Comparative Transcriptome-Based Mining of Genes Involved in the Export of Polyether Antibiotics for Titer Improvement
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Culture Conditions
2.2. Transcriptome Sequencing of BK3-25
2.3. Construction of Plasmids for Deletion and Over-Expression of Eight Candidate Genes
2.4. Conjugation between Streptomyces and E. coli
2.5. HPLC Analysis of Antibiotics
2.6. RNA Extraction and RT-qPCR Analysis
2.7. Biomass Determination under Fermentation Condition
3. Results
3.1. Transcriptome-Based Identification of Candidate Exporter Genes
3.2. All Eight Candidate Genes Were Positively Involved in Salinomycin Export
3.3. These Eight Exporter Genes Were Constitutively Expressed
3.4. SLNHY_0929 and SLNHY_1893 Improved Resistance to Salinomycin in Streptomyces lividans
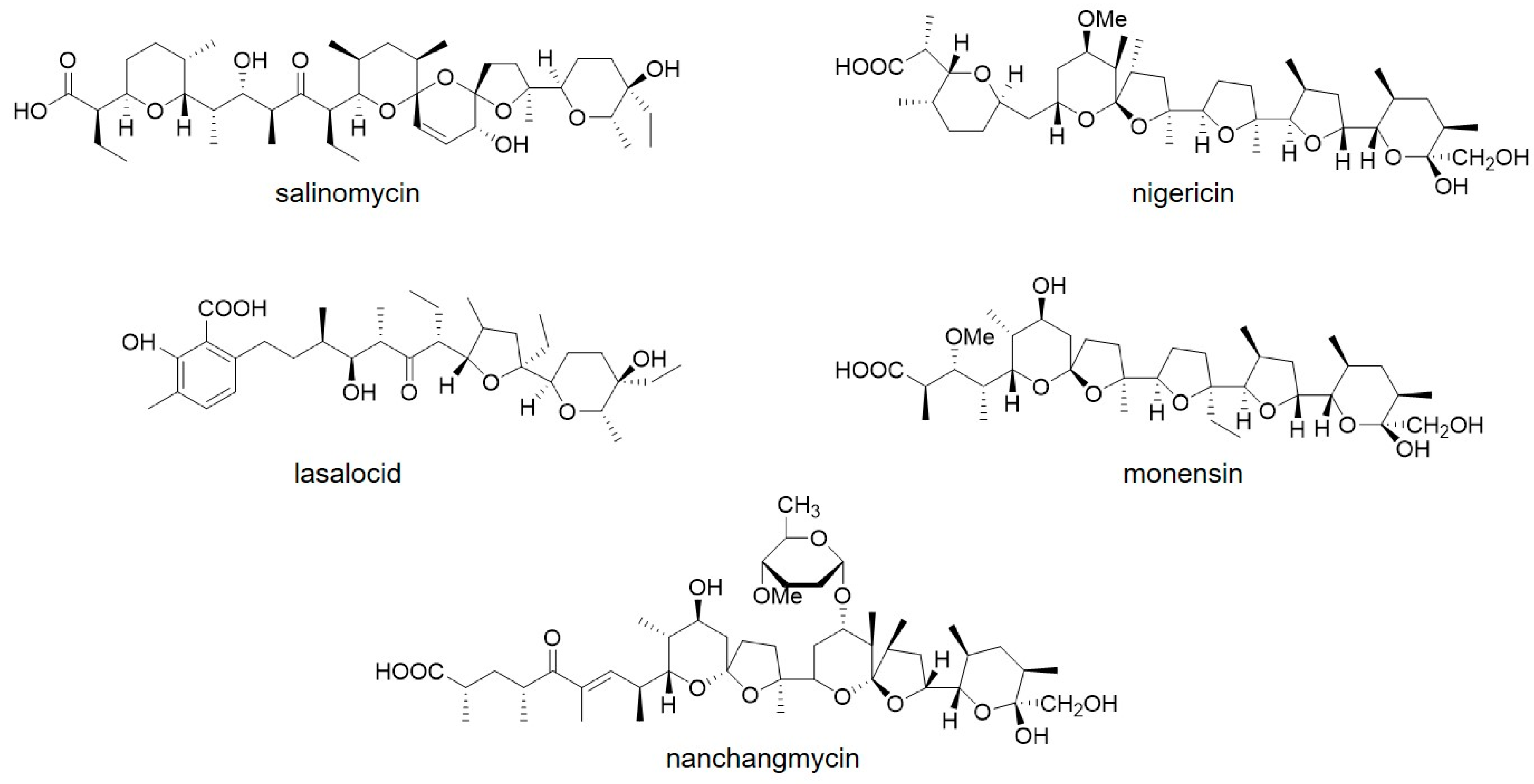
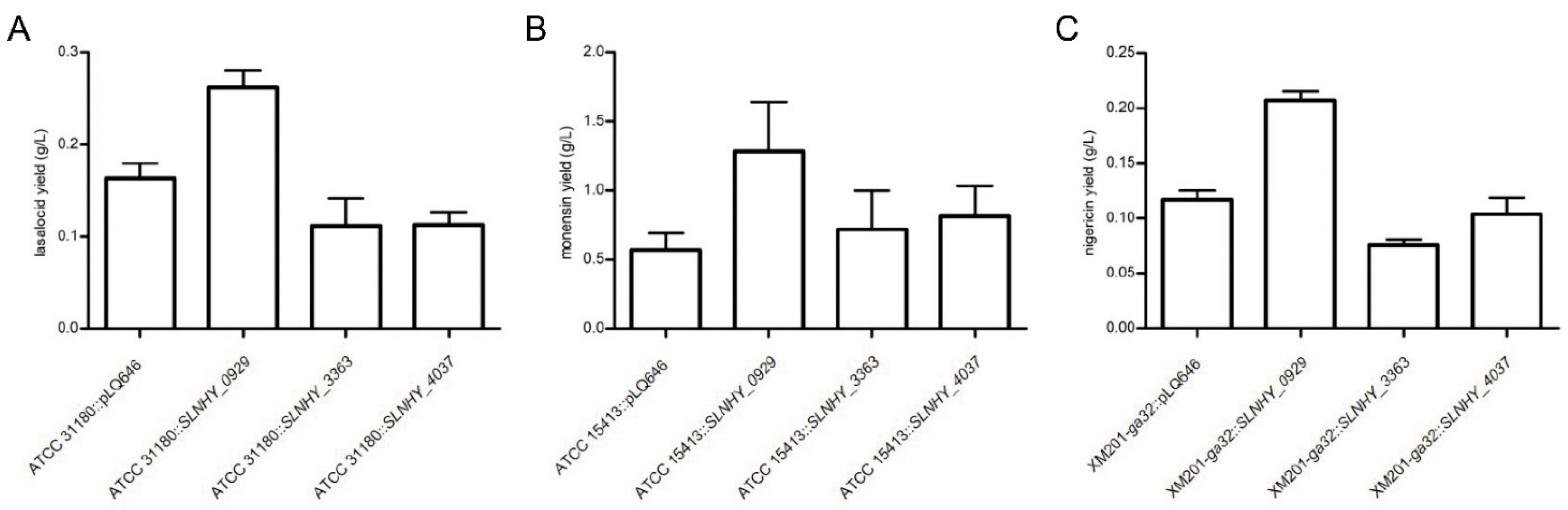
3.5. SLNHY_0929 Was a Universal Exporter for Polyether Antibiotics with Similar Structure with Salinomycin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, H.; Liu, N.; Xi, L.; Rong, X.; Ruan, J.; Huang, Y. Genetic screening strategy for rapid access to polyether ionophore producers and products in Actinomycetes. Appl. Environ. Microbiol. 2011, 77, 3433–3442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cane, D.E. Unified stereochemical model of polyether antibiotic structure and biogenesis. J. Am. Chem. Soc. 1983, 2, 3594–3600. [Google Scholar] [CrossRef]
- Yurkovich, M.E.; Tyrakis, P.A.; Hong, H.; Sun, Y.; Samborskyy, M.; Kamiya, K.; Leadlay, P.F. A late-stage intermediate in salinomycin biosynthesis is revealed by specific mutation in the biosynthetic gene cluster. ChemBioChem 2012, 13, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Wietrzyk, J.; Antoszczak, M.; Popiel, K.; Stefa, J. Synthesis, cytotoxicity and antibacterial activity of new esters of polyether antibiotic salinomycin. Eur. J. Med. Chem. 2014, 76, 435–444. [Google Scholar]
- Fuchs, D.; Daniel, V.; Sadeghi, M.; Opelz, G.; Naujokat, C. Salinomycin overcomes ABC transporter-mediated multidrug and apoptosis resistance in human leukemia stem cell-like KG-1a cells. Biochem. Biophys. Res. Commun. 2010, 394, 1098–1104. [Google Scholar] [CrossRef]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Zhang, X.; Jiang, M.; Bai, L. Enhanced salinomycin production by adjusting the supply of polyketide extender units in Streptomyces albus. Metab. Eng. 2016, 35, 129–137. [Google Scholar] [CrossRef]
- Martín, J.F.; Liras, P. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr. Opin. Microbiol. 2010, 13, 263–273. [Google Scholar] [CrossRef]
- Hopwood, D.A. MicroCommentary How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol. Microbiol. 2007, 63, 937–940. [Google Scholar] [CrossRef]
- Mak, S.; Xu, Y.; Nodwell, J.R. Micro Review The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol. Microbiol. 2014, 93, 391–402. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, H.; Kang, Q.; Liu, J.; Bai, L. Cloning and characterization of the polyether salinomycin biosynthesis gene cluster of Streptomyces albus XM211. Appl. Environ. Microbiol. 2012, 78, 994–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Jiang, C.; Zhang, B.; Bai, L. Involvement of ABC transporter genes slnTI and slnTII in salinomycin biosynthesis. Microbiol. China 2014, 41, 58–66. [Google Scholar]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Méndez, C.; Salas, J.A. The role of ABC transporters in antibiotic-producing organisms: Drug secretion and resistance mechanisms. Nucleic Acids Res. 2001, 152, 341–350. [Google Scholar] [CrossRef]
- Saier, M.H.; Reddy, V.S.; Tamang, D.G.; Västermark, Å. The transporter classification database. Nucleic Acids Res. 2014, 42, 251–258. [Google Scholar] [CrossRef]
- Omura, S.; Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Takahashi, C.; Shinose, M.; Takahashi, Y.; Horikawa, H.; Nakazawa, H.; Osonoe, T.; et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 2001, 98, 12215–12220. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Shan, Y.; Li, H. Multiple transporters are involved in natamycin efflux in Streptomyces chattanoogensis L10. Mol. Microbiol. 2017, 103, 713–728. [Google Scholar] [CrossRef] [Green Version]
- Chu, L.; Li, S.; Dong, Z.; Zhang, Y.; Jin, P.; Ye, L.; Wang, X.; Xiang, W. Mining and engineering exporters for titer improvement of macrolide biopesticides in Streptomyces. Microb. Biotechnol. 2021, 15, 1120–1132. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, C.; Bai, L. Conversion of the high-yield salinomycin producer Streptomyces albus BK3-25 into a surrogate host for polyketide production. Sci. China Life Sci. 2017, 60, 1000–1009. [Google Scholar] [CrossRef]
- Hopwood, D.A. Highlights of Streptomyces genetics. Heredity 2019, 123, 23–32. [Google Scholar] [CrossRef]
- Ling, Y.; Zeng, Z. Optimization of fermentation conditions of polyether antibiotic lasalocid by response surface method. Chin. J. Antibiot. 2017, 42, 28–32. [Google Scholar]
- Wang, X.; Ning, X.; Zhao, Q.; Kang, Q.; Bai, L. Improved PKS gene expression with strong endogenous promoter resulted in geldanamycin yield increase. Biotechnol. J. 2017, 12, 1700321. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, X.; Pang, A.; Lin, C.; Zhang, Y.; Zhang, J.; Qiao, J.; Zhao, G. Characterization of three pathway-specific regulators for high production of monensin in Streptomyces cinnamonensis. Appl. Microbiol. Biotechnol. 2017, 101, 6083–6097. [Google Scholar] [CrossRef] [PubMed]
- Leulmi, N.; Sighel, D.; Defant, A.; Khenaka, K.; Boulahrouf, A.; Mancini, I. Enhanced production and quantitative evaluation of nigericin from the Algerian soil-living Streptomyces youssoufiensis SF10 strain. Fermentation 2019, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Qu, S.; Kang, Q.; Wu, H.; Wang, L.; Bai, L. Positive and negative regulation of GlnR in validamycin A biosynthesis by binding to different loci in promoter region. Appl. Microbiol. Biotechnol. 2015, 99, 4771–4783. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Wang, J.; Xiang, S.; Feng, X.; Yang, K. An engineered strong promoter for streptomycetes. Appl. Environ. Microbiol. 2013, 79, 4484–4492. [Google Scholar] [CrossRef] [Green Version]
- Du, D.; Zhu, Y.; Wei, J.; Tian, Y.; Niu, G.; Tan, H. Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Appl. Microbiol. Biotechnol. 2013, 97, 6383–6396. [Google Scholar] [CrossRef]
- Qiu, J.; Zhuo, Y.; Zhu, D.; Zhou, X.; Zhang, L.; Bai, L.; Deng, Z. Overexpression of the ABC transporter AvtAB increases avermectin production in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 2011, 92, 337–345. [Google Scholar] [CrossRef]
- Nigam, S.K. What do drug transporters really do? Nat. Rev. Drug Discov. 2014, 14, 29–44. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.A.; Lee, S.Y. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kell, D.B.; Swainston, N.; Pir, P.; Oliver, S.G. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 2015, 33, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Migita, A.; Watanabe, M.; Hirose, Y.; Watanabe, K.; Tokiwano, T.; Kinashi, H.; Oikawa, H. Identification of a gene cluster of polyether antibiotic lasalocid from Streptomyces lasaliensis. Biosci. Biotechnol. Biochem. 2009, 73, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliynyk, M.; Stark, C.B.W.; Bhatt, A.; Jones, M.A.; Hughes-Thomas, Z.A.; Wilkinson, C.; Oliynyk, Z.; Demydchuk, Y.; Staunton, J.; Leadlay, P.F. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol. Microbiol. 2003, 49, 1179–1190. [Google Scholar] [CrossRef] [PubMed]
- Harvey, B.M.; Mironenko, T.; Sun, Y.; Hong, H.; Deng, Z.; Leadlay, P.F.; Weissman, K.J.; Haydock, S.F. Insights into polyether biosynthesis from analysis of the nigericin biosynthetic gene cluster in Streptomyces sp. DSM4137. Chem. Biol. 2007, 14, 703–714. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lin, C.Y.; Li, X.M.; Tang, Z.K.; Qiao, J.; Zhao, G.R. DasR positively controls monensin production at two-level regulation in Streptomyces cinnamonensis. J. Ind. Microbiol. Biotechnol. 2016, 43, 1681–1692. [Google Scholar] [CrossRef]
- Armando, J.W.; Boghigian, B.A.; Pfeifer, B.A. LC-MS/MS quantification of short-chain acyl-CoA’s in Escherichia coli demonstrates versatile propionyl-CoA synthetase substrate specificity. Lett. Appl. Microbiol. 2012, 54, 140–148. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, C.; Bai, L. Mechanism of salinomycin overproduction in Streptomyces albus as revealed by comparative functional genomics. Appl. Microbiol. Biotechnol. 2017, 101, 4635–4644. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A.; Hintermann, G.; Kieser, T.; Wright, H.M. Integrated DNA sequences in three streptomycetes form related autonomous plasmids after transfer to Streptomyces lividans. Plasmid 1984, 11, 1–16. [Google Scholar] [CrossRef]
- Day, L.E.; Chamberlin, J.W.; Gordee, E.Z.; Chen, S.; Gorman, M.; Hamill, R.L.; Ness, T.; Weeks, R.E.; Stroshane, R. Biosynthesis of monensin. Antimicrob. Agents Chemother. 1973, 4, 410–414. [Google Scholar] [CrossRef] [Green Version]
- Smith, P. High efficiency intergeneric conjugal transfer of plasmid DNA from Esc herichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997, 155, 223–229. [Google Scholar]
- Wang, X.; Wang, R.; Kang, Q.; Bai, L. The antitumor agent ansamitocin P-3 binds to cell division protein FtsZ in Actinosynnema pretiosum. Biomolecules 2020, 10, 699. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bai, L.; Liang, J.; Zhou, X.; Deng, Z. Two pHZ1358 derivative vectors for efficient gene knockout in Streptomyces. J. Microbiol. Biotechnol. 2010, 20, 678–682. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Wu, Y.; Zhang, X.; Kang, Q.; Yan, Y.; Bai, L. Comparative Transcriptome-Based Mining of Genes Involved in the Export of Polyether Antibiotics for Titer Improvement. Antibiotics 2022, 11, 600. https://doi.org/10.3390/antibiotics11050600
Liu X, Wu Y, Zhang X, Kang Q, Yan Y, Bai L. Comparative Transcriptome-Based Mining of Genes Involved in the Export of Polyether Antibiotics for Titer Improvement. Antibiotics. 2022; 11(5):600. https://doi.org/10.3390/antibiotics11050600
Chicago/Turabian StyleLiu, Xian, Yuanting Wu, Xiaojie Zhang, Qianjin Kang, Yusi Yan, and Linquan Bai. 2022. "Comparative Transcriptome-Based Mining of Genes Involved in the Export of Polyether Antibiotics for Titer Improvement" Antibiotics 11, no. 5: 600. https://doi.org/10.3390/antibiotics11050600
APA StyleLiu, X., Wu, Y., Zhang, X., Kang, Q., Yan, Y., & Bai, L. (2022). Comparative Transcriptome-Based Mining of Genes Involved in the Export of Polyether Antibiotics for Titer Improvement. Antibiotics, 11(5), 600. https://doi.org/10.3390/antibiotics11050600






