Abstract
The objective of this study was to determine the presence and persistence of antimicrobial-resistant enterobacteria and their clonal distribution in hospital wastewater. A descriptive cross-sectional study was carried out in wastewater from two Mexico City tertiary level hospitals. In February and March of 2020, eight wastewater samples were collected and 26 isolates of enterobacteria were recovered, 19 (73.1%) isolates were identified as E. coli, 5 (19.2%) as Acinetobacter spp. and 2 (7.7%) as Enterobacter spp. Antimicrobial susceptibility profiles were performed using the VITEK 2® automated system and bacterial identification was performed by the Matrix-Assisted Laser Desorption/Ionization-Time of Flight mass spectrometry (MALDI-TOF MS®). ESBL genes were detected by polymerase chain reaction (PCR) and clonal distributions of isolates were determined by pulsed-field gel electrophoresis (PFGE). E. coli susceptibility to different classes of antimicrobials was analyzed and resistance was mainly detected as ESBLs and fluoroquinolones. One E. coli strain was resistant to doripenem, ertapenem, imipenem and meropenem. The analysis by PCR showed the presence of specific β-lactamases resistance genes (blaKPC, blaCTX-M). The PFGE separated the E. coli isolates into 19 different patterns (A–R). PFGE results of Acinetobacter spp. showed the presence of a majority clone A. Surveillance of antimicrobial resistance through hospital wastewater is an important tool for early detection of clonal clusters of clinically important bacteria with potential for dissemination.
1. Introduction
Antimicrobial resistance (AMR) is a growing public health problem worldwide due to the presence of multidrug-resistant (MDR) bacterial pathogens in healthcare settings [1]. The World Health Assembly issued the “Global Action Plan on Antimicrobial Resistance” to address the AMR problem [2], urging countries to strengthen surveillance to direct targeted research and interventions appropriated for each country or even institutions. In 2017, the World Health Organization published a list of pathogens of priority attention, among which are carbapenem-resistant and extended-spectrum β-lactamase Enterobacteriaceae [3].
Antimicrobial-resistant enterobacteria producers of extended-spectrum β-lactamase (ESBL) are the most frequently isolated microorganisms in healthcare units; these are highlighted by the presence of several enzymes that can hydrolyze penicillin and inhibit penicillin and broad-spectrum beta-lactam antibiotics, such as third- and fourth-generation cephalosporins and monobactams. Different variants of ESBL (e.g., TEM, TLA, SHV, CTX-M) have been described with worldwide dissemination, including Mexico [4,5]. Despite the availability of a few antibiotics treatments for ESLB-producing enterobacteria, there is emerging resistance, which limits patients’ therapeutic options and is associated with high morbidity and mortality [6].
The presence of antimicrobial-resistant enterobacteria is frequently described in hospital settings; however, their presence in other locations or settings, such as community, agricultural, and environmental settings, demonstrates the ability of enterobacteria to spread beyond healthcare settings [7,8]. Some studies carried out in hospitals in Mexico report data on E. coli resistant to beta-lactams with alarming values (50%), as well as an increase in resistance to carbapenems in Enterobacter spp. isolates [9,10]. In this regard, hospital wastewater plays an essential role in the spread of antimicrobial-resistant bacteria, including Enterobacteriaceae, because wastewater carries and receives both bacteria and resistance genes and antibiotic concentrations from healthcare institutions. Therefore, investigating the presence of antimicrobial-resistant bacteria in hospital and/or community wastewater through an epidemiological surveillance system makes it possible to monitor, prevent, alert and control the spread of antibiotic resistant bacteria both in the hospital environment and in the environment [11,12,13].
Some hospitals have wastewater treatment plants (WWTPs), and different authors have described that WWTP treatments significantly reduce the abundance of bacteria. The main objective of WWTPs is to remove organic matter and most environmental microorganisms [14]. Unfortunately, WWTPs do not eliminate all antibiotic-resistant bacteria and their resistance genes, so the persistence of pathogenic bacterial after treatment is feasible. This poses a paradigm for public health epidemiological surveillance based on hospital wastewater as an alternative approach and rapid assessment due to the excretion of AMR and partially metabolized antimicrobials in patients’ urine and feces through the sewer [15,16].
In this paper we describe the antibiotic resistant and clonal profiles of Escherichia coli, Acinetobacter spp. and Enterobacter spp. in raw and treated wastewater from two hospitals.
2. Results
2.1. Enterobacteriaceae Isolates
Eight wastewater samples were collected from the influent and affluent of the WWTPs of two third-level attention hospitals in Mexico. A total of 26 enterobacteria were presumptively identified using selective chromo-agar and subsequently confirmed by MALDI-TOF MS ® and VITEK 2®; 19 (73.1%) isolates were identified as E. coli, 5 (19.2%) as Acinetobacter spp. and 2 (7.7%) as Enterobacter spp. Half of the isolates were isolated from raw wastewater and the other half were from treated wastewater. In Hospital A, eight Escherichia coli and one Entererobacter cloacae were isolated from the raw wastewater and three E. coli, one E. bugandensis and five Acinetobacter spp. (A. haemolyticus and A. modestus) were isolated from the treated wastewater. At Hospital B, four E. coli were isolated from the raw wastewater, four E. coli were isolated from treated wastewater, and no isolates of other enterobacteria were detected in this hospital.
2.2. Antimicrobial Susceptibility
Antimicrobial susceptibility results detected by VITEK 2® showed that 11 strains were resistant to ampicillin-sulbactam (MIC90 of >32 mg/L); five strains were resistant to piperacillin-tazobactam (MIC90 of >128 mg/L); and eight strains were resistant to cefoxitin, ceftazidime, ceftriaxone and cefepime (second-, third- and fourth-generation cephalosporin) (MIC90 of >64 mg/L). In addition, one E. coli strain was resistant to doripenem, ertapenem, imipenem and meropenem (MIC90 of ≤0.12, ≤0.5, ≤0.25 and ≤0.25 mg/L, respectively); eleven strains were resistant to ciprofloxacin (MIC90 of >4 mg/L) and two strains were resistant to gentamycin (MIC90 of <1 mg/L). Both Enterobacter spp. isolates were resistant to cefoxitin (MIC90 of >64 mg/L). All Acinetobacter spp. isolates were susceptible to tygecyline and colistin, and three isolates were amikacyn-resistant (MIC90 of >64 mg/L) (Table 1).

Table 1.
Phenotyping properties of E. coli, Acinetobacter spp. and Enterobacter spp. isolated from the hospital wastewater treatment plants (WTTP).
2.3. Detection of β-Lactamases Resistance Genes
The molecular analysis by PCR showed the presence of specific β-lactamases resistance genes (blaKPC, blaCTX-M); it was observed that one E. coli strains carried blaKPC, and another harbored blaCTX-M. Two isolates were also observed to harbor more than one gene (Table 1).
2.4. Molecular Typing by Pulsed-Field Gel Electrophoresis (PFGE)
The PFGE separated the E. coli isolates into 19 different patterns (A–R) without subtypes; the patterns A–K were from Hospital A and the patterns L–R were from Hospital B. The patterns A–H and L–Ñ were present in E. coli isolates from raw wastewater, whereas the patterns I–K and O–R were isolates from treated wastewater. The PFGE separated the Enterobacter spp. into two patterns (S and T): one pattern in raw wastewater and one pattern in treated wastewater from Hospital A (Figure 1).
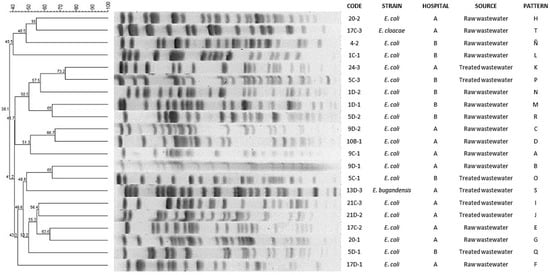
Figure 1.
Dendrogram based on pulsed-field gel electrophoresis (PFGE) patterns after digestion with enzyme Xba I of E. coli and Enterobacter spp. isolates isolated from hospital wastewater.
PFGE results of Acinetobacter spp. isolates showed the presence of a majority clone A (n = 4) and an unrelated pattern (B); these clones were detected in strains collected in treated wastewater from Hospital A (Figure 2).
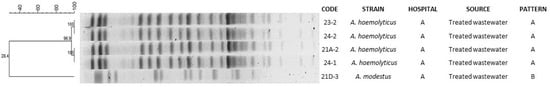
Figure 2.
Dendrogram based on pulsed-field gel electrophoresis (PFGE) patterns after digestion with enzyme Apa I of Acinetobacter spp. isolates isolated from hospital wastewater.
The PFGE patterns analysis showed similarity percentages from 73% to 38% in E. coli and Enterobacter spp. and similarity percentages that were from 100% to 28% in Acinetobacter spp.
When PFGE patterns were compared with antimicrobial susceptibility profiles, it was observed that E. coli strains with patterns F, G, I, K, L, M, O and P showed resistance to a higher number of antimicrobials tested, whereas strains with patterns A, C, E, J, N, Ñ and R were sensitive to all antimicrobials tested. The patterns T and S of Enterobater spp. had the same resistance profile. On the other hand, in Acinetobacter spp. clone A, resistance to amikacin was observed in three of the four strains tested, whereas clone B was sensitive to all antimicrobials tested.
3. Discussion
The high frequency of E. coli strains in raw and treated hospital wastewater samples, compared to other bacteria, would be expected in some extent, mainly because it is one of the bacterial species that are part of the microbiota of the digestive tract of healthy and sick humans and is widely distributed in the environment. However, its dissemination and spread in hospital wastewater highlights the risk of contamination of aquatic environments or bodies of water by possible pathogenic strains of E. coli resistant to antimicrobials [17]. Although the objective of this study was not to determine whether E. coli strains isolated from hospital wastewater were pathogenic or not, it was revealed that they were the main bacterial species showing multidrug resistance (n = 7) and there was a presence of extended-spectrum beta-lactamases in three E. coli strains tested. The E. coli resistance patterns were relatively similar between the two hospitals studied (Table 1).
Resistance of E. coli to different classes of antimicrobials, such as beta-lactams and fluoroquinolones, mainly amoxicillin and ciprofloxacin, was evident in raw and treated wastewater samples from both hospitals (A and B), which may be due to the selection pressure of antimicrobial-resistant strains in the clinical hospital environment and their persistence through the treatment processes in the wastewater treatment plants. Several studies in different countries have reported a similarly high rate of resistance among E. coli strains to beta-lactams of clinical and environmental origin [18,19]. This is associated with the antimicrobial resistance mechanism generated by beta-lactamase enzymes, encoded by resistance genes and carried on plasmids, which can be exchanged between different bacterial species by horizontal transmission of genes (HTG) and mobile genetic elements (MGE) [20].
The two isolates of the Enterobacter spp. isolated in this study were resistant to second-generation cephalosporins (Table 1). Barraud et al. [21] reported one strain of Enterobacter cloacae resistant to quinolones, beta lactams, aminoglycosides and tetracycline obtained from hospital residual water. A carbapenem-resistant strain of E. coli was detected in the raw wastewater at Hospital B. Although resistance to carbapenems due to the presence of carbapenemase enzymes has been widely described in Enterobacteriaceae, mainly in genera such as K. pneumoniae, it has also been reported in E. coli and Enterobacter spp. isolated from sewage. This is of concern because Enterobacteriaceae are responsible for infectious outbreaks and carbapenem antibiotics are generally used as a last resort in the treatment of diseases caused by bacteria resistant to other classes of antibiotics [7]. Enterobacter spp. has been claimed to be a bacterial genus that serves as a reservoir of antimicrobial resistance genes through HTG and MGE [8].
Only isolates of the Acinetobacter spp. were evaluated against colistin, and they were found to be sensitive (Table 1). Overall, the highest proportion of isolates was sensitive to aminoglycosides and fully sensitive to tigecycline and this differs from the findings of Wang et al. [22] and Zhang et al. [23], who reported one and ten strains of Acinetobacter spp., respectively, with a resistance profile to cephalosporin and carbapenems. In this work we reported three strains of A. haemolyticus with resistance to amikacin (MIC range 32–64 mg/L). Similar results were reported by Zong et al. [23], Bardhan et al. [19] and Kovacic et al. [24], who detected amikacin-resistant Acinetobacter spp. in hospital wastewater.
Although in this study, eight primers were used for the detection of β-lactamases resistance genes including a mixture of the AmpC gene, we only detected the presence of blaKPC and blaCTX-M genes in E. coli strains. Compared to K. pneumoniae isolates, blaKPC producing E. coli strains remain less frequent [25]. However, blaCTX-M producing E. coli were more frequently reported. The first report on the blaCTX-M gene was in 1989, with CTX-M being the most prevalent ESBL in the world [26]. We found that two strain of E. coli carried blaKPC and blaCTX-M, and this differs from the findings of Kutilova et al. [27], who reported a high prevalence of blaCTX-M (>95%) in strains of E. coli isolated from raw hospital wastewater. Xu et al. [25] reported similar results to ours in samples of river water where wastewater is discharged, reporting three strains of E. coli carrying blaKPC and blaCTX-M. This is in agreement with the results reported by Zumaya et al. regarding the excess use of cephalosporins and carbapenems in the medical prescription of the hospitals included in this study [28].
Molecular typing by PFGE was performed to investigate the clonal relationship between the E. coli strains, Enterobacter spp. and Acinetobacter spp. (Figure 1 and Figure 2). In our study, we detected a remarkable diversity of patterns in E. coli strains and Enterobacter spp. (n = 21); this heterogenicity of patterns has been observed in other studies, such as those published by Galvin et al. [29] and Zhao et al. [30], who found 18 PFGE patterns among the 23 isolates analyzed and 7 PFGE patters in 9 strains studied isolated from hospital WWTPs. However, it is remarkably interesting that antimicrobial resistance profiles are highly similar in the different clones. Thus, these data suggest that these antimicrobial resistance genes are highly selected in Enterobacteriaceaes in the hospital environment. Another of our important findings was that the strains of A. haemolyticus that carried the same PFGE pattern were found in treated wastewater, showing its ability to persist in its passage through the WWTP. The clonality of Acinetobacter spp. has been document in other studies. In studies carried out in Croatia and India, PFGE revealed related patterns among A. baumannii strains [19,31]. In another works in China, PFGE analysis showed pattern heterogeneity, this being different from our results [23,32].
4. Materials and Methods
4.1. Study Sites and Samples Collection
The study was conducted in two third-level hospitals in the Mexico City Metropolitan Zone in February and March 2020. One is a national referral hospital (Hospital A) with 119 beds, the other is a regional hospital (Hospital B) with 246 beds located in the State of Mexico, which provides specialized care to people from several states in the center of the country. Single samples of raw wastewater and treated wastewater were the primary sampling sources. Wastewater released from both hospitals flows directly to a wastewater treatment plant, and then to the city sewer.
Hospital wastewater samples were collected from the influent and effluent of each wastewater treatment plant (WWTP). A total of eight wastewater samples were collected over three weeks, two raw and two treated wastewater samples each from Hospital A and B. Wastewater simple samples were obtained by the “grap” technique in sterile 1 L containers [14], and transported at <4 °C to the Research Center for Infectious Disease at the National Institute of Public Health in Cuernavaca, Morelos within two hours of collection for processing.
4.2. Microbial Culturing and Identification
Microbial culturing of the Enterobacteriaceae family was performed by taking an aliquot of 100 mL of raw wastewater and 200 mL of treated wastewater in 50 mL conical bottom propylene tubes, which were centrifuged for 20 min at 5000 revolutions per minute (rpm). After decanting the supernatant, the pellet was dissolved in 10 mL of phosphate buffer solution (PBS) to homogenized by shaking.
After homogenization and prior to plating, the samples were diluted 1:1000 with PBS, the following agar plates were plated by single streaking: Hi-Crome™ ECC (HiMedia Laboratories, Mumbai; India), Koser Citrate Medium® and Hi-Crome™ Acinetobacter Agar Base (HiMedia Laboratories, Mumbai; India) inoculating 100 μL of solution. The plates were labeled with the type of wastewater, sampling location, date and sample number. Plates were incubated at 37 °C, and results were read after 18 hours. Different presumptive colonies of each bacterial species were selected based on the color patterns described by the manufacturer on chromogenic agars. They were then transferred to MacConkey agar at 37 °C for 18 h for purification and identification. The isolates were identified by mass spectrometry, using the Microflex MALDI-TOF MS® (Bruker Daltonics, Bremen, Germany) equipment.
4.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility profiling of strains was carried out by a VITEK 2® automated system (BioMe’rieux, Marcy l’Etoile, France), according to the break point established by the CLSI 2021 guidelines [33]. Fourteen antimicrobials were tested: ampicillin-sulbactam, piperacillin-tazobactam, cefoxitin, ceftazidime, ceftriaxone, cefepime, doripenem, ertapenem, imipenem, meropenem, amikacin, gentamicin, ciprofloxacin and tigecycline.
4.4. Detection of β-Lactamases Genes
The presence of β-lactamases genes in Enterobacteriaceae isolates was confirmed by PCR. The assay included primers to detected the following β-lactamases genes: blaCTX-M, blaSHV, blaAMPc, blaGES-2, blaKPC, blaOXA-48-like, blaNDM-1 and blaIMP described previously [34,35,36,37,38,39,40,41]. DNA templates were used by PCR analysis and the products were visualized in 1% agarose gel.
4.5. Molecular Typing of PFGE and Computer Fingerprint Analysis
After digestion with Xba I and Apa I endonucleases, the DNA was separated using a CHEF-DR II system (BioRad, Birmingham, UK) [42,43]. The Salmonella serotype Braenderup strain (H9812) was included in each PFGE gel as an internal control. Computer analysis of PFGE profiles was done using the Gel Compar II software, v.6.6.11 (Applied Maths, Inc.; Sint-Martens-Latem, Belgium) after visual inspection using the criteria of Tenover [44]. The Dice coefficients were calculated and were then transformed into an agglomerative cluster by the unweighted pair group method with arithmetic average (UPGMA).
5. Conclusions
Our results highlight the potential threat of extended-spectrum beta-lactamase resistance gene transmission in raw and treated hospital wastewater, as well as the risk of dissemination to environmental reservoirs such as rivers and other water bodies. The wide heterogeneity of PFGE pathways shown by E. coli strains may favor the selection of several genotypes, mainly those that showed resistance to the antibiotics tested. The clonality shown in A. haemolyticus strains isolated from treated wastewater suggests that this bacterium has a genetic potential to survive hospital WWTP treatment systems. Surveillance of antimicrobial resistance through hospital wastewater is an important tool for early detection of clonal clusters of clinically important bacteria with potential for dissemination.
Author Contributions
Conceptualization, C.M.A.-A., M.E.V.-M., M.G.-L. and P.S.-H.; Project administration C.M.A.-A. and M.E.V.-M., methodology, M.E.V.-M., M.G.-L., M.B.-d.-V., B.A.C.-Q., P.C.-J. and A.S.-G.; writing—original draft preparation, C.M.A.-A., M.E.V.-M. and M.G.-L.; writing—review and editing, C.M.A.-A., M.E.V.-M., M.G.-L., M.B.-d.-V., P.C.-J., A.P.-d.-L. and A.S.-G.; funding acquisition, C.M.A.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed by FOSISS-CONACYT-2017, number 290618; M.G.-L. is a PhD student with a scholarship from CONACYT.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. Ethical approval was obtained from the Ethic Committee of the Instituto Nacional de Salud Pública (CI: 1164).
Acknowledgments
We thank the administrative staff of the Research Center of Infectious Diseases at the National Institute of Public Health (Instituto Nacional de Salud Pública—INSP) for their efforts and valuable support, biologist Mario Sánchez-Vargas for his support in the collection of samples, the nurse Bertha García Pineda of the Instituto Nacional de Cancerología for her support in obtaining the samples, and QFB Yvonne Guadalupe Villalobos Zapata, QC Norma Irene López García and QFB Veronica Esteban Kenel of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán for their technical support in the use of MALDI-TOF MS® and PCR.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- World Health Organization Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 5 May 2020).
- World Health Organization. WHO Global Action Plan on Antimicrobial Resistance. Microbe Mag. 2015, 10, 354–355. [Google Scholar] [CrossRef]
- World Health Organization. WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR) WHO List of Critically Important Antimicrobials (CIA). In Report of the 7th Meeting; WHO: Geneva, Switzerland, 2018; ISBN 978-92-4-151552-8. [Google Scholar]
- Silva, J.; Aguilar, C.; Ayala, G.; Estrada, M.A.; Garza-Ramos, U.; Lara-Lemus, R.; Ledezma, L. TLA-1: A New Plasmid-Mediated Extended-Spectrum β-Lactamase from Escherichia Coli. Antimicrob. Agents Chemother. 2000, 44, 997–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coque, T.M.; Baquero, F.; Canton, R. Increasing Prevalence of ESBL- Producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 19044. [Google Scholar] [CrossRef] [PubMed]
- Hocquet, D.; Muller, A.; Bertrand, X. What Happens in Hospitals Does Not Stay in Hospitals: Antibiotic-Resistant Bacteria in Hospital Wastewater Systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Havenga, B.; Ndlovu, T.; Clements, T.; Reyneke, B.; Waso, M.; Khan, W. Exploring the Antimicrobial Resistance Profiles of WHO Critical Priority List Bacterial Strains. BMC Microbiol. 2019, 19, 303. [Google Scholar] [CrossRef] [Green Version]
- Daoud, Z.; Farah, J.; Sokhn, E.S.; El Kfoury, K.; Dahdouh, E.; Masri, K.; Afif, C.; Abdel-Massih, R.M.; Matar, G.M. Multidrug-Resistant Enterobacteriaceae in Lebanese Hospital Wastewater: Implication in the One Health Concept. Microb. Drug Resist. 2018, 24, 166–174. [Google Scholar] [CrossRef]
- Garza-González, E.; Franco-Cendejas, R.; Morfín-Otero, R.; Echaniz-Aviles, G.; Rojas-Larios, F.; Bocanegra-Ibarias, P.; Flores-Treviño, S.; Ponce-De-León, A.; Rodríguez-Noriega, E.; Alavez-Ramírez, N.; et al. The Evolution of Antimicrobial Resistance in Mexico during the Last Decade: Results from the INVIFAR Group. Microb. Drug Resist. 2020, 26, 1372–1382. [Google Scholar] [CrossRef]
- Ponce de León Rosales, S. Plan Universitario de Control de La Resistencia Antimicrobiana; Universidad Nacional Autónoma de México: Mexico City, Mexico, 2018; Volume 53, ISBN 9788578110796. [Google Scholar]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [Green Version]
- Gardy, J.L.; Loman, N.J. Towards a Genomics-Informed, Real-Time, Global Pathogen Surveillance System. Nat. Rev. Genet. 2018, 19, 9–20. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Woolhouse, M.E.J. Using Sewage for Surveillance of Antimicrobial Resistance. Science 2020, 367, 630–632. [Google Scholar] [CrossRef]
- Manaia, C.M.; Rocha, J.; Scaccia, N.; Marano, R.; Radu, E.; Biancullo, F.; Cerqueira, F.; Fortunato, G.; Iakovides, I.C.; Zammit, I.; et al. Antibiotic Resistance in Wastewater Treatment Plants: Tackling the Black Box. Environ. Int. 2018, 115, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Gumede, S.N.; Abia, A.L.K.; Amoako, D.G.; Essack, S.Y. Analysis of Wastewater Reveals the Spread of Diverse Extended-Spectrum β-Lactamase-Producing e. Coli Strains in Umgungundlovu District, South Africa. Antibiotics 2021, 10, 860. [Google Scholar] [CrossRef] [PubMed]
- Verburg, I.; García-Cobos, S.; Leal, L.H.; Waar, K.; Friedrich, A.W.; Schmitt, H. Abundance and Antimicrobial Resistance of Three Bacterial Species along a Complete Wastewater Pathway. Microorganisms 2019, 7, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamba, M.; Ahammad, S.Z. Sewage Treatment Effluents in Delhi: A Key Contributor of Β-Lactam Resistant Bacteria and Genes to the Environment. Chemosphere 2017, 188, 249–256. [Google Scholar] [CrossRef]
- Bréchet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater Treatment Plants Release Large Amounts of Extended-Spectrum β-Lactamase-Producing Escherichia Coli into the Environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [CrossRef]
- Bardhan, T.; Chakraborty, M.; Bhattacharjee, B. Prevalence of Colistin-Resistant, Carbapenem-Hydrolyzing Proteobacteria in Hospital Water Bodies and out-Falls of West Bengal, India. Int. J. Environ. Res. Public Health 2020, 17, 1007. [Google Scholar] [CrossRef] [Green Version]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [Green Version]
- Barraud, O.; Casellas, M.; Dagot, C.; Ploy, M.C. An Antibiotic-Resistant Class 3 Integron in an Enterobacter Cloacae Isolate from Hospital Effluent. Clin. Microbiol. Infect. 2013, 19, E306–E308. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, P.; Li, J.; Hu, X.; Lin, Y.; Yang, L.; Qiu, S.; Ma, H.; Li, P.; Song, H. An NDM-1-Producing Acinetobacter Towneri Isolate from Hospital Sewage in China. Infect. Drug Resist. 2020, 13, 1105–1110. [Google Scholar] [CrossRef] [Green Version]
- Zong, Z.; Zhang, X. BlaNDM-1-Carrying Acinetobacter Johnsonii Detected in Hospital Sewage. J. Antimicrob. Chemother. 2013, 68, 1007–1010. [Google Scholar] [CrossRef] [Green Version]
- Hrenovic, J.; Goic-Barisic, I.; Kazazic, S.; Kovacic, A.; Ganjto, M.; Tonkic, M. Carbapenem-Resistant Isolates of Acinetobacter Baumannii in a Municipal Wastewater Treatment Plant, Croatia, 2014. Eurosurveillance 2016, 21, 30195. [Google Scholar] [CrossRef]
- Xu, G.; Jiang, Y.; An, W.; Wang, H.; Zhang, X. Emergence of KPC-2-Producing Escherichia Coli Isolates in an Urban River in Harbin, China. World J. Microbiol. Biotechnol. 2015, 31, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [Green Version]
- Kutilova, I.; Medvecky, M.; Leekitcharoenphon, P.; Munk, P.; Masarikova, M.; Davidova-Gerzova, L.; Jamborova, I.; Bortolaia, V.; Pamp, S.J.; Dolejska, M. Extended-Spectrum Beta-Lactamase-Producing Escherichia Coli and Antimicrobial Resistance in Municipal and Hospital Wastewaters in Czech Republic: Culture-Based and Metagenomic Approaches. Environ. Res. 2021, 193, 110487. [Google Scholar] [CrossRef] [PubMed]
- Zumaya-Estrada, F.A.; Ponce-De-león-Garduño, A.; Ortiz-Brizuela, E.; Tinoco-Favila, J.C.; Cornejo-Juárez, P.; Vilar-Compte, D.; Sassoé-González, A.; Saturno-Hernandez, P.J.; Alpuche-Aranda, C.M. Point Prevalence Survey of Antimicrobial Use in Four Tertiary Care Hospitals in Mexico. Infect. Drug Resist. 2021, 14, 4553–4566. [Google Scholar] [CrossRef]
- Galvin, S.; Boyle, F.; Hickey, P.; Vellinga, A.; Morris, D.; Cormican, M. Enumeration and Characterization of Antimicrobial-Resistant Escherichia Coli Bacteria in Effluent from Municipal, Hospital, and Secondary Treatment Facility Sources. Appl. Environ. Microbiol. 2010, 76, 4772–4779. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Feng, Y.; Lü, X.; McNally, A.; Zong, Z. Remarkable Diversity of Escherichia Coli Carrying Mcr-1 from Hospital Sewage with the Identification of Two New Mcr-1 Variants. Front. Microbiol. 2017, 8, 2094. [Google Scholar] [CrossRef] [PubMed]
- Kovacic, A.; Music, M.S.; Dekic, S.; Tonkic, M.; Novak, A.; Rubic, Z.; Hrenovic, J.; Goic-Barisic, I. Transmission and Survival of Carbapenem-Resistant Acinetobacter Baumannii Outside Hospital Setting. Int. Microbiol. 2017, 20, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Qiu, S.; Wang, Y.; Qi, L.; Hao, R.; Liu, X.; Shi, Y.; Hu, X.; An, D.; Li, Z.; et al. Higher Isolation of NDM-1 Producing Acinetobacter Baumannii from the Sewage of the Hospitals in Beijing. PLoS ONE 2013, 8, e64857. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021; ISBN 978-1-68440-033-1. [Google Scholar]
- Cornejo-Juárez, P.; Pérez-Jiménez, C.; Silva-Sánchez, J.; Velázquez-Acosta, C.; González-Lara, F.; Reyna-Flores, F.; Sánchez-Pérez, A.; Volkow-Fernández, P. Molecular Analysis and Risk Factors for Escherichia Coli Producing Extended-Spectrum β-Lactamase Bloodstream Infection in Hematological Malignancies. PLoS ONE 2012, 7, e35780. [Google Scholar] [CrossRef]
- Rayamajhi, N.; Kang, S.G.; Lee, D.Y.; Kang, M.L.; Lee, S.I.; Park, K.Y.; Lee, H.S.; Yoo, H.S. Characterization of TEM-, SHV- and AmpC-Type β-Lactamases from Cephalosporin-Resistant Enterobacteriaceae Isolated from Swine. Int. J. Food Microbiol. 2008, 124, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of Plasmid-Mediated AmpC β-Lactamase Genes in Clinical Isolates by Using Multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weldhagen, G.F.; Prinsloo, A. Molecular Detection of GES-2 Extended Spectrum β-Lactamase Producing Pseudomonas Aeruginosa in Pretoria, South Africa. Int. J. Antimicrob. Agents 2004, 24, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Dogonchi, A.A.; Ghaemi, E.A.; Ardebili, A.; Yazdansetad, S.; Pournajaf, A. Metallo-β-lactamase-mediated Resistance among Clinical Carbapenem-resistant Pseudomonas Aeruginosa Isolates in Northern Iran: A Potential Threat to Clinical Therapeutics. Tzu Chi Med. J. 2018, 30, 90–96. [Google Scholar] [CrossRef]
- Aubron, C.; Poirel, L.; Ash, R.J.; Nordmann, P. Carbapenemase-Producing Enterobacteriaceae, U.S. Rivers. Emerg. Infect. Dis. 2005, 11, 260–264. [Google Scholar] [CrossRef]
- Bradford, P.A.; Bratu, S.; Urban, C.; Visalli, M.; Mariano, N.; Landman, D.; Rahal, J.J.; Brooks, S.; Cebular, S.; Quale, J. Emergence of Carbapenem-Resistant Klebsiella Species Possessing the Class A Carbapenem-Hydrolyzing KPC-2 and Inhibitor-Resistant TEM-30 β-Lactamases in New York City. Clin. Infect. Dis. 2004, 39, 55–60. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Zhou, H.; Li, H.; Gao, Y.; Lu, Z.; Hu, K.; Xu, B. Optimization of Pulse-Field Gel Electrophoresis for Subtyping of Klebsiella Pneumoniae. Int. J. Environ. Res. Public Health 2013, 10, 2720–2731. [Google Scholar] [CrossRef] [Green Version]
- Durmaz, R.; Otlu, B.; Koksal, F.; Hosoglu, S.; Ozturk, R.; Ersoy, Y.; Aktas, E.; Gursoy, N.C.; Caliskan, A. The Optimization of a Rapid Pulsed-Field Gel Electrophoresis Protocol for the Typing of Acinetobacter Baumannii, Escherichia Coli and Klebsiella Spp. Jpn. J. Infect. Dis. 2009, 62, 372–377. [Google Scholar]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed- Field Gel Electrophoresis: Criteria for Bacterial Strain Typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).