D.; Schaufler, K. Antibiotic-Resistant Enterobacteriaceae in Wastewater of Abattoirs. Antibiotics 2021, 10, 568. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Kreyenschmidt, J.; Schmithausen, R.M.; Sib, E.; Schmoger, S.; Käsbohrer, A.; Hammerl, J.A. Clinically Relevant Escherichia Coli Isolates from Process Waters and Wastewater of Poultry and Pig Slaughterhouses in Germany. Microorganisms 2021, 9, 698. [Google Scholar] [CrossRef]
- EPA. Wastewater Technology Fact Sheet—Disinfection for Small Systems; Water Technology Fact Sheet; US EPA: Washington, DC, USA, 2002. [Google Scholar]
- Łuczkiewicz, A.; Jankowska, K.; Fudala-Ksiazek, S.; Olańczuk-Neyman, K. Antimicrobial Resistance of Fecal Indicators in Municipal Wastewater Treatment Plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef]
- Haywood, J.; Vadlamani, G.; Stubbs, K.A.; Mylne, J.S. Antibiotic Resistance Lessons for the Herbicide Resistance Crisis. Pest Manag. Sci. 2021, 77, 3807–3814. [Google Scholar] [CrossRef]
- Van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired Antibiotic Resistance Genes: An Overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [Green Version]
- Mazel, D.; Davies, J. Antibiotic Resistance in Microbes. Cell. Mol. Life Sci. 1999, 56, 742–754. [Google Scholar] [CrossRef]
- Novo, A.; André, S.; Viana, P.; Nunes, O.C.; Manaia, C.M. Antibiotic Resistance, Antimicrobial Residues and Bacterial Community Composition in Urban Wastewater. Water Res. 2013, 47, 1875–1887. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Ge, F.; Huang, S.; Chen, M.; Wang, R. Occurrence of Veterinary Antibiotics in Animal Wastewater and Surface Water around Farms in Jiangsu Province, China. Chemosphere 2011, 82, 1408–1414. [Google Scholar] [CrossRef]
- Gotkowska-Płachta, A. The Prevalence of Virulent and Multidrug-Resistant Enterococci in River Water and in Treated and Untreated Municipal and Hospital Wastewater. Int. J. Environ. Res. Public Health 2021, 18, 563. [Google Scholar] [CrossRef]
- Zhang, Y.; Marrs, C.F.; Simon, C.; Xi, C. Wastewater Treatment Contributes to Selective Increase of Antibiotic Resistance among Acinetobacter Spp. Sci. Total Environ. 2009, 407, 3702–3706. [Google Scholar] [CrossRef]
- Agga, G.E.; Kasumba, J.; Loughrin, J.H.; Conte, E.D. Anaerobic Digestion of Tetracycline Spiked Livestock Manure and Poultry Litter Increased the Abundances of Antibiotic and Heavy Metal Resistance Genes. Front. Microbiol. 2020, 11, 614424. [Google Scholar] [CrossRef]
- Ceccarelli, D.; Hesp, A.; Van Der Goot, J.; Joosten, P.; Sarrazin, S.; Wagenaar, J.A.; Dewulf, J.; Mevius, D.J. Antimicrobial Resistance Prevalence in Commensal Escherichia Coli from Broilers, Fattening Turkeys, Fattening Pigs and Veal Calves in European Countries and Association with Antimicrobial Usage at Country Level. J. Med. Microbiol. 2020, 69, 537–547. [Google Scholar] [CrossRef]
- Innes, G.K.; Randad, P.R.; Korinek, A.; Davis, M.F.; Price, L.B.; So, A.D.; Heaney, C.D. External Societal Costs of Antimicrobial Resistance in Humans Attributable to Antimicrobial Use in Livestock. Annu. Rev. Public Health 2019, 41, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Lhermie, G.; Tauer, L.W.; Gröhn, Y.T. An Assessment of the Economic Costs to the U.S. Dairy Market of Antimicrobial Use Restrictions. Prev. Vet. Med. 2018, 160, 63–67. [Google Scholar] [CrossRef]
- Boxall, A.B.A.; Hardy, A.; Beulke, S.; Boucard, T.; Burgin, L.; Falloon, P.D.; Haygarth, P.M.; Hutchinson, T.; Kovats, R.S.; Leonardi, G.; et al. Impacts of Climate Change on Indirect Human Exposure to Pathogens and Chemicals from Agriculture. Environ. Health Perspect. 2009, 117, 508–514. [Google Scholar] [CrossRef] [Green Version]
- Hall, M.C.; Duerschner, J.; Gilley, J.E.; Schmidt, A.M.; Bartelt-Hunt, S.L.; Snow, D.D.; Eskridge, K.M.; Li, X. Antibiotic Resistance Genes in Swine Manure Slurry as Affected by Pit Additives and Facility Disinfectants. Sci. Total Environ. 2021, 761, 143287. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, E.; White, R.; Mexia, R.; Bruun, T.; Kapperud, G.; Lange, H.; Nygard, K.; Vold, L. Risk Factors for Sporadic Domestically Acquired Campylobacter Infections in Norway 2010-2011: A National Prospective Case-Control Study. PLoS ONE 2015, 10, e0139636. [Google Scholar] [CrossRef] [Green Version]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial Resistance in the Food Chain: A Review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef] [Green Version]
- Morrison, L.; Zembower, T.R. Antimicrobial Resistance. Gastrointest. Endosc. Clin. N. Am. 2020, 30, 619–635. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the Role of Vegetables in Spreading Antimicrobial-Resistant Bacteria: A Need for Quantitative Risk Assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef]
- Ulger, T.G.; Songur, A.N.; Cirak, O.; Cakiroglu, F. Role of Vegetable in Human Nutrition and Disease Prevention; IntechOpen: London, UK, 2018. [Google Scholar]
- van Hoek, A.H.A.M.; Veenman, C.; van Overbeek, W.M.; Lynch, G.; de Roda Husman, A.M.; Blaak, H. Prevalence and Characterization of ESBL- and AmpC-Producing Enterobacteriaceae on Retail Vegetables. Int. J. Food Microbiol. 2015, 204, 1–8. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Mackie, R.I.; Koike, S.; Krapac, I.G.; Lin, Y.-F.; Yannarell, A.C.; Maxwell, S.; Aminov, R.I. Fate and Transport of Antibiotic Residues and Antibiotic Resistance Genes Following Land Application of Manure Waste. J. Environ. Qual. 2009, 38, 1086–1108. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.; Jang, H.; Matthews, K.R. Effect of the Food Production Chain from Farm Practices to Vegetable Processing on Outbreak Incidence. Microb. Biotechnol. 2014, 7, 517–527. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic Resistance Genes from Livestock Waste: Occurrence, Dissemination, and Treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention (CDC) Where Resistance Spreads: Food Supply Medical Professional with Young Female Patient. Available online: https://www.cdc.gov/drugresistance/food.html (accessed on 24 April 2022).
- Centers for Disease Control and Prevention (CDC) Outbreak of Multidrug-Resistant Salmonella Infections Linked to Raw Chicken Products. Available online: https://www.cdc.gov/salmonella/infantis-10-18/index.html (accessed on 24 April 2022).
- Van Boxstael, S.; Dierick, K.; Van Huffel, X.; Uyttendaele, M.; Berkvens, D.; Herman, L.; Bertrand, S.; Wildemauwe, C.; Catry, B.; Butaye, P.; et al. Comparison of Antimicrobial Resistance Patterns and Phage Types of Salmonella Typhimurium Isolated from Pigs, Pork and Humans in Belgium between 2001 and 2006. Food Res. Int. 2012, 45, 913–918. [Google Scholar] [CrossRef]
- Walker, R.A.; Lawson, A.J.; Lindsay, E.A.; Ward, L.R.; Wright, P.A.; Bolton, F.J.; Wareing, D.R.A.; Corkish, J.D.; Davies, R.H.; Threlfall, E.J. Decreased Susceptibility to Ciprofloxacin in Outbreak-Associated Multiresistant Salmonella Typhimurium DT104. Vet. Rec. 2000, 147, 395–396. [Google Scholar] [CrossRef]
- Cody, S.H.; Abbott, S.L.; Marfin, A.A.; Schulz, B.; Wagner, P.; Robbins, K.; Mohle-Boetani, J.C.; Vugia, D.J. Two Outbreaks of Multidrug-Resistant Salmonella Serotype Typhimurium DT104 Infections Linked to Raw-Milk Cheese in Northern California. J. Am. Med. Assoc. 1999, 281, 1805–1810. [Google Scholar] [CrossRef] [Green Version]
- Ling, M.L.; Goh, K.T.; Wang, G.C.Y.; Neo, K.S.; Chua, T. An Outbreak of Multidrug-Resistant Salmonella Enterica Subsp. Enterica Serotype Typhimurium, DT104L Linked to Dried Anchovy in Singapore. Epidemiol. Infect. 2002, 128, 1–5. [Google Scholar] [CrossRef]
- Horby, P.W.; O’Brien, S.J.; Adak, G.K.; Graham, C.; Hawker, J.I.; Hunter, P.; Lane, C.; Lawson, A.J.; Mitchell, R.T.; Reacher, M.H.; et al. A National Outbreak of Multi-Resistant Salmonella Enterica Serovar Typhimurium Definitive Phage Type (DT) 104 Associated with Consumption of Lettuce. Epidemiol. Infect. 2003, 130, 169–178. [Google Scholar] [CrossRef]
- Ethelberg, S.; Sørensen, G.; Kristensen, B.; Christensen, K.; Krusell, L.; Hempel-Jørgensen, A.; Perge, A.; Nielsen, E.M. Outbreak with Multi-Resistant Salmonella Typhimurium DT104 Linked to Carpaccio, Denmark, 2005. Epidemiol. Infect. 2007, 135, 900–907. [Google Scholar] [CrossRef]
- Ellis-Iversen, J.; Seyfarth, A.M.; Korsgaard, H.; Bortolaia, V.; Munck, N.; Dalsgaard, A. Antimicrobial Resistant, E. Coli and Enterococci in Pangasius Fillets and Prawns in Danish Retail Imported from Asia. Food Control 2020, 114, 106958. [Google Scholar] [CrossRef]
- Ferreira, A.C.A.D.O.; Pavelquesi, S.L.S.; Monteiro, E.D.S.; Rodrigues, L.F.S.; Silva, C.M.D.S.; Da Silva, I.C.R.; Orsi, D.C. Prevalence and Antimicrobial Resistance of Salmonella Spp. In Aquacultured Nile Tilapia (Oreochromis Niloticus) Commercialized in Federal District, Brazil. Foodborne Pathog. Dis. 2021, 18, 778–783. [Google Scholar] [CrossRef]
- Teuber, M.; Meile, L.; Schwartz, F. Acquired Antibiotic Resistance in Lactic Acid Bacteria from Foods. Antonie Van Leeuwenhoek 1999, 76, 115–137. [Google Scholar] [CrossRef]
- Masco, L.; Van Hoorde, K.; De Brandt, E.; Swings, J.; Huys, G. Antimicrobial Susceptibility of Bifidobacterium Strains from Humans, Animals and Probiotic Products. J. Antimicrob. Chemother. 2006, 58, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Masco, L.; Baert, L.; Huys, G.; Debevere, J.; Swings, J. Prevalence and Diversity of Tetracycline Resistant Lactic Acid Bacteria and Their Tet Genes along the Process Line of Fermented Dry Sausages. Syst. Appl. Microbiol. 2003, 26, 277–283. [Google Scholar] [CrossRef]
- Kastner, S.; Perreten, V.; Bleuler, H.; Hugenschmidt, G.; Lacroix, C.; Meile, L. Antibiotic Susceptibility Patterns and Resistance Genes of Starter Cultures and Probiotic Bacteria Used in Food. Syst. Appl. Microbiol. 2006, 29, 145–155. [Google Scholar] [CrossRef]
- Resch, M.; Nagel, V.; Hertel, C. Antibiotic Resistance of Coagulase-Negative Staphylococci Associated with Food and Used in Starter Cultures. Int. J. Food Microbiol. 2008, 127, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.C.; Gebreyes, W.A.; Abley, M.J.; Harper, A.L.; Forshey, B.M.; Male, M.J.; Martin, H.W.; Molla, B.Z.; Sreevatsan, S.; Thakur, S.; et al. Methicillin-Resistant Staphylococcus Aureus in Pigs and Farm Workers on Conventional and Antibiotic-Free Swine Farms in the USA. PLoS ONE 2013, 8, e63704. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Rodríguez, F.; Mercanoglu Taban, B. A State-of-Art Review on Multi-Drug Resistant Pathogens in Foods of Animal Origin: Risk Factors and Mitigation Strategies. Front. Microbiol. 2019, 10, 2091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, D.; Sharma, P.K.; Malik, A. Prevalence and Antimicrobial Susceptibility of Drug Resistant Staphylococcus Aureus in Raw Milk of Dairy Cattle. Int. Res. J. Microbiol. 2011, 2011, 466–470. [Google Scholar]
- Woolhouse, M.E.J.; Ward, M.J. Sources of Antimicrobial Resistance. Science 2013, 341, 1460–1461. [Google Scholar] [CrossRef] [PubMed]
- Alcaine, S.D.; Warnick, L.D.; Wiedmann, M. Antimicrobial Resistance in Nontyphoidal Salmonella. J. Food Prot. 2007, 70, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E.; et al. The Global Threat of Antimicrobial Resistance: Science for Intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered from the Food Chain through National Antimicrobial Resistance Monitoring System between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; López, M.; Alvarez-Ordóñez, A. Food Processing as a Risk Factor for Antimicrobial Resistance Spread along the Food Chain. Curr. Opin. Food Sci. 2019, 30, 21–26. [Google Scholar] [CrossRef]
- Barber, D.A.; Miller, G.Y.; McNamara, P.E. Models of Antimicrobial Resistance and Foodborne Illness: Examining Assumptions and Practical Applications. J. Food Prot. 2003, 66, 700–709. [Google Scholar] [CrossRef]
- Ou, C.; Shang, D.; Yang, J.; Chen, B.; Chang, J.; Jin, F.; Shi, C. Prevalence of Multidrug-Resistant Staphylococcus Aureus Isolates with Strong Biofilm Formation Ability among Animal-Based Food in Shanghai. Food Control 2020, 112, 107106. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental Factors Influencing the Development and Spread of Antibiotic Resistance. FEMS Microbiol. Rev. 2018, 42, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Swift, B.M.C.; Bennett, M.; Waller, K.; Dodd, C.; Murray, A.; Gomes, R.L.; Humphreys, B.; Hobman, J.L.; Jones, M.A.; Whitlock, S.E.; et al. Anthropogenic Environmental Drivers of Antimicrobial Resistance in Wildlife. Sci. Total Environ. 2019, 649, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D.; Wang, J.; Fanning, S.; Mcmahon, B.J. Antimicrobial Resistance in Wildlife: Implications for Public Health. Zoonoses Public Health 2015, 62, 534–542. [Google Scholar] [CrossRef] [Green Version]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the Wild: Antibiotic Resistance Genes in Natural Environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Yuan, Y.; Liang, B.; Jiang, B.W.; Zhu, L.W.; Wang, T.C.; Li, Y.G.; Liu, J.; Guo, X.J.; Ji, X.; Sun, Y. Migratory Wild Birds Carrying Multidrug-Resistant Escherichia Coli as Potential Transmitters of Antimicrobial Resistance in China. PLoS ONE 2021, 16, e261444. [Google Scholar] [CrossRef]
- Viceconte, G.; Petrosillo, N. COVID-19 R0: Magic Number or Conundrum? Infect. Dis. Rep. 2020, 12, 8516. [Google Scholar] [CrossRef] [Green Version]
- Malvy, D.; McElroy, A.K.; de Clerck, H.; Günther, S.; van Griensven, J. Ebola Virus Disease. Lancet 2019, 393, 936–948. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Giri, S.; Wang, L.; Luo, B. SWAT Modeling of Fecal Indicator Bacteria Fate and Transport in a Suburban Watershed with Mixed Land Uses. Proc. Int. Acad. Ecol. Environ. Sci. 2018, 8, 28–46. [Google Scholar]
- Rolland, R.M.; Hausfater, G.; Marshall, B.; Levy, S.B. Antibiotic-Resistant Bacteria in Wild Primates: Increased Prevalence in Baboons Feeding on Human Refuse. Appl. Environ. Microbiol. 1985, 49, 791–794. [Google Scholar] [CrossRef] [Green Version]
- Rwego, I.B.; Isabirye-Basuta, G.; Gillespie, T.R.; Goldberg, T.L. Gastrointestinal Bacterial Transmission among Humans, Mountain Gorillas, and Livestock in Bwindi Impenetrable National Park, Uganda. Conserv. Biol. 2008, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed]
- Bonnedahl, J.; Drobni, M.; Gauthier-Clerc, M.; Hernandez, J.; Granholm, S.; Kayser, Y.; Melhus, Å.; Kahlmeter, G.; Waldenström, J.; Johansson, A.; et al. Dissemination of Escherichia Coli with CTX-M Type ESBL between Humans and Yellow-Legged Gulls in the South of France. PLoS ONE 2009, 4, e5958. [Google Scholar] [CrossRef] [PubMed]
- Thaller, M.C.; Migliore, L.; Marquez, C.; Tapia, W.; Cedeño, V.; Rossolini, G.M.; Gentile, G. Tracking Acquired Antibiotic Resistance in Commensal Bacteria of Galápagos Land Iguanas: No Man, No Resistance. PLoS ONE 2010, 5, e8989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Goethem, M.W.; Pierneef, R.; Bezuidt, O.K.I.; Van De Peer, Y.; Cowan, D.A.; Makhalanyane, T.P. A Reservoir of ‘Historical’ Antibiotic Resistance Genes in Remote Pristine Antarctic Soils. Microbiome 2018, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Tenover, F.C. Mechanisms of Antimicrobial Resistance in Bacteria. Am. J. Med. 2006, 34, S3–S10. [Google Scholar] [CrossRef]
- Fountain, J.C.; Koh, J.; Yang, L.; Pandey, M.K.; Nayak, S.N.; Bajaj, P.; Zhuang, W.J.; Chen, Z.Y.; Kemerait, R.C.; Lee, R.D.; et al. Proteome Analysis of Aspergillus Flavus Isolate-Specific Responses to Oxidative Stress in Relationship to Aflatoxin Production Capability. Sci. Rep. 2018, 8, 3430. [Google Scholar] [CrossRef] [Green Version]
- McCormick, A.; Whitney, C.; Farley, M.; Lynfield, R.; Harrison, L.H.; Bennett, N.M.; Schaffner, W.; Reingold, A.; Hadler, J.; Cieslak, P.; et al. Geographic Diversity and Temporal Trends of Antimicrobial Resistance in Streptococcus Pneumoniae in the United States. Nat. Med. 2003, 9, 424–430. [Google Scholar] [CrossRef]
- Nicholson, F.A.; Smith, S.R.; Alloway, B.J.; Carlton-Smith, C.; Chambers, B.J. An Inventory of Heavy Metals Inputs to Agricultural Soils in England and Wales. Sci. Total Environ. 2003, 311, 205–219. [Google Scholar] [CrossRef]
- Hadi, E.H. Characterization of Soil Heavy Metal Contamination in the Abandoned Mine of Zaida (High Moulouya, Morocco). Int. Res. J. Earth Sci. 2015, 3, 1–8. [Google Scholar]
- Hussey, S.J.K.; Purves, J.; Allcock, N.; Fernandes, V.E.; Monks, P.S.; Ketley, J.M.; Andrew, P.W.; Morrissey, J.A. Air Pollution Alters Staphylococcus Aureus and Streptococcus Pneumoniae Biofilms, Antibiotic Tolerance and Colonisation. Environ. Microbiol. 2017, 19, 1868–1880. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; He, S.; Huang, S.; Li, K.; Liu, Z.; Xue, G.; Sun, W. Impacts of Coexisting Antibiotics, Antibacterial Residues, and Heavy Metals on the Occurrence of Erythromycin Resistance Genes in Urban Wastewater. Appl. Microbiol. Biotechnol. 2015, 99, 3971–3980. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.S.; Peltier, G.L.; Stepanauskas, R.; McArthur, J.V. Bacterial Tolerances to Metals and Antibiotics in Metal-Contaminated and Reference Streams. FEMS Microbiol. Ecol. 2006, 58, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.; Thorsen, M.K.; Holm, P.E.; Jensen, J.; Nybroe, O.; Brandt, K.K. Cu Exposure under Field Conditions Coselects for Antibiotic Resistance as Determined by a Novel Cultivation-Independent Bacterial Community Tolerance Assay. Environ. Sci. Technol. 2010, 44, 8724–8728. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial Heavy-Metal and Antibiotic Resistance Genes in a Copper Tailing Dam Area in Northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef] [Green Version]
- Azarbad, H.; Niklińska, M.; Laskowski, R.; van Straalen, N.M.; van Gestel, C.A.M.; Zhou, J.; He, Z.; Wen, C.; Röling, W.F.M. Microbial Community Composition and Functions Are Resilient to Metal Pollution along Two Forest Soil Gradients. FEMS Microbiol. Ecol. 2015, 91, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, A.W.; Power, A.; Hansen, M.G.; Brandt, K.K.; Piliposian, G.; Appleby, P.; O’Neill, P.A.; Jones, R.T.; Sierocinski, P.; Koskella, B.; et al. Heavy Metal Pollution and Co-Selection for Antibiotic Resistance: A Microbial Palaeontology Approach. Environ. Int. 2019, 132, 105117. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, Y.; Huang, H.; Mou, L.; Ru, J.; Zhao, J.; Xiao, S. Long-Term and High-Concentration Heavy-Metal Contamination Strongly Influences the Microbiome and Functional Genes in Yellow River Sediments. Sci. Total Environ. 2018, 637–638, 1400–1412. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, L.; Ye, C.; Yu, X. Co-Selection of Antibiotic Resistance via Copper Shock Loading on Bacteria from a Drinking Water Bio-Filter. Environ. Pollut. 2018, 233, 132–141. [Google Scholar] [CrossRef]
- Shi, P.; Jia, S.; Zhang, X.X.; Zhang, T.; Cheng, S.; Li, A. Metagenomic Insights into Chlorination Effects on Microbial Antibiotic Resistance in Drinking Water. Water Res. 2013, 47, 111–120. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-Selection of Antibiotic and Metal Resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Imran, M.; Das, K.R.; Naik, M.M. Co-Selection of Multi-Antibiotic Resistance in Bacterial Pathogens in Metal and Microplastic Contaminated Environments: An Emerging Health Threat. Chemosphere 2019, 215, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; He, L.; Gao, F.; Zhang, M.; Chen, S.; Wu, D.; Liu, Y.; He, L.; Bai, H.; Ying, G. Antibiotic Resistance Genes in Surface Water and Groundwater from Mining Affected Environments. Sci. Total Environ. 2021, 772, 145516. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban Wastewater Treatment Plants as Hotspots for Antibiotic Resistant Bacteria and Genes Spread into the Environment: A Review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanauskas, R.; Glenn, T.C.; Jagoe, C.H.; Tuckfield, R.C.; Lindell, A.H.; McArthur, J.V. Elevated Microbial Tolerance to Metals and Antibiotics in Metal-Contaminated Industrial Environments. Environ. Sci. Technol. 2005, 39, 3671–3678. [Google Scholar] [CrossRef]
- Chen, S.; Li, X.; Sun, G.; Zhang, Y.; Su, J.; Ye, J. Heavy Metal Induced Antibiotic Resistance in Bacterium LSJC7. Int. J. Mol. Sci. 2015, 16, 23390–23404. [Google Scholar] [CrossRef] [Green Version]
- Kusi, J.; Scheuerman, P.R.; Maier, K.J. Antimicrobial Properties of Silver Nanoparticles May Interfere with Fecal Indicator Bacteria Detection in Pathogen Impaired Streams. Environ. Pollut. 2020, 263, 114536. [Google Scholar] [CrossRef]
- Kusi, J.; Scheuerman, P.R.; Maier, K.J. Emerging Environmental Contaminants (Silver Nanoparticles) Altered the Catabolic Capability and Metabolic Fingerprinting of Microbial Communities. Aquat. Toxicol. 2020, 228, 105633. [Google Scholar] [CrossRef]
- Liao, S.; Zhang, Y.; Pan, X.; Zhu, F.; Jiang, C.; Liu, Q.; Cheng, Z.; Dai, G.; Wu, G.; Wang, L.; et al. Antibacterial Activity and Mechanism of Silver Nanoparticles against Multidrug-Resistant Pseudomonas Aeruginosa. Int. J. Nanomed. 2019, 14, 1469–1487. [Google Scholar] [CrossRef] [Green Version]
- Mann, R.; Holmes, A.; McNeilly, O.; Cavaliere, R.; Sotiriou, G.A.; Rice, S.A.; Gunawan, C. Evolution of Biofilm-Forming Pathogenic Bacteria in the Presence of Nanoparticles and Antibiotic: Adaptation Phenomena and Cross-Resistance. J. Nanobiotechnol. 2021, 19, 291. [Google Scholar] [CrossRef]
- Lu, J.; Wang, Y.; Jin, M.; Yuan, Z.; Bond, P.; Guo, J. Both Silver Ions and Silver Nanoparticles Facilitate the Horizontal Transfer of Plasmid-Mediated Antibiotic Resistance Genes. Water Res. 2020, 169, 115229. [Google Scholar] [CrossRef]
- Stabryla, L.M.; Johnston, K.A.; Diemler, N.A.; Cooper, V.S.; Millstone, J.E.; Haig, S.J.; Gilbertson, L.M. Role of Bacterial Motility in Differential Resistance Mechanisms of Silver Nanoparticles and Silver Ions. Nat. Nanotechnol. 2021, 16, 996–1003. [Google Scholar] [CrossRef]
- Skaland, R.G.; Herrador, B.G.; Hisdal, H.; Hygen, H.O.; Hyllestad, S.; Lund, V.; White, R.; Wong, W.K.; Nygård, K. Impacts of Climate Change on Drinking Water Quality in Norway. J. Water Health 2022, 20, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Okaka, F.O.; Odhiambo, B.D.O. Relationship between Flooding and out Break of Infectious Diseasesin Kenya: A Review of the Literature. J. Environ. Public Health 2018, 2018, 5452938. [Google Scholar] [CrossRef] [Green Version]
- Saulnier, D.D.; Hanson, C.; Ir, P.; Alvesson, H.M.; von Schreeb, J. The Effect of Seasonal Floods on Health: Analysis of Six Years of National Health Data and Flood Maps. Int. J. Environ. Res. Public Health 2018, 15, 665. [Google Scholar] [CrossRef] [Green Version]
- Williamson, S. Economic Impacts of Climate Change on Colorado Contributors to the Report; The Center for Integrative Environmental Research University of Maryland: College Park, MD, USA, 2008. [Google Scholar]
- Burnham, J.P. Climate Change and Antibiotic Resistance: A Deadly Combination. Ther. Adv. Infect. Dis. 2021, 8, 2049936121991374. [Google Scholar] [CrossRef] [PubMed]
- MacFadden, D.R.; McGough, S.F.; Fisman, D.; Santillana, M.; Brownstein, J.S. Antibiotic Resistance Increases with Local Temperature. Nat. Clim. Chang. 2018, 8, 510–514. [Google Scholar] [CrossRef]
- Rodrigues, D.F.; Jaisi, D.P.; Elimelech, M. Toxicity of Functionalized Single-Walled Carbon Nanotubes on Soil Microbial Communities: Implications for Nutrient Cycling in Soil. Environ. Sci. Technol. 2013, 47, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Patz, J.; Githeko, A.; McCarty, J. Chapter 6: Climate Change and Infectious Diseases. In Climate Change and Human Health: Risks and Responses; WHO: Geneva, Switzerland, 2003. [Google Scholar]
- Treacy, J. Drinking Water Treatment and Challenges in Developing Countries. In The Relevance of Hygiene to Health in Developing Countries; Potgieter, N., Traore, A.N., Eds.; IntechOpen: London, UK, 2019. [Google Scholar]
| Surface Water | Resistant Pathogen/Gene | Country | Reference |
|---|---|---|---|
| River watershed | Shiga toxin-producing Escherichia coli | Canada | [15] |
| Lake | Enterobacteriaceae | Brazil | [16] |
| Pond | ARGs | Bangladesh | [22] |
| Lake and river | ARGs | China | [23,24] |
| Lake | ARGs | China | [25,26,27,28,29,30] |
| River | Escherichia coli and Klebsiella pneumoniae | Lebanon | [31] |
| River | ARGs | Germany | [32] |
| River/sediment | ARGs | China | [33,34,35,36] |
| River | ARGs | Brazil | [37,38] |
| Stormwater | ARGs | United States | [39] |
| River | ARGs | Sri Lanka | [40] |
| River | ARGs | China | [36,41,42] |
| River | ARGs and MDR | India | [43] |
| River | ARGs | Germany | [44] |
| Lake | E. coli, ARGs, and MDR | Sri Lanka | [45] |
| Marine /lake/river | ARGs | Puerto Rico | [17] |
| River | ARGs | South Africa | [46] |
| River | E. coli and MDR | India | [47] |
| Estuarine | ARGs | Portugal | [48] |
| Lake/river | Enterococcus faecalis, Enterococcus faecium, Enterococcus mundtii, ARGs | Serbia | [49] |
| Lake/river/sediment | MDR | Germany | [50] |
| River | ARGs | Australia and Germany | [51] |
| Lake/river/stream | ARGs and MRSA | Portugal | [18] |
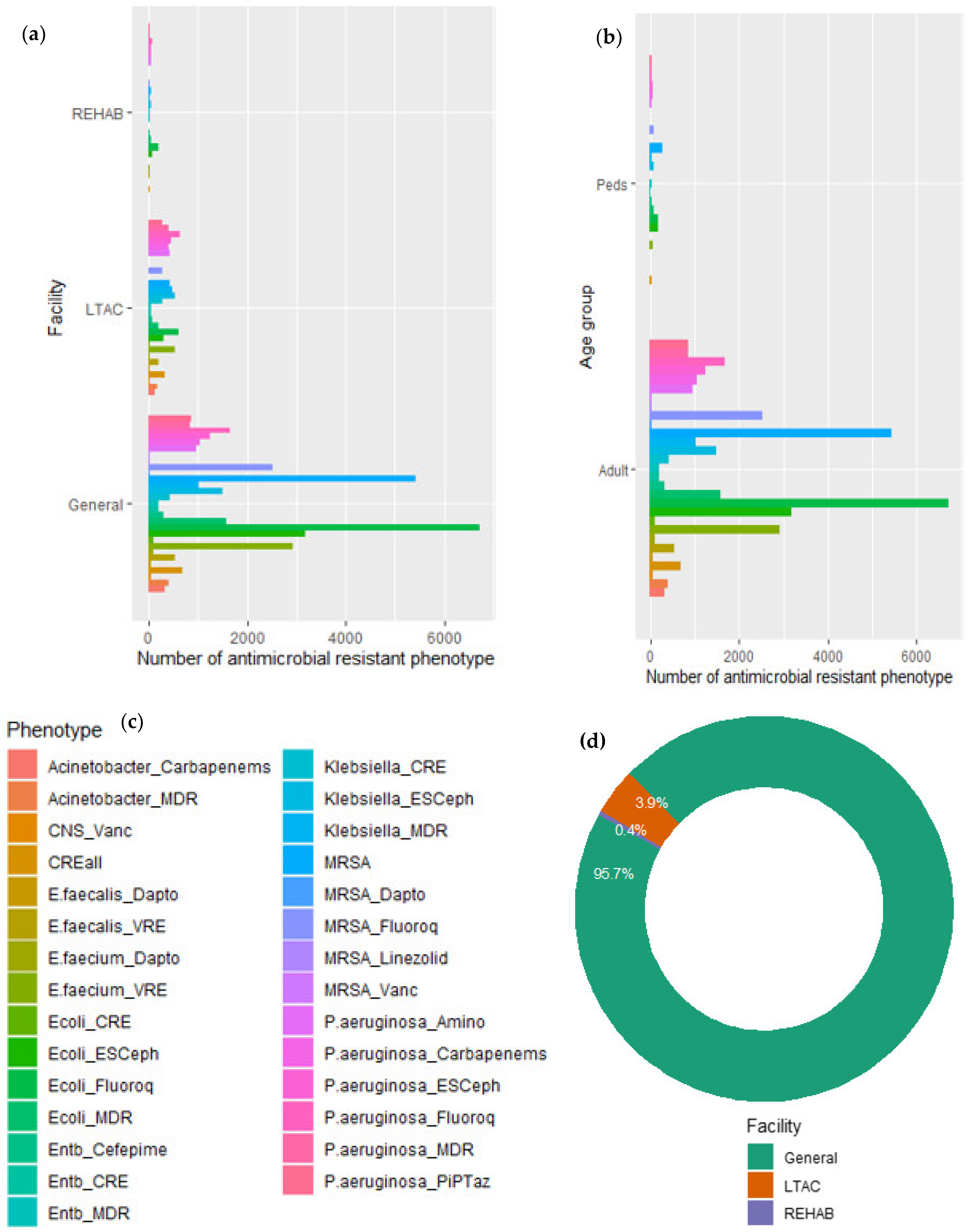
| Pathogen | Phenotype | Abbreviation | Selected Group of Antimicrobials |
|---|---|---|---|
| Escherichia coli | Carbapenem-resistant (CRE) | Ecoli_CRE | Imipenem, meropenem, doripenem, ertapenem |
| Cephalosporin-resistant | Ecoli_ESCeph | Ceftriaxone, ceftazidime, cefepime, cefotaxime | |
| Fluoroquinolone-resistant | Ecoli_Fluoroq | Ciprofloxacin, levofloxacin, moxifloxacin | |
| Multidrug-resistant (MDR) | Ecoli_MDR | Cephalosporins, fluoroquinolones, aminoglycosides, piperacillin/tazobactam | |
| Enterobacter | Carbapenem-resistant (CRE) | Entb_CRE | Imipenem, meropenem, doripenem, ertapenem |
| Cefepime-resistant | Entb_Cefepime | Cefepime | |
| Multidrug-resistant (MDR) | Entb_MDR | Cefepime, fluoroquinolones, aminoglycosides, piperacillin/tazobactam | |
| Klebsiella | Carbapenem-resistant (CRE) | Klebsiella_CRE | Imipenem, meropenem, doripenem, ertapenem |
| Cephalosporin-resistant | Klebsiella_ESCeph | Ceftriaxone, ceftazidime, cefepime, cefotaxim | |
| Multidrug-resistant (MDR) | Klebsiella_MDR | Cephalosporins, fluoroquinolones, aminoglycosides, piperacillin/tazobactam | |
| Pseudomonas aeruginosa | Carbapenem-resistant | P. aeruginosa_Carbapenems | Imipenem, meropenem, doripenem |
| Cephalosporin-resistant | P. aeruginosa_ESCeph | Ceftazidime, cefepime | |
| Fluoroquinolone-resistant | P. aeruginosa_Fluoroq | Ciprofloxacin, levofloxacin | |
| Aminoglycoside-resistant | P. aeruginosa_Amino | amikacin, gentamicin, tobramycin | |
| Piperacillin/tazobactam-resistant | P. aeruginosa_PiPTaz | Piperacillin, piperacillin/tazobactam | |
| Multidrug-resistant (MDR) | P. aeruginosa_MDR | Cephalosporins, fluoroquinolones, aminoglycosides, carbapenems, piperacillin/tazobactam | |
| Enterococcus faecium | Vancomycin-resistant (VRE) | E. faecium_VRE | Vancomycin |
| Daptomycin-resistant | E. faecium_Dapto | Daptomycin (NS) | |
| Enterococcus faecalis | Vancomycin-resistant (VRE) | E. faecalis_VRE | Vancomycin |
| Daptomycin-resistant | E. faecalis_Dapto | Daptomycin (NS) | |
| Coagulase-negative Staphylococci | Vancomycin-resistant | CNS_Vanc | Vancomycin |
| Enterobacterales | Carbapenem-resistant (CRE) | CREall | Imipenem, meropenem, doripenem, ertapenem |
| Staphylococcus aureus | Methicillin-resistant (MRSA) | MRSA | Methicillin, oxacillin, cefoxitin |
| Linezolid-resistant MRSA | MRSA_Linezolid | Linezolid | |
| Fluoroquinolone-resistant MRSA | MRSA_Fluoroq | Ciprofloxacin and/or levofloxacin | |
| Vancomycin-resistant MRSA | MRSA_Vanc | Vancomycin | |
| Daptomycin-resistant MRSA | MRSA_Dapto | Daptomycin (NS) | |
| Acinetobacter | Carbapenem-resistant | Acinetobacter_Carbapenems | Imipenem, meropenem, doripenem |
| Multidrug-resistant (MDR) | Acinetobacter_MDR | Cephalosporins, fluoroquinolones, aminoglycosides, carbapenems, piperacillin/tazobactam, ampicillin/sulbactam |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kusi, J.; Ojewole, C.O.; Ojewole, A.E.; Nwi-Mozu, I. Antimicrobial Resistance Development Pathways in Surface Waters and Public Health Implications. Antibiotics 2022, 11, 821. https://doi.org/10.3390/antibiotics11060821
Kusi J, Ojewole CO, Ojewole AE, Nwi-Mozu I. Antimicrobial Resistance Development Pathways in Surface Waters and Public Health Implications. Antibiotics. 2022; 11(6):821. https://doi.org/10.3390/antibiotics11060821
Chicago/Turabian StyleKusi, Joseph, Catherine Oluwalopeye Ojewole, Akinloye Emmanuel Ojewole, and Isaac Nwi-Mozu. 2022. "Antimicrobial Resistance Development Pathways in Surface Waters and Public Health Implications" Antibiotics 11, no. 6: 821. https://doi.org/10.3390/antibiotics11060821
APA StyleKusi, J., Ojewole, C. O., Ojewole, A. E., & Nwi-Mozu, I. (2022). Antimicrobial Resistance Development Pathways in Surface Waters and Public Health Implications. Antibiotics, 11(6), 821. https://doi.org/10.3390/antibiotics11060821






