Heteroaryl-Ethylenes as New Effective Agents for High Priority Gram-Positive and Gram-Negative Bacterial Clinical Isolates
Abstract
:1. Introduction
2. Results
2.1. Gram-Positive Clinical Strains
2.2. Gram-Negative Clinical Strains
3. Discussion
4. Materials and Methods
4.1. Characteristics of Molecule Tested in the Study
4.2. Microorganisms and Growth Conditions
4.3. Minimum Inhibitory Concentration (MIC)
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mühlberg, E.; Umstätter, F.; Kleist, C.; Domhan, C.; Mier, W.; Uhl, P. Renaissance of vancomycin: Approaches for breaking antibiotic resistance in multidrug-resistant bacteria. Can. J. Microbiol. 2020, 66, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. General principles of antibiotic resistance in bacteria. Drug Discov. Today Technol. 2014, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Karaman, R.; Jubeh, B.; Breijyeh, Z. Resistance of Gram-Positive Bacteria to Current Antibacterial Agents and Overcoming Approaches. Molecules 2020, 25, 2888. [Google Scholar] [CrossRef]
- Goll, C.; Balmer, P.; Schwab, F.; Rüden, H.; Eckmanns, T. Different trends of MRSA and VRE in a German hospital, 1999–2005. Infection 2007, 35, 245–249. [Google Scholar] [CrossRef]
- Huycke, M.M.; Sahm, D.F.; Gilmore, M.S. Multiple-drug resistant enterococci: The nature of the problem and an agenda for the future. Emerg. Infect. Dis. 1998, 4, 239–249. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Cao, B.; Liu, Y.; Gu, L.; Wang, C. Investigation of the prevalence of patients co-colonized or infected with methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci in China: A hospital-based study. Chin. Med. J. 2009, 122, 1283–1288. [Google Scholar]
- Rincón, S.; Panesso, D.; Díaz, L.; Carvajal, L.P.; Reyes, J.; Munita, J.M.; Arias, C.A. Resistance to ‘last resort’ antibiotics in Gram-positive cocci: The post-vancomycin era. Biomed. Rev. Inst. Nac. Salud 2014, 34 (Suppl. S1), 191–208. [Google Scholar] [CrossRef]
- Deurenberg, R.H.; Vink, C.; Kalenic, S.; Friedrich, A.W.; Bruggeman, C.A.; Stobberingh, E.E. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2007, 13, 222–235. [Google Scholar] [CrossRef] [Green Version]
- Schulte, R.H.; Munson, E. Staphylococcus aureus Resistance Patterns in Wisconsin: 2018 Surveillance of Wisconsin Organisms for Trends in Antimicrobial Resistance and Epidemiology (SWOTARE) Program Report. Clin. Med. Res. 2019, 17, 72–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirby, W.M. Extraction of a highly potent penicillin inactivator from penicillin resistant staphylococci. Science 1944, 99, 452–453. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.; Neumann, B.; Weber, R.E.; Kresken, M.; Wendt, C.; Bender, J.K. Thirty years of VRE in Germany—‘Expect the unexpected’: The view from the National Reference Centre for Staphylococci and Enterococci. Drug Resist. Updat. 2020, 53, 100732. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Contreras, G.A.; Murray, B.E. Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 2010, 16, 555–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, W.R.; Murray, B.E.; Rice, L.B.; Arias, C.A. Resistance in Vancomycin-Resistant Enterococci. Infect. Dis. Clin. N. Am. 2020, 34, 751–771. [Google Scholar] [CrossRef] [PubMed]
- Moxon, C.A.; Paulus, S. Beta-lactamases in Enterobacteriaceae infections in children. J. Infect. 2016, 72, S41–S49. [Google Scholar] [CrossRef] [Green Version]
- Chong, Y.; Shimoda, S.; Shimono, N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 2018, 61, 185–188. [Google Scholar] [CrossRef]
- Piperaki, E.-T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef]
- Isler, B.; Doi, Y.; Bonomo, R.A.; Paterson, D.L. New Treatment Options against Carbapenem-Resistant Acinetobacter baumannii Infections. Antimicrob. Agents Chemother. 2019, 63, e01110-18. [Google Scholar] [CrossRef] [Green Version]
- Terreni, M.; Taccani, M.; Pregnolato, M. New Antibiotics for Multidrug-Resistant Bacterial Strains: Latest Research Developments and Future Perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef]
- Shafiq, N.; Pandey, A.K.; Malhotra, S.; Holmes, A.; Mendelson, M.; Malpani, R.; Balasegaram, M.; Charani, E. Shortage of essential antimicrobials: A major challenge to global health security. BMJ Glob. Health 2021. [Google Scholar] [CrossRef] [PubMed]
- Bongiorno, D.; Musso, N.; Bonacci, P.G.; Bivona, D.A.; Massimino, M.; Stracquadanio, S.; Bonaccorso, C.; Fortuna, C.G.; Stefani, S. Heteroaryl-Ethylenes as New Lead Compounds in the Fight against High Priority Bacterial Strains. Antibiotics 2021, 10, 1034. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti.-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- EuropeanCommittee on Antimicrobial Suceptibility Testing. Available online: https://www.eucast.org/mic_distributions_and_ecoffs/ (accessed on 30. May 2022).
- Haney, E.F.; Mansour, S.C.; Hancock, R.E.W. Antimicrobial Peptides: An Introduction. Methods Mol. Biol. 2017, 1548, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Fortuna, C.G.; Barresi, V.; Bonaccorso, C.; Consiglio, G.; Failla, S.; Trovato-Salinaro, A.; Musumarra, G. Design, synthesis and in vitro antitumour activity of new heteroaryl ethylenes. Eur. J. Med. Chem. 2012, 47, 221–227. [Google Scholar] [CrossRef]
- Campanile, F.; Bongiorno, D.; Perez, M.; Mongelli, G.; Sessa, L.; Benvenuto, S.; Gona, F.; Varaldo, P.E.; Stefani, S.; Floriana Gona b AMCLI—S. aureus Survey Participants. Epidemiology of Staphylococcus aureus in Italy: First nationwide survey, 2012. J. Glob. Antimicrob. Resist. 2015, 3, 247–254. [Google Scholar] [CrossRef]
- Lazzaro, L.M.; Cassisi, M.; Stefani, S.; Campanile, F. Impact of PBP4 Alterations on β-Lactam Resistance and Ceftobiprole Non-Susceptibility Among Enterococcus faecalis Clinical Isolates. Front. Cell Infect. Microbiol. 2021, 11, 816657. [Google Scholar] [CrossRef]
- Sarti, M.; Campanile, F.; Sabia, C.; Santagati, M.; Gargiulo, R.; Stefani, S. Polyclonal Diffusion of Beta-Lactamase-Producing Enterococcus faecium. J. Clin. Microbiol. 2012, 50, 169–172. [Google Scholar] [CrossRef] [Green Version]
- Bruker Maldi-TOF/TOF Solution. Available online: https://www.bruker.com/it/products-and-solutions/mass-spectrometry/maldi-tof.html (accessed on 30 May 2022).
- Weinstein, M.P.; Lewis, J.S. The Clinical and Laboratory Standards Institute Subcommittee on Antimicrobial Susceptibility Testing: Background, Organization, Functions and Processes. J. Clin. Microbiol. 2020, 58, e01864-19. [Google Scholar] [CrossRef]
- CLSI. CLSI Supplement M100, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- European Commettee on Antimicrobial Susceptibility Testing. EUCAST Reading Guide for Broth Microdilution. 2022. Available online: https://www.eucast.org/ (accessed on 30 May 2022).
- GOLDBIO: Broth Microdilution and Disk Diffusion. 2018. Available online: https://www.goldbio.com/documents/2531/Broth%20Microdilution%20and%20Disk%20Diffusion.pdf (accessed on 30 May 2022).
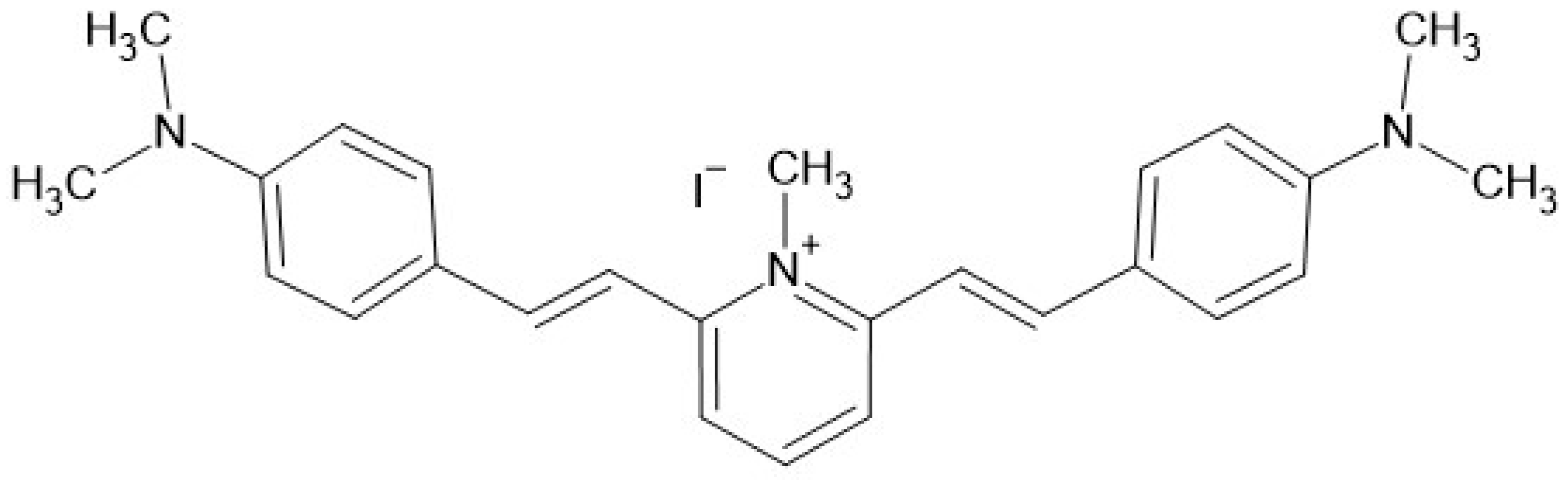
| Antimicrobial Resistance Profile | PB4 MIC | ||
|---|---|---|---|
| Strain | mg/L | µmol/L | |
| S. aureus ATCC 29213 | 0.25 | 0.49 | |
| MSSA | 1-CT | 0.12 | 0.234 |
| 2-CT | 0.12 | 0.234 | |
| 3-CT | 0.12 | 0.234 | |
| MRSA | 4-CT | 0.12 | 0.234 |
| 5-CT | 0.12 | 0.234 | |
| 6-CT | 0.12 | 0.234 | |
| 7-CT | 0.12 | 0.234 | |
| 8-CT | 0.12 | 0.234 | |
| 9-CT | 0.12 | 0.234 | |
| 10-CT | 0.12 | 0.234 | |
| 11-CT | 0.12 | 0.234 | |
| 12-CT | 0.25 | 0.49 | |
| 13-CT | 0.5 | 0.977 | |
| 14-CT | 0.5 | 0.977 | |
| Antimicrobial Resistance Profile | PB4 MIC | |||
|---|---|---|---|---|
| Strain | mg/L | µmol/L | ||
| E. faecalis ATCC 29212 | 0.5 | 0.977 | ||
| E. faecalis ATCC 51299 | 0.5 | 0.977 | ||
| E. faecalis | VRE | 15-CT | 0.25 | 0.488 |
| 16-CT | 0.25 | 0.488 | ||
| 17-CT | 0.5 | 0.977 | ||
| MDR | 18-CT | 0.25 | 0.488 | |
| NOT MDR | 19-CT | 0.25 | 0.488 | |
| 20-CT | 0.5 | 0.977 | ||
| E. faecium | VRE | 21-CT | 0.25 | 0.488 |
| 22-CT | 1 | 1.95 | ||
| Lin R | 23-CT | 0.5 | 0.977 | |
| 24-CT | 1 | 1.95 | ||
| MDR | 25-CT | 0.12 | 0.234 | |
| 26-CT | 1 | 1.95 | ||
| NOT MDR | 27-CT | 2 | 3.91 | |
| Antimicrobial Resistance Profile | PB4 MIC | ||
|---|---|---|---|
| Strain | mg/L | µmol/L | |
| E. coli ATCC 25922 | 1 | 1.95 | |
| ESBL | 28-CT | 4 | 7.82 |
| 29-CT | 16 | 31.28 | |
| 30-CT | 64 | 125.12 | |
| Susceptible | 31-CT | 4 | 7.82 |
| 32-CT | 4 | 7.82 | |
| 33-CT | 4 | 7.82 | |
| 34-CT | 4 | 7.82 | |
| 35-CT | 4 | 7.82 | |
| 36-CT | 8 | 15.64 | |
| 37-CT | 32 | 62.56 | |
| 38-CT | 32 | 62.56 | |
| 39-CT | 64 | 125.12 | |
| Antimicrobial Resistance Profile | PB4 MIC | ||
|---|---|---|---|
| Strain | mg/L | µmol/L | |
| A. baumannii ATCC 17978 | 0.5 | 0.97 | |
| CRAB | 40-CT | <0.12 | <0.23 |
| 41-CT | <0.12 | <0.23 | |
| 42-CT | <0.12 | <0.23 | |
| 43-CT | <0.12 | <0.23 | |
| 44-CT | 0.25 | 0.488 | |
| 45-CT | 0.25 | 0.488 | |
| 46-CT | 0.25 | 0.488 | |
| 47-CT | 0.25 | 0.488 | |
| 48-CT | 0.25 | 0.488 | |
| 49-CT | 0.25 | 0.488 | |
| 50-CT | 0.5 | 0.977 | |
| 51-CT | 0.5 | 0.977 | |
| 52-CT | 0.5 | 0.977 | |
| 53-CT | 1 | 1.955 | |
| PB4 MIC Value (mg/L) | |||||||||||||||
| Control Strain | MIC Value (mg/L) | Species | n. | Range | 128 | 64 | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.12 |
| S. aureus ATCC 29213 | 0.25 | S. aureus | 14 | 0.12–0.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 11 |
| E. faecalis ATCC 29212 | 0.5 | E. faecalis | 6 | 0.25–0.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 0 |
| E. faecalis ATCC 51299 VRE | 0.5 | E. faecium | 7 | 0.12–2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 | 1 | 1 |
| E. coli ATCC 25922 | 1 | E. coli | 12 | 4–64 | 0 | 2 | 2 | 1 | 1 | 6 | 0 | 0 | 0 | 0 | 0 |
| A. baumannii ATCC 17978 | 0.5 | A. baumannii | 14 | ≤0.12–1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 6 | 4 |
| PB4 MIC Value (µmol/L) | |||||||||||||||
| Control Strain | MIC Value (mg/L) | Species | n. | Range | 250.2 | 125.1 | 62.5 | 31.2 | 15.6 | 7.8 | 3.9 | 1.9 | 0.977 | 0.488 | 0.234 |
| S. aureus ATCC 29213 | 0.488 | S. aureus | 14 | 0.234–0.977 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 11 |
| E. faecalis ATCC 29212 | 0.977 | E. faecalis | 6 | 0.488–0.977 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 0 |
| E. faecalis ATCC 51299 VRE | 0.977 | E. faecium | 7 | 0.234–3.91 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 | 1 | 1 |
| E. coli ATCC 25922 | 1.9 | E. coli | 12 | 7.82–125.12 | 0 | 2 | 2 | 1 | 1 | 6 | 0 | 0 | 0 | 0 | 0 |
| A. baumannii ATCC 17978 | 0.977 | A. baumannii | 14 | ≤0.234–1.955 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 6 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bivona, D.A.; Mirabile, A.; Bonomo, C.; Bonacci, P.G.; Stracquadanio, S.; Marino, A.; Campanile, F.; Bonaccorso, C.; Fortuna, C.G.; Stefani, S.; et al. Heteroaryl-Ethylenes as New Effective Agents for High Priority Gram-Positive and Gram-Negative Bacterial Clinical Isolates. Antibiotics 2022, 11, 767. https://doi.org/10.3390/antibiotics11060767
Bivona DA, Mirabile A, Bonomo C, Bonacci PG, Stracquadanio S, Marino A, Campanile F, Bonaccorso C, Fortuna CG, Stefani S, et al. Heteroaryl-Ethylenes as New Effective Agents for High Priority Gram-Positive and Gram-Negative Bacterial Clinical Isolates. Antibiotics. 2022; 11(6):767. https://doi.org/10.3390/antibiotics11060767
Chicago/Turabian StyleBivona, Dalida Angela, Alessia Mirabile, Carmelo Bonomo, Paolo Giuseppe Bonacci, Stefano Stracquadanio, Andrea Marino, Floriana Campanile, Carmela Bonaccorso, Cosimo Gianluca Fortuna, Stefania Stefani, and et al. 2022. "Heteroaryl-Ethylenes as New Effective Agents for High Priority Gram-Positive and Gram-Negative Bacterial Clinical Isolates" Antibiotics 11, no. 6: 767. https://doi.org/10.3390/antibiotics11060767
APA StyleBivona, D. A., Mirabile, A., Bonomo, C., Bonacci, P. G., Stracquadanio, S., Marino, A., Campanile, F., Bonaccorso, C., Fortuna, C. G., Stefani, S., Musso, N., & Bongiorno, D. (2022). Heteroaryl-Ethylenes as New Effective Agents for High Priority Gram-Positive and Gram-Negative Bacterial Clinical Isolates. Antibiotics, 11(6), 767. https://doi.org/10.3390/antibiotics11060767










