Abstract
Ceftazidime-avibactam (CAV) is a new treatment option against carbapenem-resistant Klebsiella pneumoniae (CRKP) infections. However, the rapid emergence of CAV resistance mediated by KPC variants has posed a severe threat to healthcare after its clinical application. The characteristics of CAV resistance in CRKP strains needs to be determined in China. A total of 477 CRKP isolates were collected from 46 hospitals in Zhejiang Province from 2018 to 2021. The results demonstrated that CAV had a potent activity against 94.5% of all CRKP (451/477, 95% CI: 93.0–96.1%) and 86.0% of CRKP strains carrying blaKPC genes (410/477, 95% CI: 83.5–88.4%). A total of 26 CAV-resistant strains were found. Among these strains, sixteen harbored metallo-β lactamases, and two carried KPC-2 carbapenemase and mutated ompK35 and ompK36. Eight CRKP strains encoded KPC-33 or KPC-93, belonging to ST11, among which seven strains were detected in patients hospitalized in 2021 after exposure to CAV and one strain was associated with intra-hospital spread. CAV is a potent agent in vitro against CRKP strains. The rapid development of CAV resistance mediated by various KPC variants after a short period of CAV treatment has increased and brought difficulties in treating infections caused by CRKP strains, especially those belonging to ST11. The surveillance of bacterial resistance against CAV is highly recommended due to the steep development of CAV resistance and rapid evolution of KPC enzymes.
1. Introduction
The global spread of carbapenem-resistant Klebsiella pneumoniae (CRKP) is posing a serious threat to public health worldwide [1]. Previous studies have revealed that the prevalence of CRKP infection has increased noticeably in recent decades, and the mortality rate of bloodstream infection caused by CRKP strains has reached more than 70.0% [2,3]. Tigecycline and colistin are considered “last-resort antibiotics” against CRKP infections [4,5]. However, a number of reports have highlighted that the resistance to tigecycline [6,7] and colistin [8] is increasingly on the rise from mobile tigecycline resistance determinants, e.g., tmexCD1-toprJ1, and flavin-dependent mono-oxygenase tet(X) variants, and mobile colistin resistance determinants, such as mcr genes [8,9,10]. In addition, colistin treatment can cause neurotoxicity and nephrotoxicity [11].
A novel β-lactam/β-lactamase inhibitor combination called Ceftazidime-avibactam (CAV) is considered as one of the alternative therapies to treat CRKP infections. It has potent activity against CRKP with β-lactamases and extended-spectrum β-lactamases (ESBLs), Klebsiella pneumoniae carbapenemases (KPCs), AmpC, and certain oxacillinases (OXA), but not metallo-β lactamases, owing to the new non-β-lactam/β-lactamase inhibitor avibactam. It protects ceftazidime from degradation and reduces the minimum inhibitory concentration (MIC) of ceftazidime against Enterobacteriaceae producing certain β-lactamases. Therefore, CAV reverses the resistance to ceftazidime induced by ESBLs and AmpC via interacting with the active-site serine of these enzymes to form a covalent adduct reversibly [12]. A large multicenter cohort study found that CAV treatment was the sole independent predictor of survival in patients with KPC-producing K. pneumoniae (KPC-KP) bacteremia and concluded that CAV was a potential antimicrobial agent against severe KPC-KP infections [13].
As an efficacy treatment against KPC carbapenemase production, CAV was approved for the clinical treatment of complicated intra-abdominal and urinary tract infections in the USA in 2015 and was allowed in China in September 2019 [14,15]. Recently, significant increases in CAV resistance in KPC-KP strains have been observed. Mutations within the Ω-loop of KPC-2/KPC-3 enzymes were the principal resistance mechanisms observed in KPC-KP strains apart from the co-production of metallo-β lactamases [15,16,17]. The increased expression of KPC carbapenemases with porin deficiency was also related to CAV resistance in K. pneumoniae strains producing KPC-2/KPC-3 enzymes [18,19,20,21,22]. To evaluate the efficacy of this newly approved therapy and investigate the development of its resistance after two years of clinical application in Zhejiang Province, China, we collected 477 CRKP isolates from 2018 to 2021 to test the in vitro activity of CAV, and analyzed the mechanisms underlying CAV resistance in KPC-KP strains.
2. Results
2.1. Carbapenem-Resistant K. pneumoniae Isolates
A total of 477 non-duplicated CRKP strains were from Zhejiang Province, China, during 2018–2021. ST11 was the most prevalent sequence type (ST) (50.1%, 239/477, 95% CI: 46.6–53.6%), followed by ST15 (30.6%, 146/477, 95% CI: 27.4–33.8%) (Figure 1a). A total of 467 CRKP isolates carried the carbapenemase genes, including blaKPC (n = 410, 86.0%, 95% CI: 83.5–88.4%), blaOXA-232 (n = 38, 8.0%, 95% CI: 6.1–9.8%), blaNDM (n = 13, 2.7%, 95% CI: 1.6–3.9%), and blaIMP (n = 2, 0.4%, 95% CI: 0.1–1.5%). The co-occurrence of blaKPC-2 and blaNDM was detected in three strains, accounting for 0.6% (n = 3, 95% CI: 0.1–1.8%) and one strain (n = 1, 0.2%, 95% CI: 0.1–1.2%) carried both blaKPC and blaOXA-232. The KPC enzyme was the most frequently detected carbapenemase in these strains (86.0%, 410/477) (Figure 1b). The high-resistance proportions of ceftazidime, cefotaxime, cefepime, piperacillin/tazobactam, cefoperazone/sulbactam, ciprofloxacin and aztreonam among these CRKP strains were 99.2% (95% CI: 97.9–99.8%), 99.2% (95% CI: 97.9–99.8%), 97.1% (95% CI: 95.9–98.2%), 99.0% (95% CI: 97.6–99.7%), 97.9% (95% CI: 96.9–98.9%), 90.8% (95% CI: 88.8–92.8%) and 98.3% (95% CI: 97.4–99.2%), respectively. In addition, the CRKP strains manifested resistance to cefmetazole at 66.0% (95% CI: 62.8–69.3%) and amikacin at 50.3% (95% CI: 46.8–53.8%) (Table 1 and Table S1).
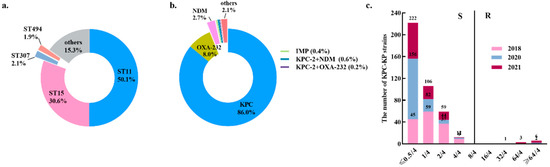
Figure 1.
The distribution of carbapenemases, MLST and MIC values of CAV among 477 CRKP strains. (a) The distribution of carbapenemases. (b) The distribution of MLST. (c) The comparison of MIC values of CAV in 477 CRKP strains collected during 2018–2021.

Table 1.
Susceptibility of 477 CRKP strains to commonly used antibiotics.
2.2. Resistance Profile of the CRKP Strains to Ceftazidime-Avibactam
MIC of CAV against 477 CRKP strains ranged from ≤0.5/4 to >64/4 μg/mL and MIC50 and MIC90 were ≤0.5/4 μg/mL and 2/4 μg/mL, respectively (Table 1; Figure 1c). A total of 26 strains (5.5%, 95% CI: 3.9–7.0%) were resistant to CAV (MIC ≥ 16/4 μg/mL) and were recovered from six administrative districts of Zhejiang Province, namely Hangzhou (n = 14, 53.8%, 95% CI: 50.4–57.3%), Jinhua (n = 4, 15.4%, 95% CI: 4.4–34.9%), Taizhou (n = 3, 11.5%, 95% CI: 2.4–30.2%), Wenzhou (n = 2, 7.7%, 95% CI: 0.9–25.1%), Ningbo (n = 2, 7.7%, 95% CI: 0.9–25.1%), and Lishui (n = 1, 3.8%, 95% CI: 0.1–19.6%) (Figure 2). Thirteen of the twenty-six CAV-resistant CRKP strains with high MICs (≥32/4 μg/mL) produced metallo-β lactamases, including NDM (n = 11, 84.6%) and IMP (n = 2, 15.4%). Co-production of NDM and KPC-2 carbapenemases was detected in three CRKP strains with CAV resistance isolated during 2018–2021 and all other CAV-resistant strains harbored the blaKPC gene. In total, 94.5% of CRKP strains (451/477, CI: 93.0–96.1%) were susceptible to CAV, including all OXA-232-producing CRKP strains (n = 38) and 400 KPC-2-producing strains.
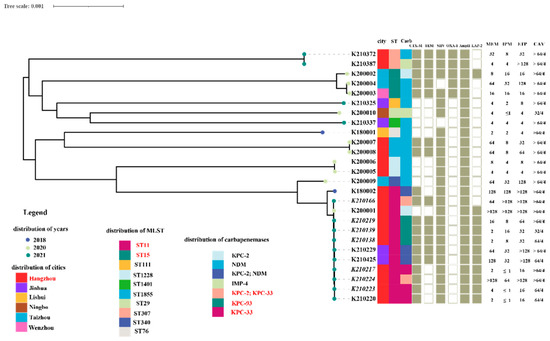
Figure 2.
The overview of microbiological and molecular characteristics of 26 CAV-resistant CRKP isolates. It consisted of phylogenetic analysis, distribution of separation time, location, MLST, carbapenemases, the other β-lactamases and the antimicrobial susceptibility of carbapenems and CAV. The names of those strains whose patients were treated with CAV are shown in italics.
Among the ten KPC-producing K. pneumoniae isolates, two carried wild-type KPC-2 β-lactamases solely, three carried blaKPC-93 and the remanent of CRKP isolates were encoded KPC-33. Among these, two CRKP strains possessed both KPC-2 and KPC-33 enzymes. All CRKP strains were recovered from patients who were treated with CAV after 23–41 days, except one, which was related to nosocomial transmission. Nine out of ten strains consistently had the ST11 clonal background, with the remaining one, numbered K200002, belonging to the ST15 clone. They were clustered into the same clade based on phylogenetic analysis. In contrast, CAV-resistant CRKP isolates producing metallo-β lactamases, including those encoding both KPC-2 and NDM carbapenemases, were genetically diverse and belonged to ST11 (n = 3), ST15 (n = 2), ST1228 (n = 2), ST1855 (n = 2), ST307 (n = 2), ST76 (n = 1), ST340 (n = 1), ST29 (n = 1), ST111 (n = 1) and ST1401 (n = 1), respectively. The phylogenetic analysis revealed that they were grouped into ten distinct clades (Figure 2).
2.3. Mechanisms of CAV Resistance in KPC-KP Strains
The KPC-33 carbapenemase carried a substitution of the aspartic acid residue at amino acid positions 179 with tyrosine (D179Y) in the Ω loop of KPC carbapenemases and were probably derived from KPC-2. This mutation impaired its capacity to hydrolyze carbapenems but enhanced the ability to resist CAV. Three KPC-33-positive strains with high MIC values of ≥64 μg/mL for CAV were susceptible to imipenem and showed intermediate susceptibility or low-level resistance to meropenem and ertapenem (meropenem MIC range from 2 to 4 µg/mL; imipenem MIC ≤ 1 µg/mL; ertapenem MIC = 16 µg/mL) (Figure 2). The co-production of KPC-2 carbapenemase in KPC-33-positive strains enabled them to highly resist carbapenems and retained a high level of resistance to CAV simultaneously (meropenem, imipenem and ertapenem, MIC ≥ 64 µg/mL) (Figure 2).
The novel KPC variant, KPC-93, shared 98.3% amino acid sequence homology with KPC-2 carbapenemases and contained a five-amino-acid insertion (Asn-Arg-Ala-pro-Asn) located between amino acids 266 and 267 compared to KPC-2, of which, the sequence from amino acids 263 to 266 was duplicated with the insertion fragment Arg-Ala-pro-Asn in KPC-93.
To understand the resistant mechanism of the two strains producing wild-type KPC-2 carbapenemases, the porin deficiency and mutations of the porins were investigated. Mutaions in ompK35 and ompK36 in these strains were observed, each exhibiting 99.2%~99.4% and 91.9%~99.5% nucleotide sequence similarity with wild-type ompK35 and ompK36, respectively. In addition, amino acid changes have occurred in these two strains for the two porins.
3. Discussion
CAV was recently approved for the treatment of KPC-KP infections around the world and in China [3,15]. However, since the application of CAV in the clinic, there has been an increasing number of reports regarding CAV resistance [15,16,17,23]. In the current study, we investigated the characteristics of CAV resistance in 477 CRKP strains in Zhejiang Province, China, during 2018–2021, including the marketing time of CAV. Our data indicate that the exposure to CAV led to the rapid evolution of KPC variants, which have become the principal mechanism underlying CAV resistance in Zhejiang Province, China.
In general, the prevalence of CAV resistance was low in CRKP strains in China. The production of metallo-β-lactamases and increased expression of the wild-type KPC-2 carbapenemases in combination with porin deficiency were the predominant mechanisms of CAV resistance before it entered the Chinese market [23,24,25]. Similarly, only two strains conferred resistance to CAV via merely carrying NDM carbapenemases and co-possessing NDM and KPC-2 carbapenemases, respectively. The CAV-resistant CRKP prevalence rate of 1.2% (95% CI: 0.1–4.2%) in our study was slightly lower than that (3.7%) in another study from China [24], indicating that CAV was indeed a potent potential substitute for carbapenems in China, as expected before the introduction into the Chinese market. However, the rapid emergence of CAV resistance in CRKP strains after exposure to CAV was observed, hinting that the application of CAV has accelerated the dispersal of CAV-resistant CRKP strains in China. Among the CAV-resistant CRKP strains identified, 53.8% were distributed in Hangzhou, probably attributing partly to the fact that CAV had been more frequently prescribed in this capital city of Zhejiang, with more tertiary care hospitals, and that a very high percentage of CRKP strains (40.0%, 191/477, 95% CI: 36.6–43.4%) were collected from Hangzhou.
Recent studies have indicated that the mutations on KPC-2 and KPC-3 carbapenemases have emerged as another primary mechanism mediating CAV resistance in KPC-KP strains. A large number of KPC variants have sprung up nearly two decades after the discovery of KPC-2 in 1996 due to the clinical application of CAV [17,23,24,26,27]. To date, 108 isoforms of KPC carbapenemases have been registered on the NCBI web site [28]. It was reported that the variations contributing to CAV resistance mainly occurred in the KPC Ω-loop region composed of 164Arg-179Asp, encircling the core of the active site of KPC carbapenemases in substrates acylation and deacylation, including KPC-31 and KPC-33, KPC-35, KPC-51, KPC-52 and KPC-57 variants [15,16,17,23,29,30,31]. The most common D179Y mutation in KPC-2 and KPC-3 enzymes reduced the inhibitory effect of avibactam and maintained ceftazidime-hydrolyzing activity, giving rise to KPC-31 and the most common KPC-33 variants in China, respectively [15,16]. So far, KPC-33-producing and CAV-resistant CRKP strains have already appeared in Shanghai, Zhejiang and Henan provinces and they tended to present low-level carbapenem resistance [15,24,32]. Shi et al. reported that the selective pressure from CAV usage resulted in the transformation of KPC-2 to KPC-33 carbapenemase, contributing to the reduced resistance to carbapenems but enhanced resistance to CAV in these strains [15]. Likewise, a total of seven strains carrying KPC variants were isolated from patients with a history of CAV usage. Three CRKP strains producing only KPC-33 in our study exhibited similar resistance profiles of carbapenem and CAV with Shi’s reports [15]. Two of them, K210217 and K210223, were isolated from patients after exposure to this drug for 23 and 41 days, respectively. Another KPC-33-positive strain, K210220, was obtained from a patient in the same hospital without a history of CAV use and the clonal relatedness was detected in these isolates with an ST11 clone background, indicating the potential for the horizontal transmission of KPC-33 carbapenemase within the hospital in a small range. Of note, the incorporation of the KPC-2 enzyme into KPC-33-producing K. pneumoniae strains, for instance, K210224 and K210166, might enhance the phenotype of carbapenem resistance, similar to previous reports [23,24], making the anti-infection treatment even worse.
The novel variant, KPC-93, with a five-amino-acid insertion (Asn-Arg-Ala-pro-Asn), was identified in our study. It mediated the modest resistance to carbapenem and high-level resistance to CAV in ST11 CRKP strains. The mutations happened in the vicinity regions of the Ω-loop and the hinge loop, consisting of amino acids 263–277. There has been another “hotspot” region of mutation (amino acids 240–243) leading to CAV resistance since 2015. This region was close to the hinge loop encompassing the active site of KPC and participated in the CAV resistance mediated by the blaKPC-14 and blaKPC-28 genes [27,33]. Thus, the KPC β-lactamases were prone to developing diverse mutations under the selection pressure of CAV, indicating that pertinent utilization and sustained surveillance were essential to prevent the emergence of novel KPC variants conferring both high resistance to CAV and high resistance to carbapenems. The ST11 clone, which often presented as the multi-drug resistance phenotype and was highly transmissible, has been reported to be the dominant epidemic genetic lineage among CRKP strains in China [34]. CAV resistance encoded by blaKPC variants has merely emerged in ST11-type CRKP strains in China [15,23,24,32] and all eight CRKP strains carrying KPC variants in the present study also belonged to the ST11 clone, indicating that this pandemic clone was the major reservoir of KPC variants and CAV resistance in China.
Herein, two CRKP isolates (K200001 and K20002) encoded the wild-type KPC-2 carbapenemase but exhibited resistance to CAV (MIC, >64/4 μg/mL). Though mutations of OmpK35 and OmpK36 were identified from these strains, it is unclear whether the mutated OmpK35 and OmpK36 contribute to their resistance to CAV as reported in other studies [21,22], which needs to be further investigated.
Although it appears that CAV is a promising antibiotic for KPC-KP infection, what is not negligible is that CAV is ineffective against metallo-β lactamase-producing strains, though the therapeutic regime of aztreonam and CAV combination might work [35,36]. In addition, one study reported that 23.0% of CRE infections recurred within 90 days after successful treatment by CAV, some even as few as 10 days [37]. So, it is still necessary to discover new inhibitors and combination antimicrobial regimens that are active against CRKP infections.
There are some limitations in this study. First, we were unable to obtain information as to whether the collected CRKP strains were from infections or colonizations for some patients due to the lack of their informed consent. Second, we do not have access to strains and data in 2019.
4. Materials and Methods
4.1. Sample Collection
Clinical CRKP strains were collected from 46 hospitals in 11 cities in Zhejiang Province, China, from 2018 to 2021. All isolates were identified via matrix-assisted laser desorption ionization–time of flight mass spectrometer (MALDI-TOF MS) (Bruker Daltonik GmbH, Bremen, Germany) and 16S rRNA gene sequencing.
4.2. Antimicrobial Susceptibility Testing and Carbapenemase-Encoding Gene Screening
Antimicrobial susceptibility testing was conducted using a broth microdilution method to assess the MIC values of 15 antibiotics (imipenem, meropenem, ertapenem, ceftazidime, cefotaxime, cefmetazole, cefepime, aztreonam, amikacin, ciprofloxacin, colistin, tigecycline, piperacillin/tazobactam, cefoperazone/sulbactam and ceftazidime/avibactam) against carbapenem-resistant K. pneumoniae isolates. The results were interpreted according to the Clinical and Laboratory Standards Institute guideline, except for tigecycline, which was interpreted based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints [38,39]. Both PCR and Sanger sequencing were performed to screen the carbapenemase-encoding genes blaNDM, blaIMP, blaKPC, blaOXA-48 and blaVIM, including blaKPC variants, as described previously [40].
4.3. Whole Genome Sequencing and Bioinformatics Analysis
Genomic DNA was extracted using a genomic DNA extraction kit (TIANGEN®, Beijing, China) and sequenced using the Illumina Hiseq 2500 platform with a 2 × 150 bp paired-end sequencing strategy [41]. De novo genome assembly was performed using SPAdes Genome Assembler version 3.11.1 (created by Anton Bankevich, et al., St. Petersburg, Russia) [42]. The annotation of assembled genome sequences was generated by the RAST tool and modified manually (created by Ross Overbeek, et al., Burr Ridge, IL, USA) [43]. Multi-locus sequence typing, antimicrobial resistance gene and virulence gene screening were conducted with Kleborate version 2.0.4 (created by Kathryn E Holt, Melbourne, Victoria, Australia) [44]. Plasmid replicons were analyzed using PlasmidFinder version 2.1 (created by Alessandra Carattoli, et al., Rome, Italy) [45]. The core genome phylogenetic tree of all CAV-resistant CRKP strains was generated using the Harvest suite (created by Todd J Treangen, et al., e, Frederick, MD, USA) [46]. The phylogenetic tree was visualized and modified using iTOL version 4 (created by Ivica Letunic and Peer Bork, Heidelberg, Germany) [47]. The wild-type ompK35 (accession number KX528047.1) and ompK36 (accession number KY086540.1) nucleotide sequences were used as the reference sequences to compare the relevant sequences of ompK35 and ompK36 of the CAV-resistant and wild-type KPC producing strains.
4.4. Statistical Analysis
The 95% confidence intervals (CI) were calculated using the approximate normal distribution method or exact probabilities method (binom.test in R), as appropriate.
5. Conclusions
In conclusion, though CAV exhibited a potent efficacy against most CRKP strains in our study, the rapid emergence of CAV resistance mediated by KPC variants among CRKP strains after exposure to CAV has brought new challenges to clinical treatment. The surveillance of CAV resistance and rational therapeutic regimens is highly suggested for clinicians to maximize the efficacy of CAV.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11060731/s1, Table S1: Antibiotic susceptibility profiles of 26 CAV-resistant strains in this study.
Author Contributions
Conceptualization and writing—review and editing, G.C. and R.Z.; methodology, C.L. and Y.W.; validation, L.H., Y.Z. (Yanyan Zhang) and Q.S.; formal analysis and writing—original draft preparation, C.L.; investigation, C.L., Y.Z. (Yanyan Zhang), Q.S., J.L. and Y.Z. (Yu Zeng); data curation, C.L., N.D., C.C. and Z.S.; funding acquisition, G.C. and R.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (R.Z., Grant # 81772250; R.Z., Grant # 81861138052; and G.C., Grant # 81871705).
Institutional Review Board Statement
This study was reviewed and approved by the Ethics Committee of Second Affiliated Hospital of Zhejiang University (2020-319).
Informed Consent Statement
Not applicable.
Data Availability Statement
The draft genome sequences of all CAV-resistant CRKP strains were deposited in the GenBank database under the BioProject number: PRJNA774095.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef] [PubMed]
- Sabino, S.; Soares, S.; Ramos, F.; Moretti, M.; Zavascki, A.P.; Rigatto, M.H. A Cohort Study of the Impact of Carbapenem-Resistant Enterobacteriaceae Infections on Mortality of Patients Presenting with Sepsis. mSphere 2019, 4, e00052-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauck, C.; Cober, E.; Richter, S.; Perez, F.; Salata, R.; Kalayjian, R.; Watkins, R.; Scalera, N.; Doi, Y.; Kaye, K.; et al. Spectrum of excess mortality due to carbapenem-resistant Klebsiella pneumoniae infections. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016, 22, 513–519. [Google Scholar] [CrossRef] [Green Version]
- Livermore, D. Tigecycline: What is it, and where should it be used? J. Antimicrob. Chemother. 2005, 56, 611–614. [Google Scholar] [CrossRef] [Green Version]
- Nang, S.; Azad, M.; Velkov, T.; Zhou, Q.; Li, J. Rescuing the Last-Line Polymyxins: Achievements and Challenges. Pharmacol. Rev. 2021, 73, 679–728. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Du, P.; Du, Y.; Sun, H.; Zhang, P.; Wan, Y.; Lin, Q.; Fanning, S.; Cui, S.; Wu, Y. Detection of plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from pork, Sichuan and Shandong Provinces, China, February 2019. Eurosurveill 2019, 24, 1900340. [Google Scholar] [CrossRef] [Green Version]
- Haller, S.; Kramer, R.; Becker, K.; Bohnert, J.A.; Eckmanns, T.; Hans, J.B.; Hecht, J.; Heidecke, C.-D.; Hübner, N.-O.; Kramer, A.; et al. Extensively drug-resistant Klebsiella pneumoniae ST307 outbreak, north-eastern Germany, June to October 2019. Eurosurveillance 2019, 24, 1900734. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Wyres, K.L.; Lam, M.M.C. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.; Walsh, T.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhanel, G.G.; Lawson, C.D.; Adam, H.; Schweizer, F.; Zelenitsky, S.; Lagace-Wiens, P.R.; Denisuik, A.; Rubinstein, E.; Gin, A.S.; Hoban, D.J.; et al. Ceftazidime-avibactam: A novel cephalosporin/beta-lactamase inhibitor combination. Drugs 2013, 73, 159–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tumbarello, M.; Trecarichi, E.; Corona, A.; De Rosa, F.; Bassetti, M.; Mussini, C.; Menichetti, F.; Viscoli, C.; Campoli, C.; Venditti, M.; et al. Efficacy of Ceftazidime-Avibactam Salvage Therapy in Patients with Infections Caused by Klebsiella pneumoniae Carbapenemase-producing K. pneumoniae. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2019, 68, 355–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yahav, D.; Giske, C.; Grāmatniece, A.; Abodakpi, H.; Tam, V.; Leibovici, L. New β-Lactam-β-Lactamase Inhibitor Combinations. Clin. Microbiol. Rev. 2020, 34, e00115-20. [Google Scholar] [CrossRef]
- Shi, Q.; Yin, D.; Han, R.; Guo, Y.; Zheng, Y.; Wu, S.; Yang, Y.; Li, S.; Zhang, R.; Hu, F. Emergence and Recovery of Ceftazidime-avibactam Resistance in blaKPC-33-Harboring Klebsiella pneumoniae Sequence Type 11 Isolates in China. Clin. Infect. Dis. 2020, 71, S436–S439. [Google Scholar] [CrossRef]
- Antonelli, A.; Giani, T.; Di Pilato, V.; Riccobono, E.; Perriello, G.; Mencacci, A.; Rossolini, G. KPC-31 expressed in a ceftazidime/avibactam-resistant Klebsiella pneumoniae is associated with relevant detection issues. J. Antimicrob. Chemother. 2019, 74, 2464–2466. [Google Scholar] [CrossRef]
- Venditti, C.; Nisii, C.; Ballardini, M.; Meledandri, M.; Di Caro, A. Identification of L169P mutation in the omega loop of KPC-3 after a short course of ceftazidime/avibactam. J. Antimicrob. Chemother. 2019, 74, 2466–2467. [Google Scholar] [CrossRef]
- Raisanen, K.; Koivula, I.; Ilmavirta, H.; Puranen, S.; Kallonen, T.; Lyytikainen, O.; Jalava, J. Emergence of ceftazidime-avibactam-resistant Klebsiella pneumoniae during treatment, Finland, December 2018. Eurosurveillance 2019, 24, 1900256. [Google Scholar] [CrossRef] [Green Version]
- Giddins, M.J.; Macesic, N.; Annavajhala, M.K.; Stump, S.; Khan, S.; McConville, T.H.; Mehta, M.; Gomez-Simmonds, A.; Uhlemann, A.C. Successive Emergence of Ceftazidime-Avibactam Resistance through Distinct Genomic Adaptations in blaKPC-2-Harboring Klebsiella pneumoniae Sequence Type 307 Isolates. Antimicrob. Agents Chemother. 2018, 62, e02101-17. [Google Scholar] [CrossRef] [Green Version]
- Shields, R.K.; Chen, L.; Cheng, S.; Chavda, K.D.; Press, E.G.; Snyder, A.; Pandey, R.; Doi, Y.; Kreiswirth, B.N.; Nguyen, M.H.; et al. Emergence of Ceftazidime-Avibactam Resistance Due to Plasmid-Borne blaKPC-3 Mutations during Treatment of Carbapenem-Resistant Klebsiella pneumoniae Infections. Antimicrob. Agents Chemother. 2017, 61, e02097-16. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Ding, B.; Ye, M.; Wang, P.; Bi, Y.; Wu, S.; Xu, X.; Guo, Q.; Wang, M. High ceftazidime hydrolysis activity and porin OmpK35 deficiency contribute to the decreased susceptibility to ceftazidime/avibactam in KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2017, 72, 1930–1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galani, I.; Antoniadou, A.; Karaiskos, I.; Kontopoulou, K.; Giamarellou, H.; Souli, M. Genomic characterization of a KPC-23-producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime-avibactam. Clin. Microbiol. Infect. 2019, 25, 763.e765–763.e768. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chen, W.; Li, H.; Li, L.; Zou, X.; Zhao, J.; Lu, B.; Li, B.; Wang, C.; Li, H.; et al. Phenotypic and genotypic analysis of KPC-51 and KPC-52, two novel KPC-2 variants conferring resistance to ceftazidime/avibactam in the KPC-producing Klebsiella pneumoniae ST11 clone background. J. Antimicrob. Chemother. 2020, 75, 3072–3074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Shi, Q.; Hu, H.; Hong, B.; Wu, X.; Du, X.; Akova, M.; Yu, Y. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin. Microbiol. Infect. 2020, 26, 124.e121–124.e124. [Google Scholar] [CrossRef] [Green Version]
- Coppi, M.; Di Pilato, V.; Monaco, F.; Giani, T.; Conaldi, P.; Rossolini, G. Ceftazidime-Avibactam Resistance Associated with Increased blaKPC-3 Gene Copy Number Mediated by pKpQIL Plasmid Derivatives in Sequence Type 258. Antimicrob. Agents Chemother. 2020, 64, e01816-19. [Google Scholar] [CrossRef]
- Mueller, L.; Masseron, A.; Prod’Hom, G.; Galperine, T.; Greub, G.; Poirel, L.; Nordmann, P. Phenotypic, biochemical and genetic analysis of KPC-41, a KPC-3 variant conferring resistance to ceftazidime-avibactam and exhibiting reduced carbapenemase activity. Antimicrob. Agents Chemother. 2019, 63, e01111-19. [Google Scholar] [CrossRef] [Green Version]
- Oueslati, S.; Iorga, B.I.; Tlili, L.; Exilie, C.; Zavala, A.; Dortet, L.; Jousset, A.B.; Bernabeu, S.; Bonnin, R.A.; Naas, T. Unravelling ceftazidime/avibactam resistance of KPC-28, a KPC-2 variant lacking carbapenemase activity. J. Antimicrob. Chemother. 2019, 74, 2239–2246. [Google Scholar] [CrossRef]
- Bacterial Antimicrobial Resistance Reference Gene Database. Available online: https://www.ncbi.nlm.nih.gov/pathogens/refgene/#KPC (accessed on 20 May 2022).
- Gaibani, P.; Campoli, C.; Lewis, R.; Volpe, S.; Scaltriti, E.; Giannella, M.; Pongolini, S.; Berlingeri, A.; Cristini, F.; Bartoletti, M.; et al. In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J. Antimicrob. Chemother. 2018, 73, 1525–1529. [Google Scholar] [CrossRef] [Green Version]
- Venditti, C.; Butera, O.; Meledandri, M.; Balice, M.; Cocciolillo, G.; Fontana, C.; D’Arezzo, S.; De Giuli, C.; Antonini, M.; Capone, A.; et al. Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2021, 27, 1040.e1041–1040.e1046. [Google Scholar] [CrossRef]
- Galani, I.; Karaiskos, I.; Angelidis, E.; Papoutsaki, V.; Galani, L.; Souli, M.; Antoniadou, A.; Giamarellou, H. Emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in KPC-2-producing Klebsiella pneumoniae of sequence type 39 during treatment. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2021, 40, 219–224. [Google Scholar] [CrossRef]
- Li, D.; Li, K.; Dong, H.; Ren, D.; Gong, D.; Jiang, F.; Shi, C.; Li, J.; Zhang, Q.; Yan, W.; et al. Ceftazidime-Avibactam Resistance in Klebsiella pneumoniae Sequence Type 11 Due to a Mutation in Plasmid-Borne blaKPC-2 to blaKPC-33, in Henan, China. Infect. Drug Resist. 2021, 14, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Chavda, K.D.; Wei, J.; Zou, C.; Marshall, S.H.; Dhawan, P.; Wang, D.; Bonomo, R.A.; Kreiswirth, B.N.; Chen, L. A Ceftazidime-Avibactam-Resistant and Carbapenem-Susceptible Klebsiella pneumoniae Strain Harboring bla (KPC-14) Isolated in New York City. mSphere 2020, 5, e00775-20. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Xiao, T.; David, S.; Wang, Q.; Zhou, Y.; Guo, L.; Aanensen, D.; Holt, K.; Thomson, N.; Grundmann, H.; et al. Novel Subclone of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 with Enhanced Virulence and Transmissibility, China. Emerg. Infect. Dis. 2020, 26, 289–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobson, C.A.; Bonacorsi, S.; Fahd, M.; Baruchel, A.; Cointe, A.; Poey, N.; Jacquier, H.; Doit, C.; Monjault, A.; Tenaillon, O.; et al. Successful Treatment of Bacteremia Due to NDM-1-Producing Morganella morganii with Aztreonam and Ceftazidime-Avibactam Combination in a Pediatric Patient with Hematologic Malignancy. Antimicrob. Agents Chemother. 2019, 63, e02463-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davido, B.; Fellous, L.; Lawrence, C.; Maxime, V.; Rottman, M. Ceftazidime-Avibactam and Aztreonam, an Interesting Strategy to Overcome beta-Lactam Resistance Conferred by Metallo-beta-Lactamases in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2017, 61, e01008-17. [Google Scholar] [CrossRef] [Green Version]
- Shields, R.K.; Potoski, B.A.; Haidar, G.; Hao, B.; Doi, Y.; Chen, L.; Press, E.G.; Kreiswirth, B.N.; Clancy, C.J.; Nguyen, M.H. Clinical Outcomes, Drug Toxicity, and Emergence of Ceftazidime-Avibactam Resistance Among Patients Treated for Carbapenem-Resistant Enterobacteriaceae Infections. Clin. Infect. Dis. 2016, 63, 1615–1618. [Google Scholar] [CrossRef] [Green Version]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Thirtieth Edition: M100; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 9.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf (accessed on 20 May 2022).
- Zhang, R.; Liu, L.; Zhou, H.; Chan, E.; Li, J.; Fang, Y.; Li, Y.; Liao, K.; Chen, S. Nationwide Surveillance of Clinical Car-bapenem-resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine 2017, 19, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Xie, M.; Dong, N.; Lin, D.; Yang, X.; Wong, M.H.Y.; Chan, E.W.-C.; Chen, S. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 2018, 7, gix132. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2013, 42, D206–D214. [Google Scholar] [CrossRef]
- Lam, M.M.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).