Inactivation of a New Potassium Channel Increases Rifampicin Resistance and Induces Collateral Sensitivity to Hydrophilic Antibiotics in Mycobacterium smegmatis
Abstract
1. Introduction
2. Results
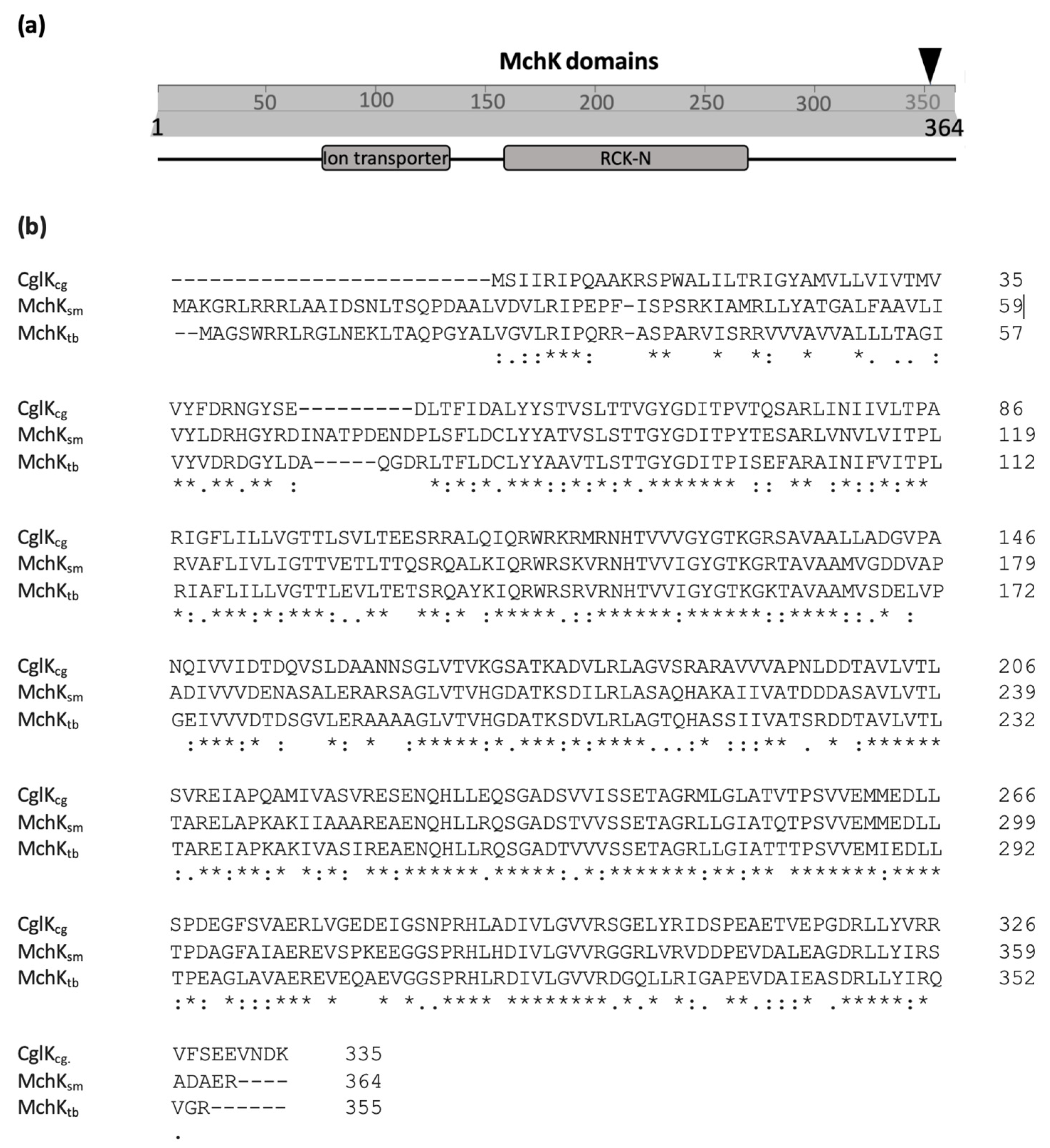
2.1. Identification of the mchK Gene as a Modulator of Rifampicin Resistance
2.2. Role of the mchK Gene in Collateral Susceptibility of M. smegmatis
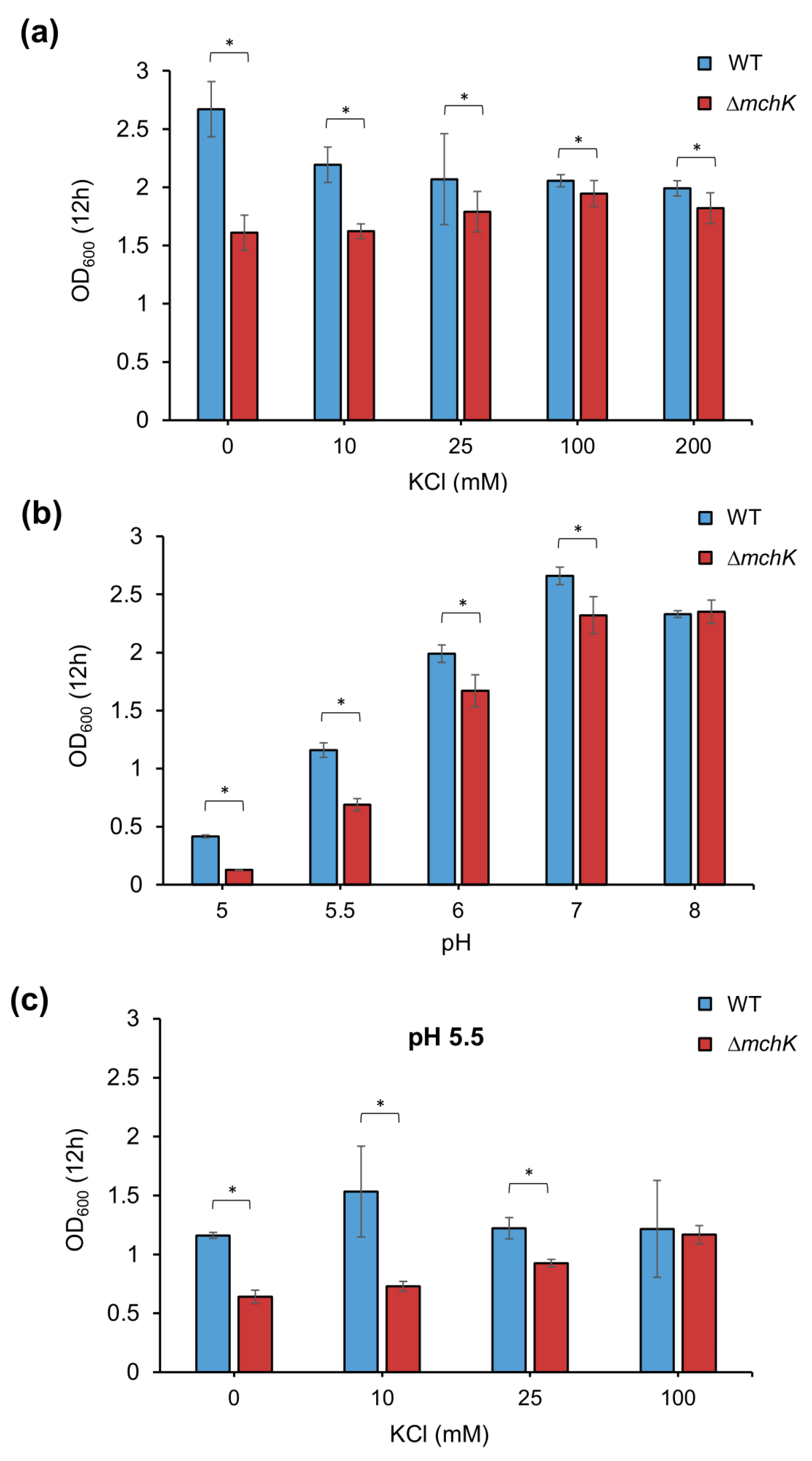
2.3. Effect of mchK Deletion on Growth and K+ Requirement
2.4. The mchK Gene and pH Homeostasis
2.5. The mchK Gene and Membrane Potential
3. Discussion
3.1. MchK as a Putative K+ Transport Protein
3.2. Potassium Transport via MchK Is Necessary for M smegmatis Growth at Acidic pH
3.3. Role of MchK in Charge Balance and Membrane Potential
3.4. MchK and Antibiotic Susceptibility
4. Materials and Methods
4.1. Bacteria, Media and Growth Conditions
4.2. Minimal Inhibitory Concentration (MIC) Determination and Survival Rates to Antibiotics
4.3. Screening of M. smegmatis φMycoMarT7 Insertion Library and Identification of the Insertion Site
4.4. Generation of an In-Frame mchK Knockout Mutant Strain
4.5. Complementation of the ΔmchK Mutant Strain
4.6. Membrane Potential Assay
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2020. 2020. Available online: https://www.who.int/publications-detail-redirect/9789240013131 (accessed on 10 December 2020).
- Nguyen, L.; Thompson, C.J. Foundations of antibiotic resistance in bacterial physiology: The mycobacterial paradigm. Trends Microbiol. 2006, 14, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A.; Imboden, P.; Marchesi, F.; Matter, L.; Schopfer, K.; Bodmer, T.; Lowrie, D.; Colston, M.J.; Cole, S. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 1993, 341, 647–650. [Google Scholar] [CrossRef]
- Riska, P.F.; Jacobs, W.R.; Alland, D. Molecular determinants of drug resistance in tuberculosis. Int. J. Tuberc. Lung Dis. 2000, 4, S4–S10. [Google Scholar] [PubMed]
- Zhang, Y.; Yew, W.W. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 2009, 13, 1320–1330. [Google Scholar] [PubMed]
- Ohno, H.; Koga, H.; Kohno, S.; Higashiyama, Y.; Miyazaki, Y.; Ogawa, K.; Yanagihara, K.; Yamamoto, Y.; Noda, T.; Miyamoto, J.; et al. Evaluation for rapid detection of rifampicin-resistant mycobacterium tuberculosis by polymerase chain reaction-single strand conformation polymorphism. Kekkaku 1994, 69, 773–778. [Google Scholar]
- Lelovic, N.; Mitachi, K.; Yang, J.; Lemieux, M.R.; Ji, Y.; Kurosu, M. Application of Mycobacterium smegmatis as a surrogate to evaluate drug leads against Mycobacterium tuberculosis. J. Antibiot. 2020, 73, 780–789. [Google Scholar] [CrossRef]
- Castañeda-García, A.; Do, T.T.; Blázquez, J. The K+ uptake regulator TrkA controls membrane potential, pH homeostasis and multidrug susceptibility in Mycobacterium smegmatis. J. Antimicrob. Chemother. 2011, 66, 1489–1498. [Google Scholar] [CrossRef]
- Beagle, S.D.; Lockless, S.W. Unappreciated Roles for K+ Channels in Bacterial Physiology. Trends Microbiol. 2021, 29, 942–950. [Google Scholar] [CrossRef]
- Follmann, M.; Becker, M.; Ochrombel, I.; Ott, V.; Krämer, R.; Marin, K. Potassium transport in Corynebacterium glutamicum is facilitated by the putative channel protein CglK, which is essential for pH homeostasis and growth at acidic pH. J. Bacteriol. 2009, 191, 2944–2952. [Google Scholar] [CrossRef]
- Choe, S. Potassium channel structures. Nat. Rev. Neurosci. 2002, 3, 115–121. [Google Scholar] [CrossRef]
- Stautz, J.; Hellmich, Y.; Fuss, M.F.; Silberberg, J.M.; Devlin, J.R.; Stockbridge, R.B.; Hänelt, I. Molecular Mechanisms for Bacterial Potassium Homeostasis. J. Mol. Biol. 2021, 433, 166968. [Google Scholar] [CrossRef] [PubMed]
- Kashket, E.R.; Barker, S.L. Effects of potassium ions on the electrical and pH gradients across the membrane of Streptococcus lactis cells. J. Bacteriol. 1977, 130, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Bakker, E.P.; Mangerich, W.E. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J. Bacteriol. 1981, 147, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Resnick, M.; Schuldiner, S.; Bercovier, H. Bacterial membrane potential analyzed by spectrofluorocytometry. Curr. Microbiol. 1985, 12, 183–185. [Google Scholar] [CrossRef]
- Booth, I.R. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 1985, 49, 359–378. [Google Scholar] [CrossRef]
- Epstein, W. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 2003, 75, 293–320. [Google Scholar]
- Dinnbier, U.; Limpinsel, E.; Schmid, R.; Bakker, E.P. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 1988, 150, 348–357. [Google Scholar] [CrossRef]
- Epstein, W. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Rev. 1986, 2, 73–78. [Google Scholar] [CrossRef]
- Kroll, R.G.; Booth, I.R. The role of potassium transport in the generation of a pH gradient in Escherichia coli. Biochem. J. 1981, 198, 691–698. [Google Scholar] [CrossRef]
- Dashper, S.G.; Reynolds, E.C. pH regulation by Streptococcus mutans. J. Dent. Res. 1992, 71, 1159–1165. [Google Scholar] [CrossRef]
- Mitchell, P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol. Rev. Camb. Philos Soc. 1966, 41, 445–502. [Google Scholar] [CrossRef] [PubMed]
- Clare, J.J. Targeting ion channels for drug discovery. Discov. Med. 2010, 9, 253–260. [Google Scholar] [PubMed]
- Girgis, H.S.; Hottes, A.K.; Tavazoie, S. Genetic Architecture of Intrinsic Antibiotic Susceptibility. PLoS ONE 2009, 4, e5629. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Hinz, A.; Bauerle, E.; Angermeyer, A.; Juhaszova, K.; Kaneko, Y.; Singh, P.K.; Manoil, C. Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. USA 2009, 106, 14570–14575. [Google Scholar] [CrossRef]
- Gries, C.M.; Bose, J.L.; Nuxoll, A.S.; Fey, P.D.; Bayles, K.W. The Ktr potassium transport system in Staphylococcus aureus and its role in cell physiology, antimicrobial resistance and pathogenesis. Mol. Microbiol. 2013, 89, 760–773. [Google Scholar] [CrossRef]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Harms, C.; Domoto, Y.; Celik, C.; Rahe, E.; Stumpe, S.; Schmid, R.; Nakamura, T.; Bakker, E.P. Identification of the ABC protein SapD as the subunit that confers ATP dependence to the K+-uptake systems TrkH and TrkG from Escherichia coli K-12. Microbiology 2001, 147, 2991–3003. [Google Scholar] [CrossRef]
- Su, C.-C.; Li, M.; Gu, R.; Takatsuka, Y.; McDermott, G.; Nikaido, H.; Yu, E.W. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 2006, 188, 7290–7296. [Google Scholar] [CrossRef]
- Eicher, T.; Brandstätter, L.; Pos, K.M. Structural and functional aspects of the multidrug efflux pump AcrB. Biol. Chem. 2009, 390, 693–699. [Google Scholar] [CrossRef]
- Steel, H.C.; Matlola, N.M.; Anderson, R. Inhibition of potassium transport and growth of mycobacteria exposed to clofazimine and B669 is associated with a calcium-independent increase in microbial phospholipase A2 activity. J. Antimicrob. Chemother. 1999, 44, 209–216. [Google Scholar] [CrossRef][Green Version]
- Parish, T.; Stoker, N.G. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 2000, 146, 1969–1975. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.K.; de la Cruz, V.F.; Fuerst, T.R.; Burlein, J.E.; Benson, L.A.; Bennett, L.T.; Bansal, G.P.; Young, J.F.; Lee, M.H.; Hatfull, G.F.; et al. New use of BCG for recombinant vaccines. Nature 1991, 351, 456–460. [Google Scholar] [CrossRef] [PubMed]


| MICs (µg/mL) | ||||
|---|---|---|---|---|
| Antibiotic | Molecular Weight | LogS a | mc2 155 | ∆mchK |
| novobiocin | 612 | −4.80 | 32 | 64 |
| rifampicin | 822 | −4.09 | 2 | 32 |
| ofloxacin | 361 | −2.40 | 0.32 | 0.32 |
| ciprofloxacin | 331 | −2.39 | 0.32 | 0.32 |
| ethionamid | 166 | −2.30 | 64 | 8 |
| ethambutol | 204 | −1.43 | 2 | 2 |
| amikacin | 585 | −1.07 | 0.8 | 0.4 |
| streptomycin | 581 | −0.96 | 0.8 | 0.2 |
| kanamycin | 484 | −0.72 | 3.2 | 0.8 |
| isoniazid | 137 | −0.59 | 128 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Do, T.T.; Rodríguez-Beltran, J.; Cebrián-Sastre, E.; Rodríguez-Rojas, A.; Castañeda-García, A.; Blázquez, J. Inactivation of a New Potassium Channel Increases Rifampicin Resistance and Induces Collateral Sensitivity to Hydrophilic Antibiotics in Mycobacterium smegmatis. Antibiotics 2022, 11, 509. https://doi.org/10.3390/antibiotics11040509
Do TT, Rodríguez-Beltran J, Cebrián-Sastre E, Rodríguez-Rojas A, Castañeda-García A, Blázquez J. Inactivation of a New Potassium Channel Increases Rifampicin Resistance and Induces Collateral Sensitivity to Hydrophilic Antibiotics in Mycobacterium smegmatis. Antibiotics. 2022; 11(4):509. https://doi.org/10.3390/antibiotics11040509
Chicago/Turabian StyleDo, Thi Thuy, Jerónimo Rodríguez-Beltran, Esmeralda Cebrián-Sastre, Alexandro Rodríguez-Rojas, Alfredo Castañeda-García, and Jesús Blázquez. 2022. "Inactivation of a New Potassium Channel Increases Rifampicin Resistance and Induces Collateral Sensitivity to Hydrophilic Antibiotics in Mycobacterium smegmatis" Antibiotics 11, no. 4: 509. https://doi.org/10.3390/antibiotics11040509
APA StyleDo, T. T., Rodríguez-Beltran, J., Cebrián-Sastre, E., Rodríguez-Rojas, A., Castañeda-García, A., & Blázquez, J. (2022). Inactivation of a New Potassium Channel Increases Rifampicin Resistance and Induces Collateral Sensitivity to Hydrophilic Antibiotics in Mycobacterium smegmatis. Antibiotics, 11(4), 509. https://doi.org/10.3390/antibiotics11040509







