Biofilm Formation of Staphylococcus aureus from Pets, Livestock, and Wild Animals: Relationship with Clonal Lineages and Antimicrobial Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Bacterial Isolates
2.2. Biofilm Formation Assay
2.2.1. Biofilm Formation Assay
2.3. Confocal Laser Scanning Microscopy
2.4. Effect of Antimicrobials on 24 h-Old Biofilms
Effect of Antimicrobials on Metabolic Activity
2.5. Statistical Analysis
3. Results
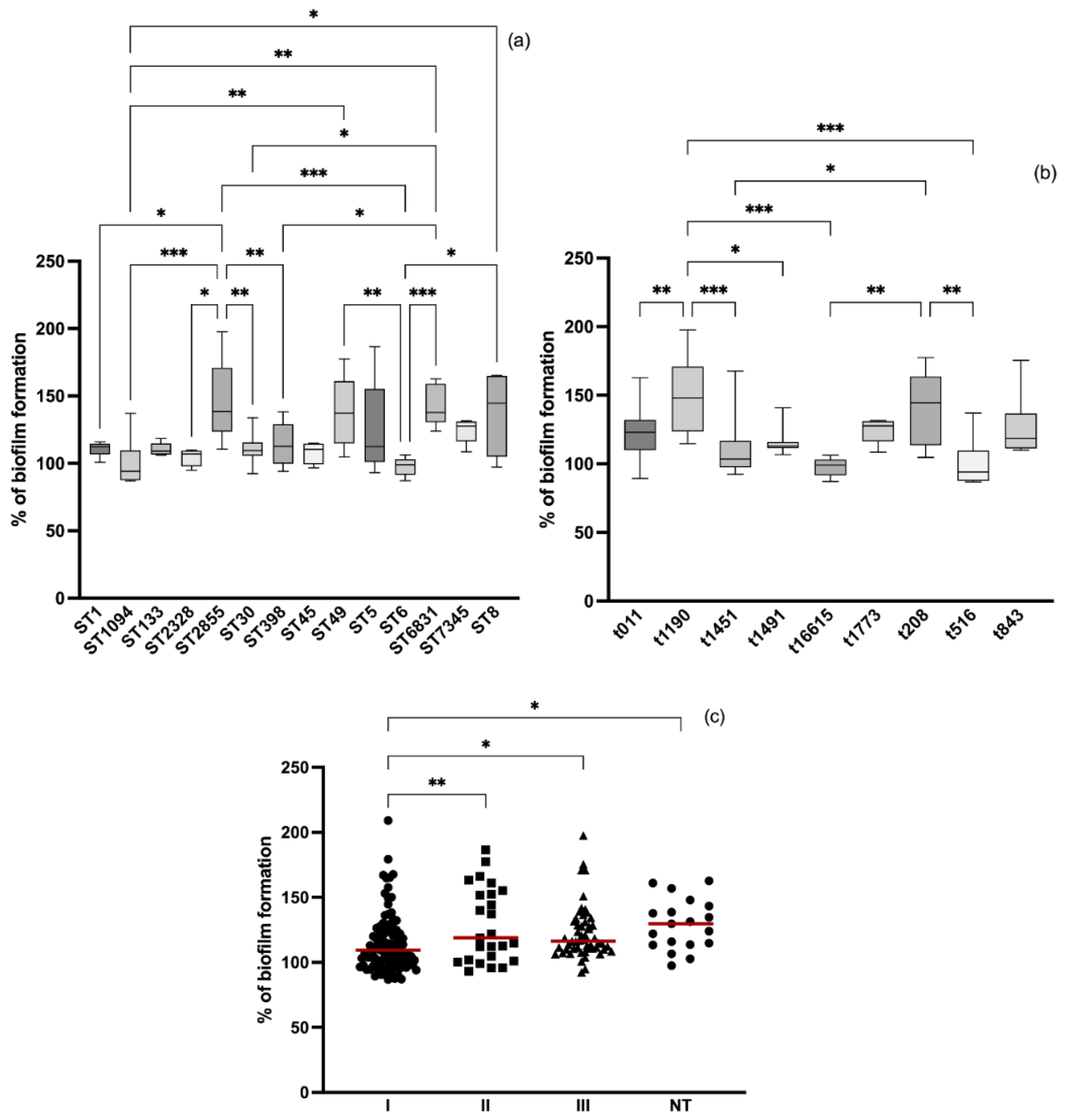
3.1. Biofilm Formation
3.2. CLSM Analysis
3.3. Antimicrobial Resistance and Biofilm Formation
3.4. Relation between Molecular Typing and Biofilm Formation
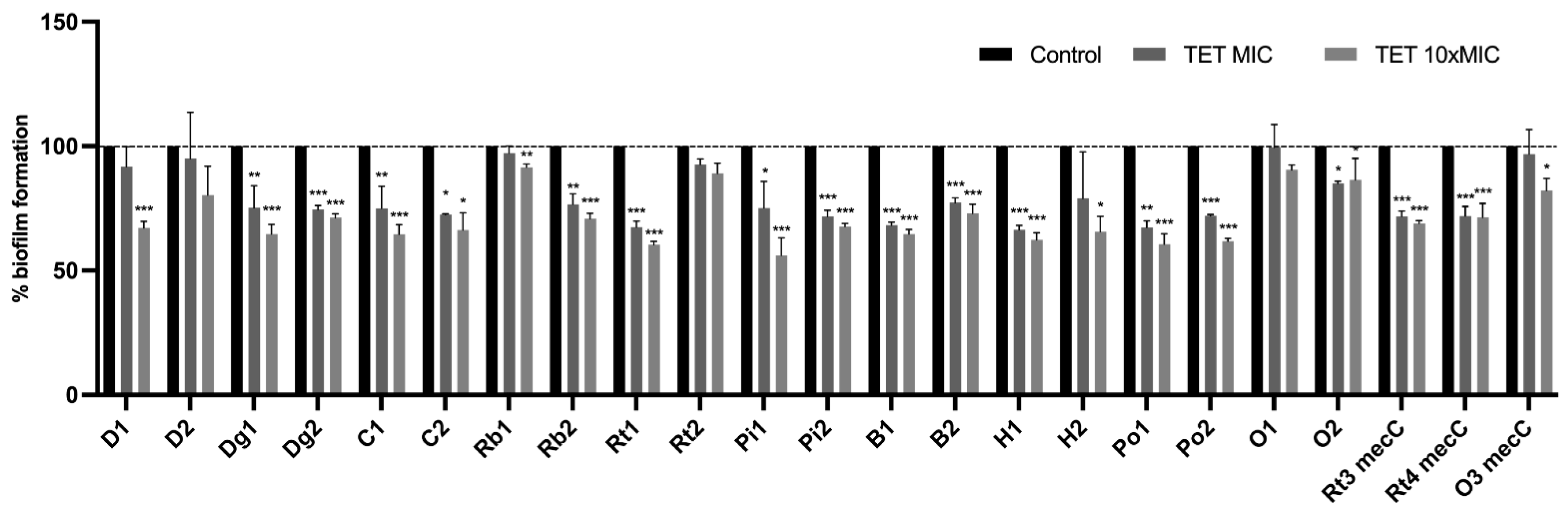
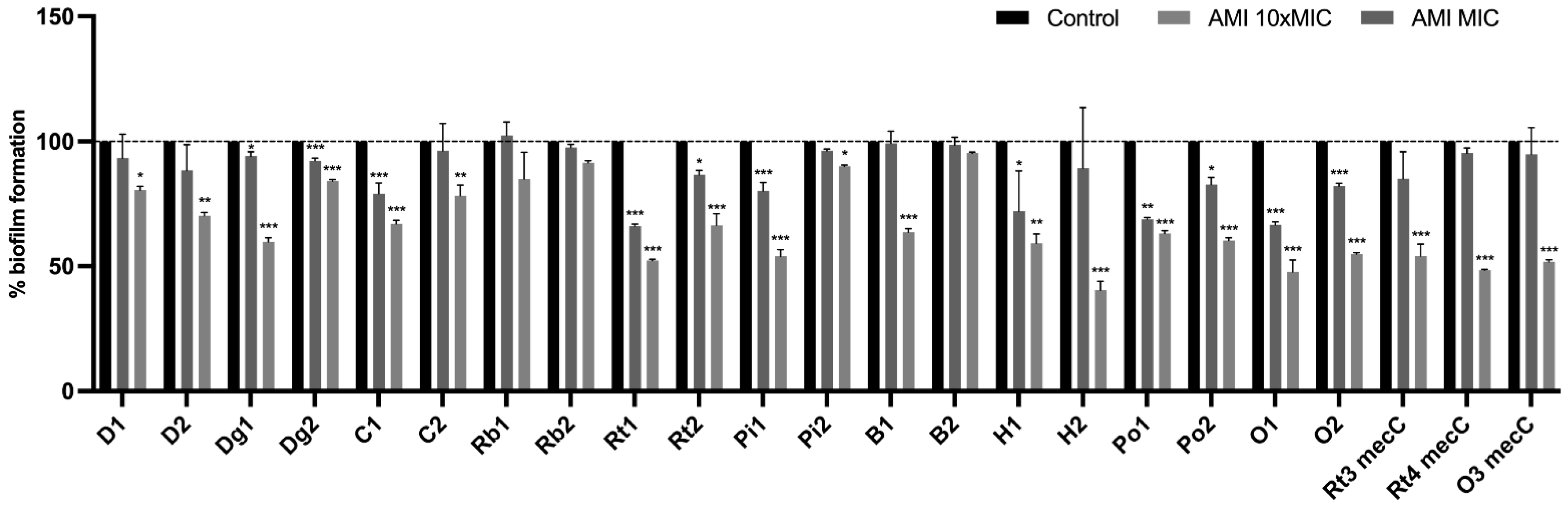
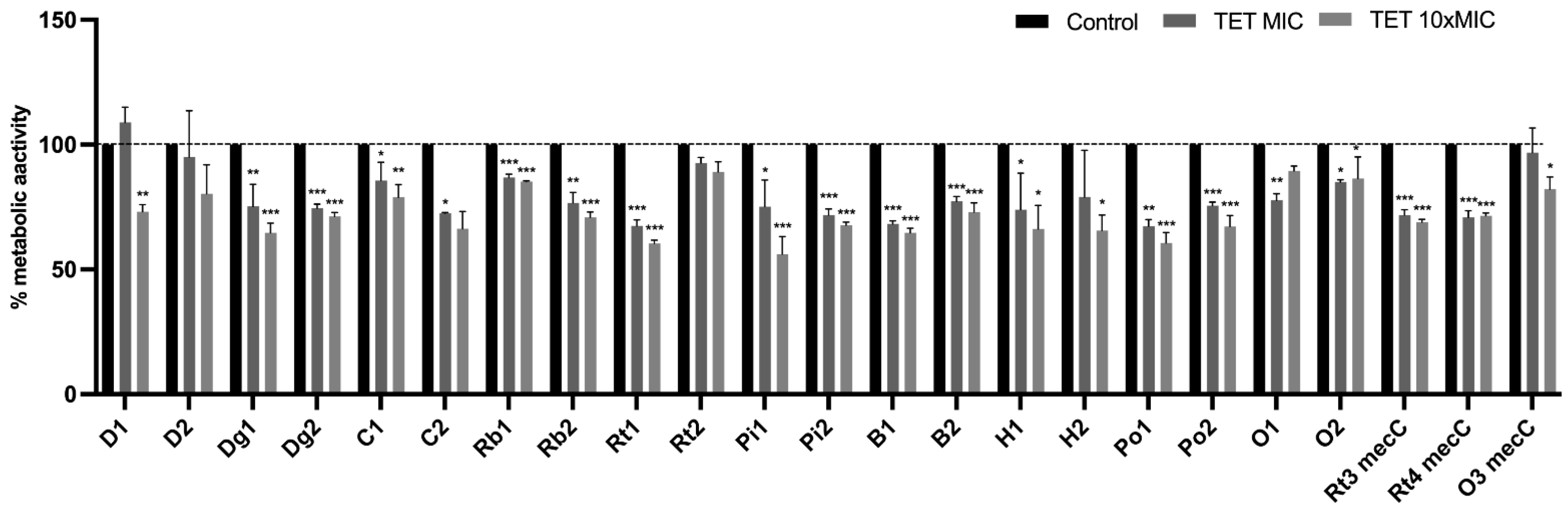
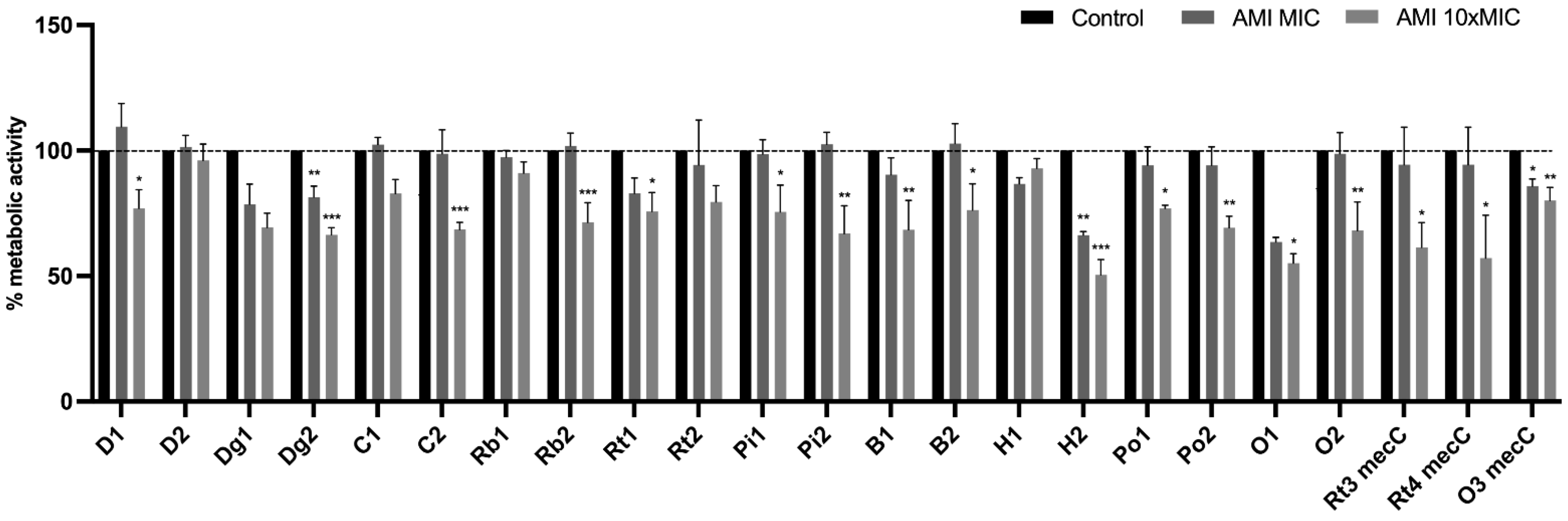
3.5. Effect of Antimicrobials on 24 h-Old Biofilms
Effect of Antimicrobials on Metabolic Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bruce, S.A.; Smith, J.T.; Mydosh, J.L.; Ball, J.; Needle, D.B.; Gibson, R.; Andam, C.P. Shared antibiotic resistance and virulence genes in Staphylococcus aureus from diverse animal hosts. Sci. Rep. 2022, 12, 4413. [Google Scholar] [CrossRef] [PubMed]
- Olaniyi, R.; Pozzi, C.; Grimaldi, L.; Bagnoli, F. Staphylococcus aureus-associated skin and soft tissue infections: Anatomical localization, epidemiology, therapy and potential prophylaxis. In Staphylococcus aureus; Springer: Berlin/Heidelberg, Germany, 2016; pp. 199–227. [Google Scholar]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Claudia, L.; Andreas, P.; Bernhard, K. Staphylococcus aureus Colonization of the Human Nose and Interaction with Other Microbiome Members. Microbiol. Spectr. 2019, 7, 1–10. [Google Scholar]
- Haag, A.F.; Fitzgerald, J.R.; Penadés, J.R. Staphylococcus aureus in Animals. Gram-Positive Pathog. 2019, 7, 731–746. [Google Scholar]
- Silva, V.; Pereira, J.E.; Maltez, L.; Ferreira, E.; Manageiro, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity of methicillin-resistant staphylococci among wild Lepus granatensis: First detection of mecA-MRSA in hares. FEMS Microbiol. Ecol. 2020, 96, fiz204. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Lopes, A.F.; Soeiro, V.; Caniça, M.; Manageiro, V.; Pereira, J.E.; Maltez, L.; Capelo, J.L.; Igrejas, G.; Poeta, P. Nocturnal Birds of Prey as Carriers of Staphylococcus aureus and Other Staphylococci: Diversity, Antimicrobial Resistance and Clonal Lineages. Antibiotics 2022, 11, 240. [Google Scholar] [CrossRef]
- Silva, V.; Vieira-Pinto, M.; Saraiva, C.; Manageiro, V.; Reis, L.; Ferreira, E.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Prevalence and Characteristics of Multidrug-Resistant Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) CC398 Isolated from Quails (Coturnix Coturnix Japonica) Slaughtered for Human Consumption. Animals 2021, 11, 2038. [Google Scholar] [CrossRef] [PubMed]
- Held, J.; Gmeiner, M.; Mordmüller, B.; Matsiégui, P.-B.; Schaer, J.; Eckerle, I.; Weber, N.; Matuschewski, K.; Bletz, S.; Schaumburg, F. Bats are rare reservoirs of Staphylococcus aureus complex in Gabon. Infect. Genet. Evol. 2017, 47, 118–120. [Google Scholar] [CrossRef]
- Dastmalchi Saei, H.; Panahi, M. Genotyping and antimicrobial resistance of Staphylococcus aureus isolates from dairy ruminants: Differences in the distribution of clonal types between cattle and small ruminants. Arch. Microbiol. 2020, 202, 115–125. [Google Scholar] [CrossRef]
- Espinosa-Gongora, C.; Chrobak, D.; Moodley, A.; Bertelsen, M.F.; Guardabassi, L. Occurrence and distribution of Staphylococcus aureus lineages among zoo animals. Vet. Microbiol. 2012, 158, 228–231. [Google Scholar] [CrossRef]
- Silva, V.; Alfarela, C.; Caniça, M.; Manageiro, V.; Nóvoa, M.; Leiva, B.; Kress, M.; Capelo, J.L.; Poeta, P.; Igrejas, G. A One Health Approach Molecular Analysis of Staphylococcus aureus Reveals Distinct Lineages in Isolates from Miranda Donkeys (Equus asinus) and Their Handlers. Antibiotics 2022, 11, 374. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Grúa, E.; Pérez-Fuentes, S.; Viana, D.; Cardells, J.; Lizana, V.; Aguiló, J.; Selva, L.; Corpa, J.M. Marked presence of methicillin-resistant Staphylococcus aureus in wild lagomorphs in Valencia, Spain. Animals 2020, 10, 1109. [Google Scholar] [CrossRef] [PubMed]
- Abbott, Y.; Leggett, B.; Rossney, A.S.; Leonard, F.C.; Markey, B.K. Isolation rates of meticillin-resistant Staphylococcus aureus in dogs, cats and horses in Ireland. Vet. Rec. 2010, 166, 451–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiser-Thom, S.; Gerber, V.; Collaud, A.; Hurni, J.; Perreten, V. Prevalence and WGS-based characteristics of Staphylococcus aureus in the nasal mucosa and pastern of horses with equine pastern dermatitis. BMC Vet. Res. 2022, 18, 79. [Google Scholar] [CrossRef]
- Molineri, A.I.; Camussone, C.; Zbrun, M.V.; Archilla, G.S.; Cristiani, M.; Neder, V.; Calvinho, L.; Signorini, M. Antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis: Systematic review and meta-analysis. Prev. Vet. Med. 2021, 188, 105261. [Google Scholar] [CrossRef]
- Nazia, M.K.K.; Durrani, N.U.; Kamboh, A.A.; Lakho, S.A.; Rind, R.; Abro, S.H.; Soomro, N.M. Prevalence of septic arthritis caused by Staphylococcus aureus in poultry birds at Tandojam. Pakistan J. Anim. Health Prod. 2015, 3, 73–77. [Google Scholar]
- Rosell, J.M.; De la Fuente, L.F. Mastitis on Rabbit Farms: Prevalence and Risk Factors. Animals 2018, 8, 98. [Google Scholar] [CrossRef] [Green Version]
- Silva, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity and genetic lineages of environmental staphylococci: A surface water overview. FEMS Microbiol. Ecol. 2020, 96, fiaa191. [Google Scholar] [CrossRef]
- King, J.M.; Kulhankova, K.; Stach, C.S.; Vu, B.G.; Salgado-Pabón, W. Phenotypes and Virulence among Staphylococcus aureus USA100, USA200, USA300, USA400, and USA600 Clonal Lineages. Msphere 2022, 1, e00071-16. [Google Scholar] [CrossRef] [Green Version]
- Sieber, R.N.; Larsen, A.R.; Urth, T.R.; Iversen, S.; Møller, C.H.; Skov, R.L.; Larsen, J.; Stegger, M. Genome investigations show host adaptation and transmission of LA-MRSA CC398 from pigs into Danish healthcare institutions. Sci. Rep. 2019, 9, 18655. [Google Scholar] [CrossRef] [Green Version]
- Ceballos, S.; Aspiroz, C.; Ruiz-Ripa, L.; Reynaga, E.; Azcona-Gutiérrez, J.M.; Rezusta, A.; Seral, C.; Antoñanzas, F.; Torres, L.; López, C.; et al. Epidemiology of MRSA CC398 in hospitals located in Spanish regions with different pig-farming densities: A multicentre study. J. Antimicrob. Chemother. 2019, 74, 2157–2161. [Google Scholar] [CrossRef] [PubMed]
- Puvača, N.; Chantal, B. Welfare and legal aspects of making decisions on medical treatments of pet animals. Pravo-Teor. I Praksa 2020, 37, 55–64. [Google Scholar] [CrossRef]
- Lika, E.; Puvača, N.; Jeremić, D.; Stanojević, S.; Shtylla Kika, T.; Cocoli, S.; de Llanos Frutos, R. Antibiotic Susceptibility of Staphylococcus Species Isolated in Raw Chicken Meat from Retail Stores. Antibiotics 2021, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Morar, A.; Ban-Cucerzan, A.; Herman, V.; Tîrziu, E.; Sallam, K.I.; Abd-Elghany, S.M.; Imre, K. Multidrug Resistant Coagulase-Positive Staphylococcus aureus and Their Enterotoxins Detection in Traditional Cheeses Marketed in Banat Region, Romania. Antibiotics 2021, 10, 1458. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Microbiol. Spectr. 2018, 6, 4–6. [Google Scholar] [CrossRef]
- Silva, V.; Capelo, J.L.; Igrejas, G.; Poeta, P. Molecular Mechanisms of Antimicrobial Resistance in Staphylococcus aureus Biofilms BT-Emerging Modalities in Mitigation of Antimicrobial Resistance; Akhtar, N., Singh, K.S., Prerna, Goyal, D., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 291–314. ISBN 978-3-030-84126-3. [Google Scholar]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef] [Green Version]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Liang, Y.; Chen, L.; Wang, W.; Wang, J.; Li, B.; Li, L.; Chen, D.; Xu, Z. Formation and development of Staphylococcus biofilm: With focus on food safety. J. Food Saf. 2017, 37, e12358. [Google Scholar] [CrossRef]
- Kang, M.; Ko, Y.-P.; Liang, X.; Ross, C.L.; Liu, Q.; Murray, B.E.; Höök, M. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. J. Biol. Chem. 2013, 288, 20520–20531. [Google Scholar] [CrossRef] [Green Version]
- Silva, V.; Almeida, L.; Gaio, V.; Cerca, N.; Manageiro, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Biofilm Formation of Multidrug-Resistant MRSA Strains Isolated from Different Types of Human Infections. Pathogens 2021, 10, 970. [Google Scholar] [CrossRef]
- Kumaran, D.; Taha, M.; Yi, Q.; Ramirez-Arcos, S.; Diallo, J.-S.; Carli, A.; Abdelbary, H. Does Treatment Order Matter? Investigating the Ability of Bacteriophage to Augment Antibiotic Activity against Staphylococcus aureus Biofilms. Front. Microbiol. 2018, 9, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speziale, P.; Geoghegan, J.A. Biofilm formation by staphylococci and streptococci: Structural, functional, and regulatory aspects and implications for pathogenesis. Front. Cell. Infect. Microbiol. 2015, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Malone, M.; Bjarnsholt, T.; McBain, A.J.; James, G.A.; Stoodley, P.; Leaper, D.; Tachi, M.; Schultz, G.; Swanson, T.; Wolcott, R.D. The prevalence of biofilms in chronic wounds: A systematic review and meta-analysis of published data. J. Wound Care 2017, 26, 20–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef]
- Pedersen, R.R.; Krömker, V.; Bjarnsholt, T.; Dahl-Pedersen, K.; Buhl, R.; Jørgensen, E. Biofilm Research in Bovine Mastitis. Front. Vet. Sci. 2021, 8, 449. [Google Scholar] [CrossRef]
- Milivojevic, D.; Šumonja, N.; Medić, S.; Pavic, A.; Moric, I.; Vasiljevic, B.; Senerovic, L.; Nikodinovic-Runic, J. Biofilm-forming ability and infection potential of Pseudomonas aeruginosa strains isolated from animals and humans. Pathog. Dis. 2018, 76, fty041. [Google Scholar] [CrossRef]
- Shah, M.S.; Qureshi, S.; Kashoo, Z.; Farooq, S.; Wani, S.A.; Hussain, M.I.; Banday, M.S.; Khan, A.A.; Gull, B.; Habib, A.; et al. Methicillin resistance genes and in vitro biofilm formation among Staphylococcus aureus isolates from bovine mastitis in India. Comp. Immunol. Microbiol. Infect. Dis. 2019, 64, 117–124. [Google Scholar] [CrossRef]
- Santos, V.; Gomes, A.; Ruiz-Ripa, L.; Mama, O.M.; Sabença, C.; Sousa, M.; Silva, V.; Sousa, T.; Vieira-Pinto, M.; Igrejas, G.; et al. Methicillin-Resistant Staphylococcus aureus CC398 in Purulent Lesions of Piglets and Fattening Pigs in Portugal. Microb. Drug Resist. 2020, 26, 850–856. [Google Scholar] [CrossRef]
- Silva, V.; de Sousa, T.; Gómez, P.; Sabença, C.; Vieira-Pinto, M.; Capita, R.; Alonso-Calleja, C.; Torres, C.; Capelo, J.L.; Igrejas, G.; et al. Livestock-Associated Methicillin-Resistant Staphylococcus aureus (MRSA) in Purulent Subcutaneous Lesions of Farm Rabbits. Foods 2020, 9, 439. [Google Scholar] [CrossRef] [Green Version]
- Silva, V.; Gabriel, S.I.; Borrego, S.B.; Tejedor-Junco, M.T.; Manageiro, V.; Ferreira, E.; Reis, L.; Caniça, M.; Capelo, J.L.; Igrejas, G.; et al. Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal. Animals 2021, 11, 1537. [Google Scholar] [CrossRef]
- Oniciuc, E.-A.; Cerca, N.; Nicolau, A.I. Compositional Analysis of Biofilms Formed by Staphylococcus aureus Isolated from Food Sources. Front. Microbiol. 2016, 7, 390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Machado, C.; Capita, R.; Riesco-Peláez, F.; Alonso-Calleja, C. Visualization and quantification of the cellular and extracellular components of Salmonella Agona biofilms at different stages of development. PLoS ONE 2018, 13, e0200011. [Google Scholar] [CrossRef]
- Silva, V.; Caniça, M.; Ferreira, E.; Vieira-Pinto, M.; Saraiva, C.; Pereira, J.E.; Capelo, J.L.; Igrejas, G.; Poeta, P. Multidrug-Resistant Methicillin-Resistant Coagulase-Negative Staphylococci in Healthy Poultry Slaughtered for Human Consumption. Antibiotics 2022, 11, 365. [Google Scholar] [CrossRef]
- EUCAST. EUCAST Reading Guide for Broth Microdilution; Version 4.0. 2022. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2022_manuals/Reading_guide_BMD_v_4.0_2022.pdf (accessed on 23 February 2022).
- Cascioferro, S.; Carbone, D.; Parrino, B.; Pecoraro, C.; Giovannetti, E.; Cirrincione, G.; Diana, P. Therapeutic Strategies To Counteract Antibiotic Resistance in MRSA Biofilm-Associated Infections. ChemMedChem 2021, 16, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Kern, Z.T.; Jacob, M.E.; Gilbertie, J.M.; Vaden, S.L.; Lyle, S.K. Characteristics of Dogs with Biofilm-Forming Escherichia Coli Urinary Tract Infections. J. Vet. Intern. Med. 2018, 32, 1645–1651. [Google Scholar] [CrossRef] [Green Version]
- Matuszewska, M.; Murray, G.G.R.; Harrison, E.M.; Holmes, M.A.; Weinert, L.A. The Evolutionary Genomics of Host Specificity in Staphylococcus aureus. Trends Microbiol. 2020, 28, 465–477. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, D.; Shi, L.; Cai, R.; Li, C.; Yan, H. Association Between agr Type, Virulence Factors, Biofilm Formation and Antibiotic Resistance of Staphylococcus aureus Isolates from Pork Production. Front. Microbiol. 2018, 9, 1876. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Filipello, V.; Di Ciccio, P.A.; Pitozzi, A.; Ghidini, S.; Scali, F.; Ianieri, A.; Zanardi, E.; Losio, M.N.; Simon, A.C. Assessment of the antibiotic resistance profile, genetic heterogeneity and biofilm production of Methicillin-Resistant Staphylococcus aureus (MRSA) isolated from the Italian swine production chain. Foods 2020, 9, 1141. [Google Scholar] [CrossRef]
- Abdullahi, U.F.; Igwenagu, E.; Mu’azu, A.; Aliyu, S.; Umar, M.I. Intrigues of biofilm: A perspective in veterinary medicine. Vet. World 2016, 9, 12–18. [Google Scholar] [CrossRef]
- Pinto, J.B.; Rossatto, F.C.P.; Martins, P.D.; Frazzon, A.P.G. Genetic relationships and virulence factors in Staphylococcus aureus isolated from raw poultry in South Brazil. Ann. Microbiol. 2015, 65, 1933–1940. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Liang, L.; Xu, X.; Zhou, G. Prevalence, genetic characterization and biofilm formation in vitro of Staphylococcus aureus isolated from raw chicken meat at retail level in Nanjing, China. Food Control 2018, 86, 11–18. [Google Scholar] [CrossRef]
- Szweda, P.; SCHIELMAnn, M.; Milewski, S.; FRAnKOWSKA, A.; JAKuBCZAK, A. Biofilm production and presence of ica and bap genes in Staphylococcus aureus strains isolated from cows with mastitis in the eastern Poland. Pol. J. Microbiol. 2012, 61, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Thiran, E.; Di Ciccio, P.A.; Graber, H.U.; Zanardi, E.; Ianieri, A.; Hummerjohann, J. Biofilm formation of Staphylococcus aureus dairy isolates representing different genotypes. J. Dairy Sci. 2018, 101, 1000–1012. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Li, J.; Qiao, M.; Meng, D.; Meng, Q.; Qiao, J.; Zhang, X.; Wang, L.; Cai, K.; Zhang, J. Characteristic profiles of biofilm, enterotoxins and virulence of Staphylococcus aureus isolates from dairy cows in Xinjiang Province, China. J. Vet. Sci. 2019, 20, 1138054. [Google Scholar] [CrossRef] [Green Version]
- Silveira, D.R.; de Moraes, T.P.; Kaefer, K.; Bach, L.G.; de Oliveira Barbosa, A.; Moretti, V.D.; de Menezes, P.Q.; de Medeiros, U.S.; da Silva, T.T.; Bandarra, P.M. MRSA and enterobacteria of one health concern in wild animals undergoing rehabilitation. Res. Soc. Dev. 2021, 10, e34810111809. [Google Scholar] [CrossRef]
- Osman, K.M.; Amer, A.M.; Badr, J.M.; Helmy, N.M.; Elhelw, R.A.; Orabi, A.; Bakry, M.; Saad, A.S.A. Antimicrobial Resistance, Biofilm Formation and mecA Characterization of Methicillin-Susceptible S. aureus and Non-S. aureus of Beef Meat Origin in Egypt. Front. Microbiol. 2016, 7, 222. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Sun, H.; Yao, K.; Cai, J.; Ren, Y.; Chi, Y. The Prevalence, Antibiotic Resistance and Biofilm Formation of Staphylococcus aureus in Bulk Ready-To-Eat Foods. Biomolecules 2019, 9, 524. [Google Scholar] [CrossRef] [Green Version]
- Neopane, P.; Nepal, H.P.; Shrestha, R.; Uehara, O.; Abiko, Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int. J. Gen. Med. 2018, 11, 25. [Google Scholar] [CrossRef]
- Ou, C.; Shang, D.; Yang, J.; Chen, B.; Chang, J.; Jin, F.; Shi, C. Prevalence of multidrug-resistant Staphylococcus aureus isolates with strong biofilm formation ability among animal-based food in Shanghai. Food Control 2020, 112, 107106. [Google Scholar] [CrossRef]
- Eyoh, A.B.; Toukam, M.; Atashili, J.; Fokunang, C.; Gonsu, H.; Lyonga, E.E.; Mandi, H.; Ikomey, G.; Mukwele, B.; Mesembe, M. Relationship between multiple drug resistance and biofilm formation in Staphylococcus aureus isolated from medical and non-medical personnel in Yaounde, Cameroon. Pan Afr. Med. J. 2014, 17, 186. [Google Scholar] [CrossRef] [PubMed]
- Jimi, S.; Miyazaki, M.; Takata, T.; Ohjimi, H.; Akita, S.; Hara, S. Increased drug resistance of meticillin-resistant Staphylococcus aureus biofilms formed on a mouse dermal chip model. J. Med. Microbiol. 2017, 66, 542. [Google Scholar] [CrossRef] [PubMed]
- Gaire, U.; Thapa Shrestha, U.; Adhikari, S.; Adhikari, N.; Bastola, A.; Rijal, K.R.; Ghimire, P.; Banjara, M.R. Antibiotic Susceptibility, Biofilm Production, and Detection of mecA Gene among Staphylococcus aureus Isolates from Different Clinical Specimens. Diseases 2021, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Lin, Z.; Hu, X.; Yao, W.; Bai, B.; Wang, H.; Li, D.; Chen, Z.; Cheng, H.; Pan, W.; et al. Biofilm formation in erythromycin-resistant Staphylococcus aureus and the relationship with antimicrobial susceptibility and molecular characteristics. Microb. Pathog. 2018, 124, 47–53. [Google Scholar] [CrossRef]
- Chen, L.; Tang, Z.-Y.; Cui, S.-Y.; Ma, Z.-B.; Deng, H.; Kong, W.-L.; Yang, L.-W.; Lin, C.; Xiong, W.-G.; Zeng, Z.-L. Biofilm Production Ability, Virulence and Antimicrobial Resistance Genes in Staphylococcus aureus from Various Veterinary Hospitals. Pathogens 2020, 9, 264. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Grúa, E.; Pérez-Fuentes, S.; Muñoz-Silvestre, A.; Viana, D.; Fernández-Ros, A.B.; Sanz-Tejero, C.; Corpa, J.M.; Selva, L. Characterization of Livestock-Associated Methicillin-Resistant Staphylococcus aureus Isolates Obtained From Commercial Rabbitries Located in the Iberian Peninsula. Front. Microbiol. 2018, 9, 1812. [Google Scholar] [CrossRef] [Green Version]
- Naicker, P.R.; Karayem, K.; Hoek, K.G.P.; Harvey, J.; Wasserman, E. Biofilm formation in invasive Staphylococcus aureus isolates is associated with the clonal lineage. Microb. Pathog. 2016, 90, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Vergara, A.; Normanno, G.; Di Ciccio, P.; Pedonese, F.; Nuvoloni, R.; Parisi, A.; Santagada, G.; Colagiorgi, A.; Zanardi, E.; Ghidini, S.; et al. Biofilm Formation and Its Relationship with the Molecular Characteristics of Food-Related Methicillin-Resistant Staphylococcus aureus (MRSA). J. Food Sci. 2017, 82, 2364–2370. [Google Scholar] [CrossRef]
- Amaral, M.M.; Coelho, L.R.; Flores, R.P.; Souza, R.R.; Silva-Carvalho, M.C.; Teixeira, L.A.; Ferreira-Carvalho, B.T.; Figueiredo, A.M.S. The predominant variant of the Brazilian epidemic clonal complex of methicillin-resistant Staphylococcus aureus has an enhanced ability to produce biofilm and to adhere to and invade airway epithelial cells. J. Infect. Dis. 2005, 192, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Croes, S.; Deurenberg, R.H.; Boumans, M.-L.L.; Beisser, P.S.; Neef, C.; Stobberingh, E.E. Staphylococcus aureus biofilm formation at the physiologic glucose concentration depends on the S. aureus lineage. BMC Microbiol. 2009, 9, 229. [Google Scholar] [CrossRef] [Green Version]
- Vitale, M.; Galluzzo, P.; Buffa, P.G.; Carlino, E.; Spezia, O.; Alduina, R. Comparison of Antibiotic Resistance Profile and Biofilm Production of Staphylococcus aureus Isolates Derived from Human Specimens and Animal-Derived Samples. Antibiotics 2019, 8, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.-X.; Li, Y.; Yang, X.-Q.; Su, H.-Y.; Wang, Q.; Zhang, Z.-H.; Liu, Y.-C.; Tian, C.-L.; Cui, C.-C.; Liu, M.-C. In vitro Antibiotic Susceptibility, Virulence Genes Distribution and Biofilm Production of Staphylococcus aureus Isolates from Bovine Mastitis in the Liaoning Province of China. Infect. Drug Resist. 2020, 13, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Pichette-Jolette, S.; Millette, G.; Demontier, E.; Bran-Barrera, D.; Cyrenne, M.; Ster, C.; Haine, D.; Keefe, G.; Malouin, F.; Roy, J.P. Partial prediction of the duration and the clinical status of Staphylococcus aureus bovine intramammary infections based on the phenotypic and genotypic analysis of isolates. Vet. Microbiol. 2019, 228, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Kwiecinski, J.M.; Jacobsson, G.; Horswill, A.R.; Josefsson, E.; Jin, T. Biofilm formation by Staphylococcus aureus clinical isolates correlates with the infection type. Infect. Dis. 2019, 51, 446–451. [Google Scholar] [CrossRef]
- Cha, J.-O.; Yoo, J.I.; Yoo, J.S.; Chung, H.-S.; Park, S.-H.; Kim, H.S.; Lee, Y.S.; Chung, G.T. Investigation of Biofilm Formation and its Association with the Molecular and Clinical Characteristics of Methicillin-resistant Staphylococcus aureus. Osong Public Health Res. Perspect. 2013, 4, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Cafiso, V.; Bertuccio, T.; Santagati, M.; Demelio, V.; Spina, D.; Nicoletti, G.; Stefani, S. agr-Genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2007, 51, 220–227. [Google Scholar] [CrossRef] [Green Version]
- Jordan, S.C.; Hall, P.R.; Daly, S.M. Nonconformity of biofilm formation in vivo and in vitro based on Staphylococcus aureus accessory gene regulator status. Sci. Rep. 2022, 12, 1251. [Google Scholar] [CrossRef]
- Derakhshan, S.; Navidinia, M.; Haghi, F. Antibiotic susceptibility of human-associated Staphylococcus aureus and its relation to agr typing, virulence genes, and biofilm formation. BMC Infect. Dis. 2021, 21, 627. [Google Scholar] [CrossRef]
- Suresh, M.K.; Biswas, R.; Biswas, L. An update on recent developments in the prevention and treatment of Staphylococcus aureus biofilms. Int. J. Med. Microbiol. 2019, 309, 1–12. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, P.; Maira-Litran, T.; McBain, A.J.; Rickard, A.H.; Whyte, F.W. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 2002, 46, 203–256. [Google Scholar]
- Boles, L.R.; Awais, R.; Beenken, K.E.; Smeltzer, M.S.; Haggard, W.O.; Jessica, A.J. Local Delivery of Amikacin and Vancomycin from Chitosan Sponges Prevent Polymicrobial Implant-Associated Biofilm. Mil. Med. 2018, 183, 459–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baishya, R.; Bhattacharya, A.; Mukherjee, M.; Lahiri, D.; Banerjee, S. Establishment of a simple reproducible model for antibiotic sensitivity pattern study of biofilm forming Staphylococcus aureus. Mater. Today Proc. 2016, 3, 3461–3466. [Google Scholar] [CrossRef]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 2010, 65, 1955–1958. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: An in vitro study. J. Med. Microbiol. 2009, 58, 1067–1073. [Google Scholar] [CrossRef] [Green Version]
- Conlon, B.P.; Rowe, S.E.; Lewis, K. Persister cells in biofilm associated infections. Biofilm-Based Healthc.-Assoc. Infect. 2015, 831, 1–9. [Google Scholar]
- Chopra, S.; Harjai, K.; Chhibber, S. Antibiotic susceptibility of ica -positive and ica -negative MRSA in different phases of biofilm growth. J. Antibiot. 2015, 68, 15–22. [Google Scholar] [CrossRef]
- Carvalhais, V.; Pérez-Cabezas, B.; Oliveira, C.; Vitorino, R.; Vilanova, M.; Cerca, N. Tetracycline and rifampicin induced a viable but nonculturable state in Staphylococcus epidermidis biofilms. Future Microbiol. 2017, 13, 27–36. [Google Scholar] [CrossRef]
- Gaio, V.; Cerca, N. Cells released from S. epidermidis biofilms present increased antibiotic tolerance to multiple antibiotics. PeerJ 2019, 7, e6884. [Google Scholar] [CrossRef] [Green Version]
- Yamada, S.; Sugai, M.; Komatsuzawa, H.; Matsumoto, A. Suppressed localization of a major autolysin on Staphylococcus aureus treated with tetracycline. J. Electron. Microsc. 2001, 50, 359–364. [Google Scholar] [CrossRef]
- Ledala, N.; Wilkinson, B.J.; Jayaswal, R.K. Effects of oxacillin and tetracycline on autolysis, autolysin processing and atl transcription in Staphylococcus aureus. Int. J. Antimicrob. Agents 2006, 27, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liang, Y.; Lin, S.; Chen, D.; Li, B.; Li, L.; Deng, Y. Crystal Violet and XTT Assays on Staphylococcus aureus Biofilm Quantification. Curr. Microbiol. 2016, 73, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.M.; Balkis, M.; Chandra, J.; Mukherjee, P.K.; Ghannoum, M.A. Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J. Clin. Microbiol. 2003, 41, 506–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerca, N.; Martins, S.; Cerca, F.; Jefferson, K.K.; Pier, G.B.; Oliveira, R.; Azeredo, J. Comparative assessment of antibiotic susceptibility of coagulase-negative staphylococci in biofilm versus planktonic culture as assessed by bacterial enumeration or rapid XTT colorimetry. J. Antimicrob. Chemother. 2005, 56, 331–336. [Google Scholar] [CrossRef] [PubMed]
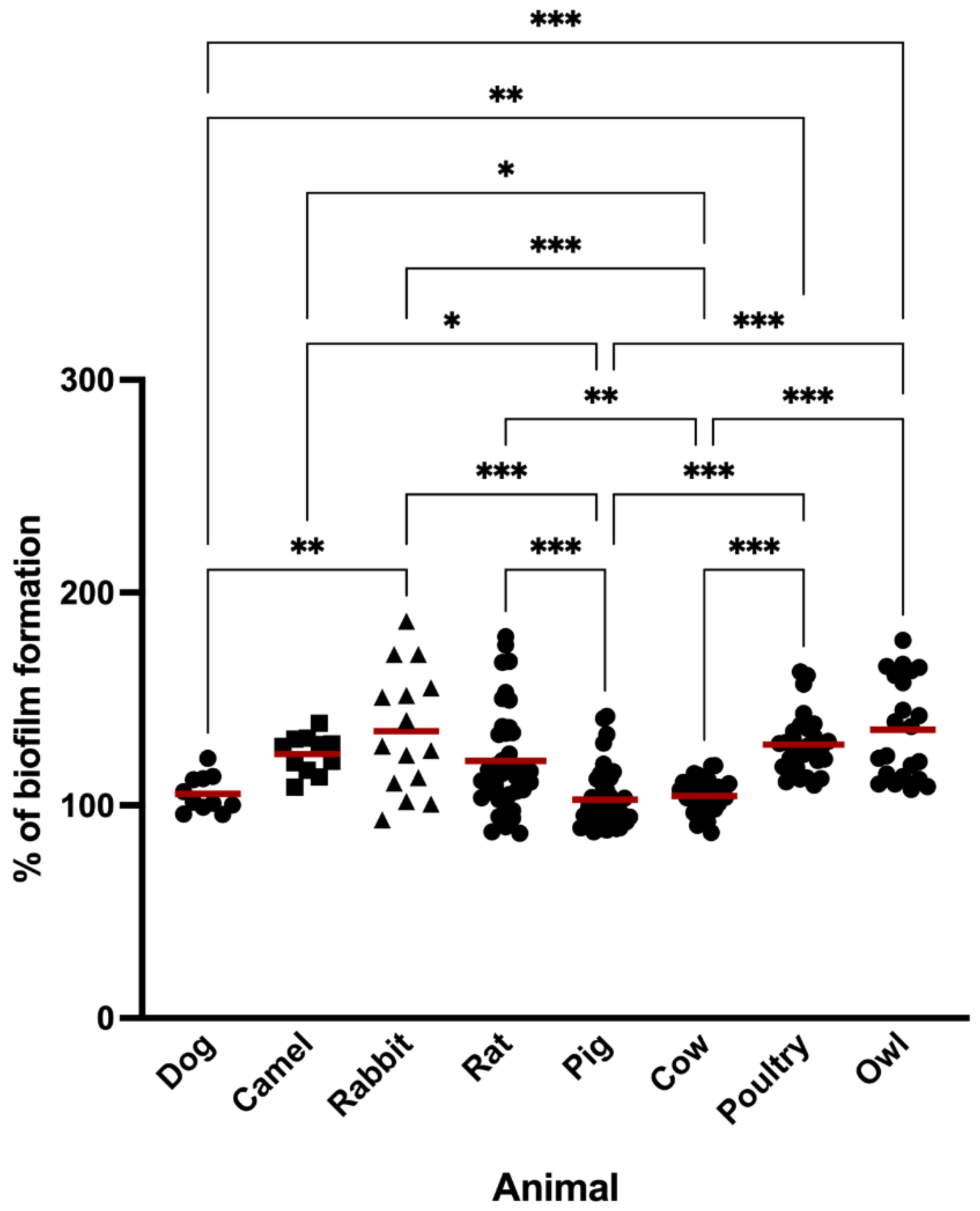
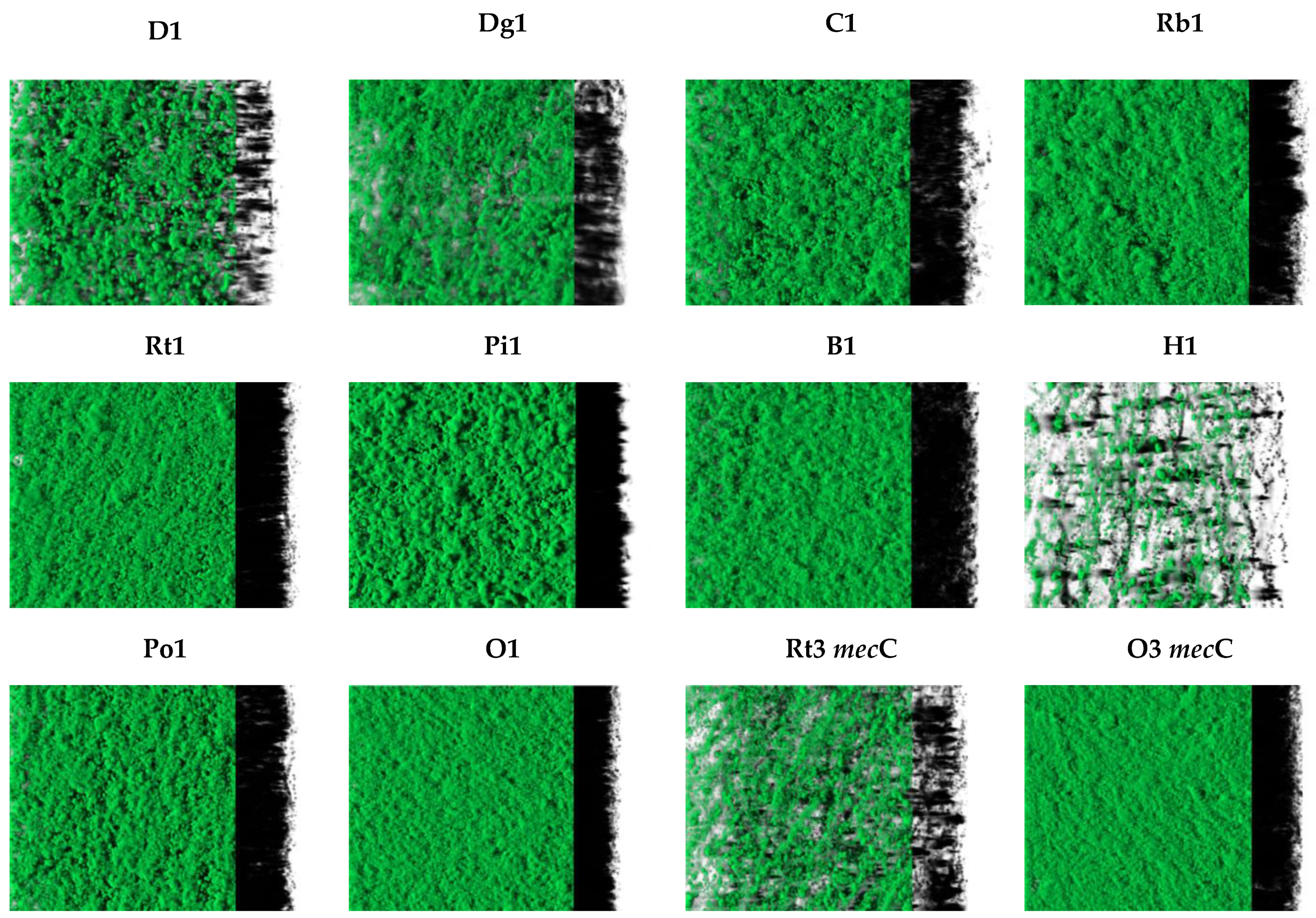
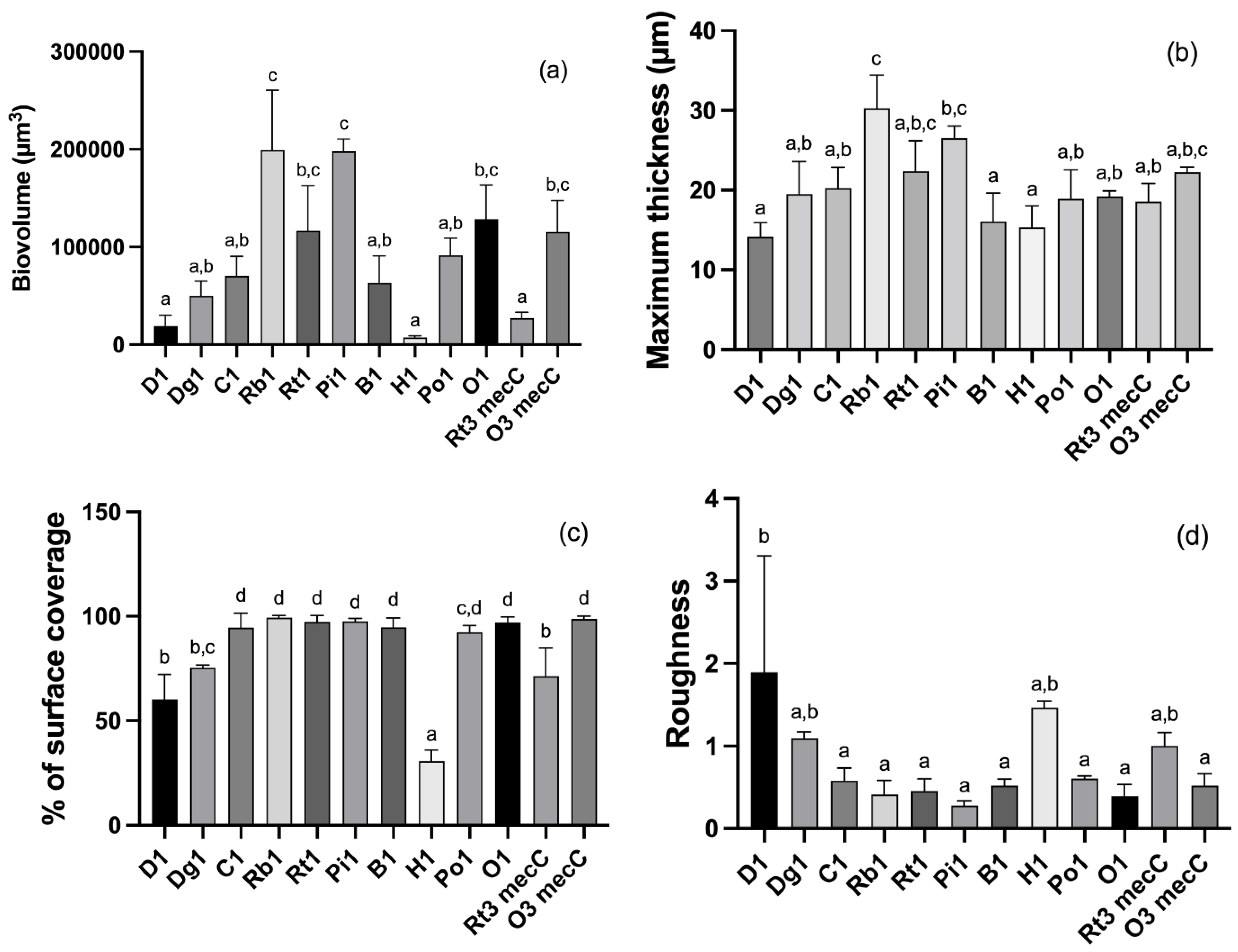
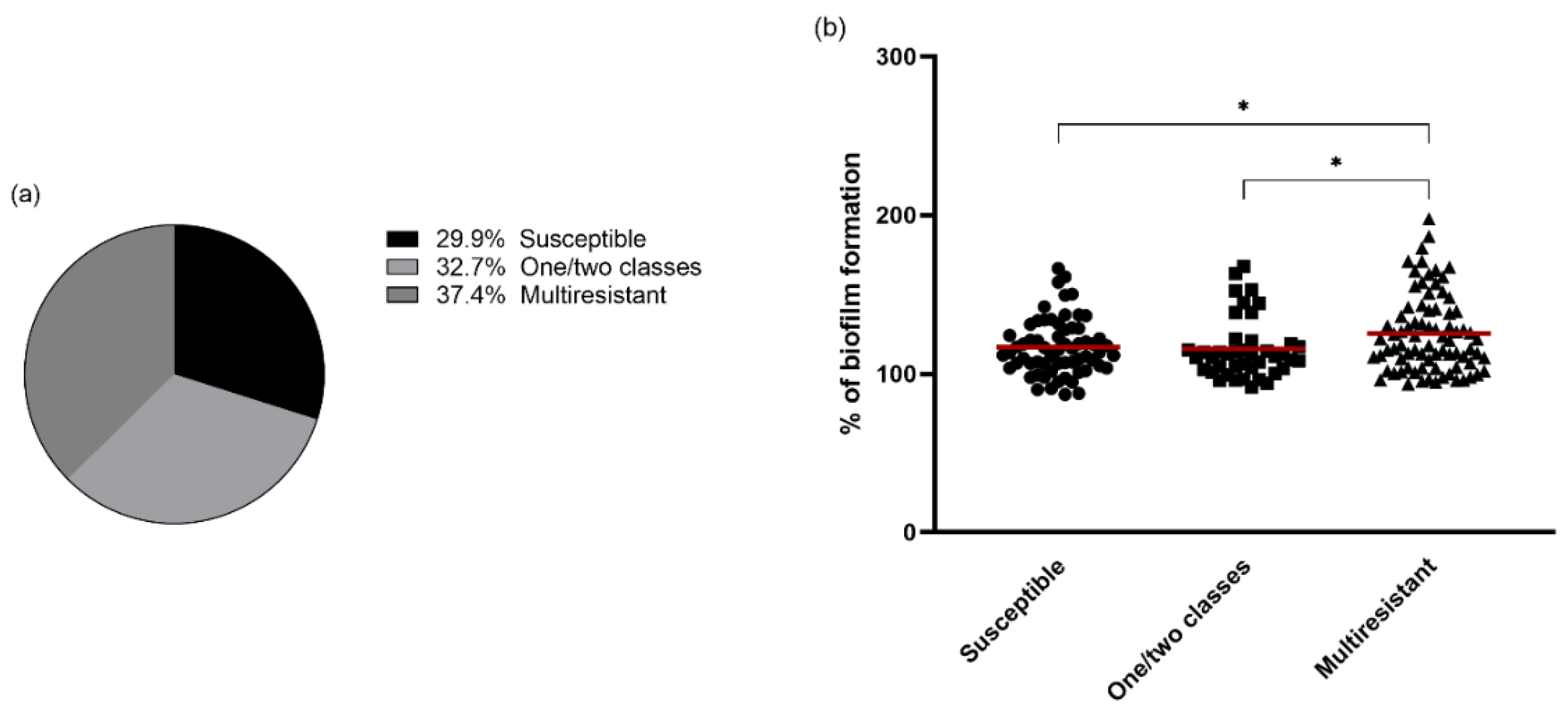
| Isolate | % of Biofilm Formation | Phenotypic Resistance/Susceptibility |
|---|---|---|
| D1 | 152.34% | PEN, KAN |
| D2 | 104.72% | KAN, TET |
| Dg1 | 122.09% | PEN |
| Dg2 | 95.86% | ERY |
| C1 | 138.52% | CIP |
| C2 | 108.54% | Susceptible |
| Rb1 | 197.61% | PEN, FOX, ERY, CD, CIP |
| Rb2 | 93.10% | PEN, FOX, ERY, CD, CIP, FD |
| Rt1 | 179.31% | PEN, FOX, ERY, CIP |
| Rt2 | 86.85% | Susceptible |
| Pi1 | 141.86% | PEN, CN, TOB, KAN, TET, C, CIP |
| Pi2 | 87.51% | PEN, FOX, ERY, CD, KAN, TET, C, CIP |
| B1 | 118.64% | Susceptible |
| B2 | 87.18% | FD |
| H1 | 147.98% | PEN, FOX, ERY, CD, CN |
| H2 | 114.81% | PEN, FOX, ERY, CD |
| Po1 | 162.72% | PEN, FOX, CN, TOB, KAN, ERY, CD, TET, CIP |
| Po2 | 109.58% | PEN, FOX, CIP, ERY, CD, TET, FD |
| O1 | 209.05% | Susceptible |
| O2 | 107.51% | Susceptible |
| Rt3 mecC | 127.90% | PEN, FOX |
| Rt4 mecC | 155.22% | PEN, FOX |
| O3 mecC | 110.02% | PEN, FOX |
| Antimicrobial | Resistant M ± SD | Susceptible M ± SD | p |
|---|---|---|---|
| Penicillin | 119.878 ± 22.831 | 116.624 ± 23.576 | 0.310 |
| Cefoxitin | 122.015 ± 24.605 | 115.7175 ± 20.661 | 0.023 |
| Ciprofloxacin | 126.632 ± 23.554 | 113.707 ± 19.932 | <0.001 |
| Gentamicin | 123.539 ± 23.002 | 116.666 ± 22.044 | 0.036 |
| Tobramycin | 122.309 ± 21.526 | 117.651 ± 23.469 | 0.262 |
| Kanamycin | 123.997 ± 22.816 | 116.4612 ± 22.03 | 0.022 |
| Erythromycin | 124.634 ± 25.630 | 114.832 ± 19.881 | 0.001 |
| Clindamycin | 124.634 ± 25.630 | 115.536 ± 21.380 | 0.004 |
| Tetracycline | 119.381 ± 20.812 | 118.031 ± 24.293 | 0.689 |
| Chloramphenicol | 109.437 ± 15.221 | 119.551 ± 23.763 | 0.027 |
| Fusidic acid | 121.917 ± 23.568 | 118.219 ± 23.164 | 0.552 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, V.; Correia, E.; Pereira, J.E.; González-Machado, C.; Capita, R.; Alonso-Calleja, C.; Igrejas, G.; Poeta, P. Biofilm Formation of Staphylococcus aureus from Pets, Livestock, and Wild Animals: Relationship with Clonal Lineages and Antimicrobial Resistance. Antibiotics 2022, 11, 772. https://doi.org/10.3390/antibiotics11060772
Silva V, Correia E, Pereira JE, González-Machado C, Capita R, Alonso-Calleja C, Igrejas G, Poeta P. Biofilm Formation of Staphylococcus aureus from Pets, Livestock, and Wild Animals: Relationship with Clonal Lineages and Antimicrobial Resistance. Antibiotics. 2022; 11(6):772. https://doi.org/10.3390/antibiotics11060772
Chicago/Turabian StyleSilva, Vanessa, Elisete Correia, José Eduardo Pereira, Camino González-Machado, Rosa Capita, Carlos Alonso-Calleja, Gilberto Igrejas, and Patrícia Poeta. 2022. "Biofilm Formation of Staphylococcus aureus from Pets, Livestock, and Wild Animals: Relationship with Clonal Lineages and Antimicrobial Resistance" Antibiotics 11, no. 6: 772. https://doi.org/10.3390/antibiotics11060772
APA StyleSilva, V., Correia, E., Pereira, J. E., González-Machado, C., Capita, R., Alonso-Calleja, C., Igrejas, G., & Poeta, P. (2022). Biofilm Formation of Staphylococcus aureus from Pets, Livestock, and Wild Animals: Relationship with Clonal Lineages and Antimicrobial Resistance. Antibiotics, 11(6), 772. https://doi.org/10.3390/antibiotics11060772









