Abstract
Acute and chronic lower airway disease still represent a major cause of morbidity and mortality on a global scale. With the steady rise of multidrug-resistant respiratory pathogens, such as Pseudomonas aeruginosa and Klebsiella pneumoniae, we are rapidly approaching the advent of a post-antibiotic era. In addition, potentially detrimental novel variants of respiratory viruses continuously emerge with the most prominent recent example being severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). To this end, alternative preventive and therapeutic intervention strategies will be critical to combat airway infections in the future. Chronic respiratory diseases are associated with alterations in the lung and gut microbiome, which is thought to contribute to disease progression and increased susceptibility to infection with respiratory pathogens. In this review we will focus on how modulating and harnessing the microbiome may pose a novel strategy to prevent and treat pulmonary infections as well as chronic respiratory disease.
1. Introduction
Acute and chronic lower airway diseases are among the leading causes of death worldwide and the third most common cause of death in the United States. In 2021, infectious and chronic respiratory diseases accounted for more than 550,000 deaths in the US [1]. SARS-CoV-2 alone was the 3rd leading cause of death, outpacing all other causes with the exception of heart disease and malignant neoplasms [1]. Direct medical expenditures for the treatment of lower respiratory tract disease as well as indirect costs from the loss of productivity represent a considerable socioeconomic burden [2,3]. These conditions are becoming increasingly challenging to manage due to the continuous emergence of multi-drug resistant (MDR) bacteria and novel viral variants that promote infection and exacerbation of chronic lung disease. In the absence of effective countermeasures, it is estimated that infection with MDR strains will be responsible for 317,000 deaths each year in the US by 2050, a more than 10-fold increase to current numbers, underlining the urgent need for alternative prevention and intervention strategies [4,5]. The human body harbors trillions of microorganisms and comprises an intricate network of bacteria, archaea, protozoa and fungi as well as bacteriophages and eukaryotic viruses. It is estimated that the number of microorganisms that colonize the human body is at least equal to the number of somatic cells and that 500–1000 bacterial species inhabit our mucosal surfaces and the skin at any given time [6,7,8,9]. These microbes continuously interact with each other and with the human host, and this tightly regulated interplay is essential for the development and priming of the immune system and the maintenance of homeostasis [10,11,12]. When this symbiosis is perturbed due to alterations in the diversity and composition of the microbiome, the resulting dysbiosis can predispose to or exacerbate disease locally and at distal body sites. Dysbiosis can be induced by a variety of environmental factors such as treatment with antibiotics, diet and lifestyle or can be a consequence of chronic inflammation, infections or metabolic disorders [9,13,14,15,16,17,18,19]. Advances in culture-independent high-throughput bacterial sequencing approaches that enable large-scale taxonomic profiling of bacterial communities in patients and animal models, in combination with immunological and multi-omics datasets as well as mechanistic studies in germ-free rodent models, strongly suggest that aberrant microbiome alterations are a key driver of disease. A lot of these insights were gained in the course of the Human Microbiome Project (HMP), a 10-year interdisciplinary worldwide effort to characterize the composition of the healthy human microbiome at different body sites, analyze the relationship between microbiome changes and disease, and provide resources and novel technologies to broadly facilitate these studies in the scientific community [8,20,21,22]. Importantly, these studies also revealed a bidirectional cross-talk between the gut and lung microbiota, and growing experimental and clinical evidence show that changes in the intestinal microbiota can influence clinical outcomes of respiratory infections and chronic lung disease [23,24]. Since our understanding of other members of the human microbiome is still limited, we will briefly summarize implications of lung and intestinal commensal bacteria in lower respiratory tract infections and chronic lung disease and review how manipulation of the local and distal microbiota may aid in preventing disease while also serving as a source of novel drugs or drug targets for treatment.
2. The Lung and Gut Microbiome and Their Implications in Respiratory Disease
2.1. The Gut Microbiota
The gut microbiome performs many essential functions to maintain host homeostasis, including the breakdown of complex carbohydrates, synthesis of vitamins, maintenance of mucosal barrier integrity and protection against pathogens [25]. In addition, intestinal bacteria play an important role in the development and priming of local and systemic immunity [11]. The human gastrointestinal (GI) tract harbors more than 1014 bacteria and it is estimated that the genomic information contained in these bacteria outnumbers the genetic information contained in the human genome by at least 100-fold [26,27]. The healthy intestine contains >1000 bacterial species and despite a high degree of interindividual variability, can be reduced to a core microbiome that is dominated by taxa derived from the phyla Firmicutes, Bacteroidetes and to a lesser extent Proteobacteria, Actinobacteria and Verrucomicrobia [28,29,30,31,32]. As a consequence of oxygen availability, pH value and the presence of antimicrobial peptides (AMP) and bile acids, the composition and density of the microbiome varies greatly from the proximal to the distal GI tract. It gradually increases from ~102 colony forming units/gram [CFU/g] luminal content in the acidic, sparsely populated bile acid- and AMP-rich environment of the proximal small intestine and reaches its highest density in the colon (~1011 CFU/g) [30]. Due to the higher availability of oxygen and the selective pressure provided by natural antimicrobials, the small intestine is mainly populated by fast-growing facultative anaerobic bacteria of the Lactobacillaceae and Enterococcoceae families, while the colon is dominated by fermentative anaerobes of the Bacteroidaceae and Clostridia families [30]. Colonization of the GI tract is induced during birth upon contact with the vaginal microbiota and once established remains relatively stable over time [9,30]. However, diet, lifestyle, and disease status can profoundly alter the microbiota and induce a dysbiotic state [13,14,15,16,19,33]. Microbiome-wide association studies (MWAS) in combination with mechanistic studies in germ-free or microbiota-depleted animals established the causal relationship between intestinal dysbiosis and the pathogenesis of a wide array of human diseases including not only gastrointestinal conditions but also systemic manifestations such as obesity, type 2 diabetes as well as allergic asthma and respiratory infections [34,35].
2.2. The Lung Microbiota
While it is well established that the upper respiratory tract (URT) of healthy individuals is continuously colonized by microbes, due to technical limitations, the lung environment was historically considered to be sterile. Recent advances in culture-independent bacterial sequencing approaches have now revealed that the lower respiratory tract (LRT) harbors a unique microbiome that plays a key role in promoting pulmonary homeostasis and may facilitate the priming of local immune cell populations [36]. In addition, the respiratory tract microbiome has emerged as a critical modulator of immune responses to respiratory infections and the pathogenesis of chronic lung disease [37,38,39,40]. Compared to the large intestine, the LRT colonization density in healthy individuals is relatively low, with 103–105 CFU/g lung tissue [41]. Under homeostatic conditions, the most abundant bacterial phyla in the human lung are Bacteroidetes (45–50%, genus Prevotella), Firmicutes (30–35%, genera Streptococcus, Veillonella) and to a lesser extent Proteobacteria (10–15%, genera Haemophilus, Neisseria), Actinobacteria (5%, genus Corynebacterium) and Fusobacteria (5%, genus Fusobacteria) [41,42,43,44,45,46,47,48,49]. The lung microbiota is transient and maintained by continuous translocation of microorganisms from the URT and subsequent clearance by innate lung immune mechanisms, rather than local expansion of lung-resident bacteria [36,44,46]. This balance between migration and elimination is often perturbed during respiratory disease, which leads to overgrowth of bacteria with a competitive advantage and consequently a loss of microbial diversity. Structural alterations of the small airways and excessive mucus production are a hallmark of COPD, asthma and cystic fibrosis (CF). The resulting airway obstruction disrupts mucociliary clearance and the excess mucus may additionally support colonization with potential pathogens such as Pseudomonas aeruginosa, which is associated with increased mortality in CF and COPD [50,51,52,53,54,55]. In addition, defective clearance of bacteria by alveolar macrophages and/or airway neutrophils is observed in COPD [56,57,58,59,60], asthma [61,62], CF [63], idiopathic pulmonary fibrosis (IPF) [64] and after respiratory viral infections [65,66,67]. In accordance, patients suffering from chronic lung diseases exhibit elevated bacterial loads in their lungs, with increased abundance of potentially pathogenic proteobacteria including Haemophilus, Moraxella and Pseudomonas spp. [36,40,68,69,70]. Altogether, defects in mucociliary clearance as well as airway macrophage and neutrophil dysfunction may further contribute to disease progression and exacerbation by predisposing patients to bacterial infections.
2.3. The Gut-Lung Axis in Respiratory Disease
Increasing experimental and clinical evidence suggests that the gut microbiota and the lung are engaged in a continuous bidirectional cross-talk, termed the gut-lung axis and that dysbiosis at either site can contribute to the development and progression of distal diseases (Figure 1) [71,72]. Using animal models, it was shown that this dialogue is mainly facilitated by structural bacterial ligands, such as lipopolysaccharide (LPS), that bind and activate pattern recognition receptors (PRR) on host cells [38,73,74,75]. Microbiome-derived metabolites also play a critical role in this process [65,76,77,78,79]. In addition, the migration of activated immune cells from the intestine to the lung was described to aid in the defense against pulmonary helminth and bacterial infections [80,81]. Importantly, these gut-lung protective mechanisms could be directly stimulated by the intestinal microbiota and were abrogated in germ-free and antibiotic-treated mice, resulting in increased susceptibility to a wide array of bacterial and viral respiratory infections as well as chronic respiratory diseases [24,65,75,76,81,82,83,84,85,86].
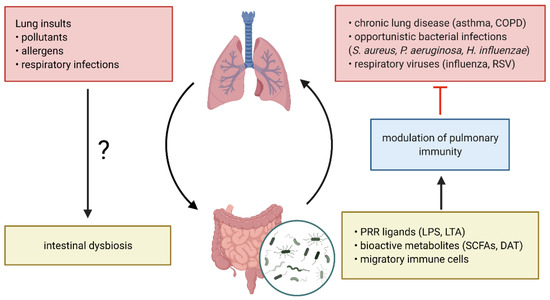
Figure 1.
The gut-lung axis in respiratory disease. The continuous cross-talk between the gut and lung is facilitated by structural bacterial pattern recognition receptor (PRR) ligands including bacterial cell wall components, gut commensal-derived metabolites and migratory immune cells. Bacterial PRR ligands and metabolites are released in the circulation and bind to their respective receptors on pulmonary immune and/or epithelial cells, thereby modulating immunity to respiratory pathogens during chronic lung disease. Pulmonary insults can induce intestinal dysbiosis, however the underlying mechanisms are not well understood. COPD, chronic obstructive pulmonary disease; RSV, respiratory syncytial virus; PRR, pattern recognition receptor; LPS, lipopolysaccharide; LTA, lipoteichoic acid; SCFAs, short-chain fatty acids; DAT, desaminotyrosine. Figure created with BioRender.com.
Up to 50% of inflammatory bowel disease (IBD) patients and one third of individuals with irritable bowel syndrome (IBS) exhibit pulmonary manifestations ranging from subclinical aberrations to chronic lung disease [84,87]. The intestinal symptoms in IBD precede the lung phenotype, indicating that impaired lung function may be a consequence of a dysbiotic gut [88]. Furthermore, reduced intestinal microbiota diversity following antibiotic treatment during the first months of life as well as low levels of LPS and the microbiota-derived short-chain fatty acid (SCFA) metabolites are associated with the development of childhood asthma [89,90,91,92,93,94,95,96]. In accordance, genetically modified mice that are unable to inactivate LPS exhibit reduced pulmonary type 2 immune responses in a mouse model of allergic airway inflammation. This effect was ablated in mice treated with antibiotics and could be restored upon intrarectal administration of LPS, indicating a protective role of intestinal-derived LPS in allergic asthma [97]. Recent evidence suggests that SCFAs, the end products of dietary fiber fermentation by colonic commensals, are key modulators of the gut-lung axis and exert potent anti-inflammatory functions both locally and systemically. Oral supplementation with dietary fibers, SCFAs or SCFA-producing bacteria ameliorated pulmonary inflammation in mouse models of allergic asthma, further underlining the important role of the gut-lung axis in shaping lung immunity [76,78,86,98,99,100,101]. Cigarette smoke and aging are major risk factors for COPD development and are associated with shifts in gut microbial communities towards a decrease in alpha diversity as well as an increase in the Firmicute/Bacteroidetes ratio, established markers of intestinal dysbiosis [40,102,103,104]. Similar to allergic airway inflammation, intake of fermentable dietary fiber was associated with improved lung function in COPD patients and ameliorated lung pathology and inflammation in mouse models of COPD [105,106]. Furthermore, transfer of fecal matter from COPD patients into mice induced lung inflammation and accelerated the development of COPD-like pathology in respective animal models, further establishing a link of the gut microbiome to chronic lung disease etiology [107]. Importantly, the gut-lung axis is bidirectional, and it is well established that chronic lower airway diseases are often accompanied by manifestations in the gastrointestinal tract. For example, asthma patients exhibit structural changes in their intestinal mucosa, and COPD is often accompanied by chronic gastrointestinal tract diseases, intestinal microbiome alterations and lower levels of SCFAs compared to healthy controls [82,107,108,109,110,111]. However, the mechanisms behind these changes are only beginning to be understood and whether the differences in the gut microbiota are a cause and/or a consequence of chronic respiratory disease is often hard to discern and needs to be further addressed.
Besides chronic disease, intestinal dysbiosis was linked to the development of acute lower respiratory tract infections in children and evidence from mouse models show that the gut microbiota plays a key role in the defense against pulmonary pathogens [112]. Accordingly, germ-free and antibiotic-treated mice exhibit increased susceptibility to influenza virus [71,72,75,79] and respiratory syncytial virus (RSV) infections [83] and worse outcomes following lung infections with Streptococcus pneumoniae [38,67,74,81,113], Klebsiella pneumoniae [38,74,85], Pseudomonas aeruginosa [114,115] and Staphylococcus aureus [116]. Importantly, the administration of PRR agonists or the transfer of fecal material from control mice was sufficient to restore pulmonary immunocompetence and reversed susceptibility to lung infections in dysbiotic mice [38,72,74,75,85]. Furthermore, supplementation with dietary fibers and the metabolites SCFAs or desaminotyrosine (DAT) conferred protection against viral respiratory infections [79,83,117]. On the contrary, infections with influenza virus induce transient changes in the intestinal microbiota composition as well as gastroenteritis-like symptoms in mice and humans which cannot be explained by intestinal tropism of the virus [65,118,119,120,121,122,123,124,125]. These changes in the intestinal microbiota following influenza infection further predisposed the host to secondary enteric infections in mouse models of co-infection [121,124]. Influenza virus-mediated intestinal dysbiosis was also shown to feed back on the lung, leading to decreased bactericidal activity of alveolar macrophages and predisposing to pneumococcal pneumonia [89]. Importantly, alveolar macrophage function could be restored by oral SCFA supplementation [65]. Shifts in the intestinal microbiota towards increased abundance of Bacteroidetes and a decrease in Firmicutes were observed in a mouse model of RSV infection and specific microbial profiles were linked to disease severity in RSV-infected children [122,126]. Intestinal manifestations are also frequently reported in individuals infected with SARS-CoV-2, however, as opposed to influenza virus, there is clear evidence that SARS-CoV-2 is able to productively infect gastrointestinal epithelial cells and thereby directly causes intestinal dysbiosis [127,128].
3. Microbiome-Based Prevention and Intervention Strategies in Respiratory Disease
Given the implications of the intestinal microbiome in respiratory diseases, modulating the composition or activity of the resident microbiome by supplementation with bacterial substrates (prebiotics), live bacteria (probiotics, fecal microbial transplantation) or inanimate bacterial preparations (postbiotics) has been proposed as a novel prevention and intervention strategy for infectious and chronic lung disease. In addition, the selective depletion of opportunistic pathogens by antibacterial monoclonal antibodies or bacteriophages has shown promising results in preventing bacterial pneumonia in at-risk patients and both will be further discussed below and are summarized in Figure 2.
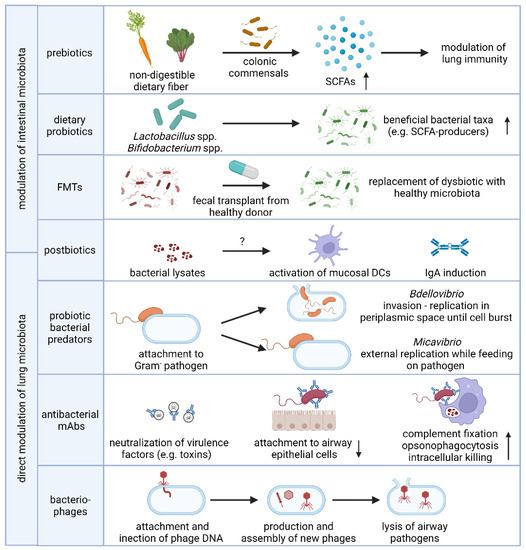
Figure 2.
Microbiome-based strategies to prevent and treat respiratory disease. Modulation of the composition and/or activity of the intestinal microbiota to promote lung immunity aids in the defense against respiratory bacterial infections and management of chronic lung disease. Intake of non-digestible dietary fiber drives the production of the bioactive SCFA metabolites by colonic commensal bacteria while the use of probiotics aims to modify the composition of the microbiome towards beneficial bacterial taxa, including SCFA-producers. The precise immunomodulatory mechanism of intranasally or orally administered postbiotics remain elusive but may be in part due to mucosal DC and IgA induction. In addition, the lung microbiota can be directly modulated by selectively eliminating bacterial pathogens using probiotic predatory bacteria, antibacterial human mAbs or strain-specific bacteriophages. Abbreviations: SCFAs, short-chain fatty acids; FMT, fecal microbiota transplant; DC, dendritic cell; IgA, immunoglobulin A; mAb, monoclonal antibody. Figure created with BioRender.com.
3.1. Supplementation with Prebiotics, Live Bacteria or Postbiotics
3.1.1. Prebiotics
According to the International Scientific Association of Probiotics and Prebiotics (ISAPP), a prebiotic is “a substrate that is selectively utilized by host microorganisms to confer a health benefit to the host” [129]. Beneficial effects conferred by prebiotics are associated with modulation of the composition and/or activity of the commensal microbiota and include the expansion of beneficial bacterial taxa as well as the production of anti-inflammatory bacterial metabolites. The most well-studied and recognized prebiotics are non-digestible dietary fibers such as fructo-oligosaccharides, galacto-oligosaccharides, inulin and pectin, which are complex carbohydrates that are metabolized to SCFAs by commensal bacteria in the colon via anaerobic fermentation. Epidemiological and preclinical data strongly suggests a causative relationship between dietary fiber intake and lung health [130,131]. The generic Western diet contains low amounts of fermentable fiber and the associated gut microbiota harbors decreased microbial diversity, lower abundances of SCFA-generating bacteria and is associated with reduced lung function [132,133,134]. In accordance, increased dietary fiber intake positively correlated with pulmonary function and is associated with an up to 50% reduction in respiratory-related death rate [133,135,136,137]. It was suggested that the observed beneficial effects may be at least partly due to the biological functions of SCFAs such as acetate, propionate and butyrate. SCFAs exert important local physiological functions by stimulating intestinal epithelial cell turnover, promoting intestinal barrier integrity and serving as a vital energy source for colonocytes [138]. Furthermore, SCFAs exhibit anti-inflammatory and immunomodulatory functions either locally or at peripheral sites through activation of the G protein-coupled receptors (GPCRs) on epithelial cells and/or immune cells [139,140,141]. Several studies using mouse models showed that SCFAs promote the development and differentiation of extrathymic regulatory T cells (Tregs) [100,142,143,144] and can restrain allergic airway inflammation by impairing dendritic cells to drive TH2 effector functions and suppressing group 2 innate lymphoid cell (ILC2) activity [76,78,86]. In addition, SCFAs can promote lung immunity by modulating immune cell hematopoiesis in the bone marrow [78,115,117]. Immunomodulatory mechanisms of SCFAs in animal models are described in more detail in Table 1. Propionate and acetate are mainly produced by members of the Bacteroidetes, and Actinobacteria phyla such as Bacteroides spp. and Bifidobacterium spp., respectively, which can in addition to SCFAs, also generate lactate during dietary fiber fermentation [145,146]. The mucin-degrading bacteria Akkermansia muciniphila (Verrucomicrobia phylum) also produces both propionate and acetate [147] and Firmicutes, particularly Faecalibacterium prausnitzii (clostridial cluster IV), Eubacterium rectale and Roseburia spp. are the main butyrate-producing bacteria in the human intestine [146,148]. Anaerostipes spp. (clostridial cluster XIVa) and other colonic acetate- and lactate-utilizers can additionally generate butyrate through cross-feeding interactions [148]. Humans and mice consuming a diet high in fiber exhibited an elevated ratio of beneficial SCFA-producing Bacteroidetes to Firmicutes in the gut, which corresponded to higher local and systemic SCFA concentrations [78,100].
Consumption of a high-fiber diet or SCFA supplementation can ameliorate airway inflammation and improve lung function in asthmatic subjects and was protective in animal models of allergic airway inflammation [76,78,86,98,100,101,149,150]. Similar observations were made in COPD patients and animal models of COPD where high fiber intake was associated with a significant increase in lung function and decrease in COPD development and progression [105,106,151,152,153,154,155]. Other studies demonstrated that dietary fiber and SCFA supplementation was protective in experimental infection models of influenza and RSV as well as bacterial pneumonia and secondary bacterial infections following viral infections [83,117,156,157,158]. Direct evidence that the protective effects observed stem from SCFAs was obtained in studies using SCFA receptor-deficient mice which are more susceptible to bacterial pneumonia as well as respiratory viral infections, and exhibit exacerbated allergic airway inflammation in asthma models. However, the precise molecular mechanisms on how SCFAs mediate their beneficial effects are not yet fully understood. Dietary intake of fermentable fibers to promote the growth and activity of SCFA-producing gut bacteria and thereby remotely modulate lung immunity is a relatively easy and promising strategy to limit the onset and progression of chronic airway disease and may also confer protection against respiratory bacterial and viral infections. Clinical trials are underway to assess the potential effect of soluble dietary intake of inulin in COPD patients [159], and supplementation of asthmatics with a prebiotic that selectively promotes the growth and development of Bifidobacteria [160].

Table 1.
Role of commensal-derived metabolites in mouse models of infections and chronic lung disease.
Table 1.
Role of commensal-derived metabolites in mouse models of infections and chronic lung disease.
| Metabolite | Disease Model | Immunomodulatory Effect | References |
|---|---|---|---|
| DAT | IAV |
| [79] |
| acetate | RSV |
| [83] |
| acetate | IAV + S. pneumoniae |
| [65] |
| acetate | S. pneumoniae |
| [161] |
| acetate | K. pneumoniae |
| [158] |
| butyrate | IAV |
| [117] |
| butyrate | heat exposure + IAV |
| [156] |
| butyrate | K. pneumoniae |
| [157] |
| acetate | HDM |
| [100] |
| butyrate acetate propionate | vancomycin + OVA, papain |
| [76] |
| butyrate | OVA |
| [98] |
| propionate | HDM |
| [78] |
| butyrate | IL-33 |
| [86] |
Abbreviations: DAT, desaminotyrosine; IAV, influenza A virus; RSV, respiratory syncytial virus; HDAC, histone deacetylase; HDM, house dust mite; OVA, ovalbumin; DC, dendritic cell; mLN, mediastinal lymph node; ILC2, group 2 innate lymphoid cell.
3.1.2. Fecal Microbiota Transplantation and Probiotics
Fecal microbiota transplantation (FMT) from healthy donors has been successfully used to treat patients with recurrent intestinal Clostridioides difficile infections and induce remission in ulcerative colitis patients [162]. These initial successes have sparked interest for the potential application of FMTs in extraintestinal diseases that are associated with gut dysbiosis including respiratory conditions. However, besides a proposed clinical trial administering FMTs to SARS-CoV-2 patients to potentially attenuate cytokine storm and pulmonary inflammation, the efficacy of FMTs for prevention or treatment of respiratory conditions has not yet been addressed in the clinic [163]. After fecal transplants contaminated with drug-resistant Escherichia coli resulted in the severe illness and death of clinical trial participants, safety concerns to enhance the donor screening process were voiced [164]. This incident and other safety issues have led efforts to focus on the development of defined and well-characterized formulations of fecal-derived live bacterial consortia or the use of known probiotic strains.
The ISAAP defines probiotics are “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” [165]. Probiotics are one of the most commonly consumed dietary supplements in the United States despite their efficacy being highly debated for many indications including respiratory diseases, and no probiotic has yet been approved as a live biotherapeutic agent [166]. However, favorable effects on the outcomes of respiratory infections and chronic airway disease were reported in some studies, mainly using animal models and in the context of upper respiratory tract infections in humans [167]. Due to their critical roles in immune maturation, their ability to generate bioactive metabolites and their critical role in the maintenance of intestinal barrier function and homeostasis, the most commonly used probiotics for preventive and therapeutic purposes are members of the Bifidobacterium (e.g., B. breve, B. animalis, B. longum) and Lactobacillus (e.g., L. rhamnosus GG, L. paracasei, L. casei, L. plantarum) genera, and to a lesser extent Streptococcus thermophilus and Enterococcus faecium [168]. Gut dysbiosis in early life characterized by reduced abundance and/or changes in in the composition of gut Bifidobacteria and Lactobacilli has been associated with allergic sensitization and the development of asthma [96,169]. Several studies suggest that preventive and therapeutic administration of probiotics may confer protection in animal models of allergic asthma. Mice that were supplemented with certain strains of Lactobacilli (e.g., L. rhamnosus GG, L. reuteri, L. johnsonii), Bifidobacteria (e.g., B. lactis, B. breve) or Enterococcus faecalis exhibited dampened allergic airway inflammation and airway hyperreactivity as well as decreased expression of pulmonary type 2 cytokines (IL-4, IL-5, IL-13), while anti-inflammatory cytokines such as TGF-β and regulatory T cells were induced [170,171,172,173,174,175]. Although these studies point to a potential protective role of probiotics, human studies testing the efficacy of supplementation with probiotics to prevent or treat asthma are so far inconclusive. Similar observations were made in cigarette smoke-induced COPD animal models where therapeutic and preventive dietary supplementation with L. rhamnosus or colonization with the commensal Parabacteroides goldsteinii ameliorated pulmonary inflammation and pathology [176,177]. However, there is no clinical evidence of a beneficial effect of probiotics on the onset or progression of COPD in humans.
The use of probiotics to prevent viral and bacterial lower respiratory tract infections yielded promising results in animal models. Oral supplementation or intranasal administration of the probiotics Lactobacilli or Bifidobacteria was protective in mouse models of influenza virus, RSV and mouse pneumonia virus infection [173,178,179,180]. Furthermore, dietary supplementation with L. casei, L. rhamnosus or B. longum enhanced lung clearance of P. aeruginosa, S. pneumoniae or K. pneumoniae, respectively, and ameliorated pulmonary inflammation [181,182,183]. Several clinical studies assessed whether dietary supplementation with probiotics leads to a reduction in the incidence and duration of acute respiratory tract infections. Meta-analyses showed a modest effect in reducing the duration and incidence of upper respiratory tract infections compared to placebo controls [184]. Efficacy of probiotics in specifically preventing lower respiratory tract infections has only been addressed in a few studies, including a clinical trial in high-risk ICU patients where daily oropharyngeal and gastric administration of L. rhamnosus significantly reduced the incidence of VAP compared to a placebo control [185]. In addition, COVID-19 patients orally receiving a multi-strain probiotic cocktail exhibited a decreased risk of developing respiratory failure compared to standard of care alone [186]. While these data are promising, further clinical evidence is needed to support the preventive or therapeutic use of probiotics in lower respiratory tract disease.
Besides the increasing the abundance of beneficial bacteria, dysbiosis can be counteracted by limiting the outgrowth of pathogenic bacteria. One potential approach to attack respiratory pathogens is the probiotic use of predatory bacteria from the genera Bdellovibrio spp. or Micavibrio spp. which specifically target and prey on other Gram-negative bacteria. Bdellovibrio attach to their prey, hydrolyze outer cell wall components and penetrate the periplasmic space, where they replicate and eventually burst the cell envelope of the host bacteria to start a new life cycle [187]. Bdellovibrio exhibit a broad host range, including major respiratory pathogens [115]. Importantly, B. bacteriovorus, the most extensively studied bacterial predator, was able to kill planktonic cultures as well as biofilm-embedded multidrug (MDR)- and extensively drug-resistant (XDR) clinical isolates of the opportunistic respiratory pathogens K. pneumoniae, E. coli, Acinetobacter baumannii, and Pseudomonas aeruginosa [188,189,190]. Another well-studied bacterial predator, Micavibrio aeruginosavorus, exhibits a more narrow host range than B. bacteriovorus and rather than invading its prey, M. aeruginosavorus attaches irreversibly to the prey cell surface and feeds on it while replicating externally, ultimately resulting in the death of the infected cells. M. aeruginosavorus shows potent in vitro killing activity when co-cultured with the respiratory pathogens P. aeruginosa, K. pneumoniae and E. coli [187]. Importantly, intranasal administration of both B. bacteriovorus and M. aeruginosavorus significantly reduced K. pneumoniae lung burden in a rat pneumonia model, had no adverse effect on the host in rodent models, and were cleared within days by innate immune mechanisms, encouraging their potential use to treat bacterial pneumonia in humans [191,192,193]. In addition, the administration of B. bacteriovorus generally exhibits low immunogenicity in vertebrate models due to the unique LPS composition, and no adverse effects were observed upon intravenous injection, ingestion or topical application in several independent studies [191,192,193,194,195]. Furthermore, B. bacteriovorus is present and abundant in the duodenum and ileum of healthy human individuals with significantly reduced abundance in Crohn’s disease and celiac disease patients, indicating a potential role of B. bacteriovorus in the maintenance of intestinal homeostasis [196]. While it is well established that predatory bacteria exhibit potent in vitro killing of Gram-negative respiratory pathogens, and no known adverse physiological effects were reported upon administration in animal models, further in vivo studies addressing safety concerns and effects on the host microbiome are needed. Predatory bacteria may also be for single use only due to the potential development of adaptive host immune responses.
3.1.3. Postbiotics
Per ISAAP, postbiotics refer to “preparations of inanimate microorganisms and/or their components that confers health benefits in the host” that must stem from defined microorganisms [197]. Although they might be present in postbiotic preparations, substantially purified bacterial metabolites and molecules such as SCFAs, exopolysaccharides and proteins do not qualify as postbiotics due to the absence of cellular biomass [197]. The most commonly used postbiotics in the context of respiratory tract diseases are mixtures of whole and/or fractionated bacterial lysates of common respiratory pathogens which have been shown to reduce the frequency of acute recurrent respiratory infections [198,199,200,201,202]. Bacterial lysates were proposed to exert immunomodulatory functions by activating mucosal dendritic cells and stimulating mucosal pathogen-specific IgA responses, however the precise molecular mechanisms underlying their potentially beneficial clinical effects are not understood [203]. Polyvalent bacterial lysates such as Broncho-Vaxom or Ismigen are currently used for prophylactic purposes in the management of respiratory tract infections and COPD exacerbations. Modestly favorable outcomes were reported from a limited number of clinical trials. However, due to the relatively small sample sizes of these studies and contradictory results from similar studies, there is a clear need for more robust clinical trials to assess the preventive or therapeutic efficacy of these postbiotics.
3.2. Prevention and Treatment of Lower Respiratory Tract Infections by Selective Depletion of Opportunistic Bacterial Pathogens
Opportunistic respiratory pathogens including P. aeruginosa, S. aureus, K. pneumoniae, S. pneumoniae, M. catarrhalis and H. influenzae are commonly found in the human upper respiratory tract. While these bacteria are known to asymptomatically colonize healthy individuals, they can cause severe lower airway infections and exacerbations in patients suffering from chronic respiratory diseases. In addition, they are major etiological agents of hospital-acquired pneumonia (HAP) including ventilator-associated pneumonia (VAP), which due to their high incidence and mortality rates are a predominant cause of death among hospital infections. Treatment of these infections is further complicated by multi-drug resistant bacterial strains that are commonly isolated from the lower airways of HAP and VAP patients. Therefore, selective depletion of these bacteria in at-risk patient populations as well as therapeutic intervention by antibiotic-independent strategies to prevent and control potentially detrimental lung infections represents a critical area of research for the future management of multi-drug resistant lower airway infections.
3.2.1. Antibacterial Monoclonal Antibodies
The development and use of monoclonal human antibodies (mAbs) that target and inactivate bacteria, their virulence factors and/or toxins is widely considered as one of the most promising antibiotic-independent approaches to combat infectious diseases [204]. Antibacterial mAbs exhibit several advantages over the use of conventional antibiotics. Due to their narrow target specificity, they exhibit no known adverse effects on the host microbiota and are less likely to induce widespread resistance [205]. Recent advances in human mAb technologies now allow for the design of polyvalent mAbs that can exert multiple mechanisms of action, including inactivation of virulence factors as well as complement the deposition and subsequent innate immune activation to further aid in bacterial clearance [206]. With a general half-life of several weeks, which can be further extended by introducing amino-acid substitutions in the Fc region to increase the binding to the neonatal Fc receptor, a single injection of a human mAb may be sufficient to provide protection against infection as opposed to multiple regimens of antibiotics a day [207,208,209]. While only three antibacterial antibodies targeting the exotoxins of Clostridioides difficile, Clostridium botulinum or Bacillus anthracis have been approved for clinical use so far, multiple mAbs for the management of lower respiratory tract infections are currently in clinical development [210,211]. S. aureus and P. aeruginosa are the leading causes of bacterial nosocomial pneumonia, including VAP, and are associated with significant mortality and morbidity [212,213,214]. Colonization of the upper airways with S. aureus or P. aeruginosa is a known risk factor for the development of VAP, and modulation of oropharyngeal colonization was shown to be effective in preventing VAP development [215,216]. Due to the high frequency of multi-drug resistant isolates, both pathogens are prime targets for the development of monoclonal antibodies to treat and prevent respiratory infections. Gremubamab (MEDI3902; AstraZeneca) is a bispecific human IgG1 mAb that selectively binds to the P. aeruginosa exopolysaccharide Psl and the type 3 secretion system (T3SS) protein PcrV, both highly conserved virulence factors, and was developed to prevent nosocomial P. aeruginosa pneumonia in high-risk patients [206,217,218]. Binding to Psl promotes complement fixation and opsonophagocytic bacterial killing by neutrophils as well as preventing the attachment of P. aeruginosa to airway epithelial cells whereas targeting PcrV inactivates the T3SS and enhances intracellular killing bacteria following phagocytosis [206,219]. Prophylactic as well as therapeutic administration of Gremubamab proved highly protective in rodent models of acute P. aeruginosa pneumonia as well as a rabbit model of VAP [206,220,221,222]. The effectiveness of MEDI3902 as a preventive measure for VAP in adult P. aeruginosa-colonized ICU patients was assessed in a phase 2 proof-of-concept study. Although the primary efficacy endpoint of reduction in Pa VAP incidence was not met in the total study population, risk reduction was observed in patients with lower baseline inflammation [223]. In addition to Gremubamab, the monoclonal human IgM antibody panobacumab (AR-101, Aerumab; Aridis Pharmaceuticals) is in clinical development for counteracting P. aeruginosa HAP. Panobacumab targets lipopolysaccharide from the highly prevalent P. aeruginosa serotype O11, which accounts for more than 20% of all nosocomial P. aeruginosa pneumonia cases [224]. Binding of Panobacumab to surface LPS results in complement-mediated clearance of P. aeruginosa by host phagocytes in vitro and the therapeutic administration of Panobacumab reduced bacterial burden and alleviated lung inflammation in a mouse model of acute P. aeruginosa lung infection [225,226]. Following promising results from a phase IIa study, Panobacumab is currently in late clinical development as an adjunctive therapy to standard of care antibiotics for hospital-acquired pneumonia [227].
Two antibodies are currently under clinical investigation for the prevention and treatment of nosocomial S. aureus pneumonia. Suvratoxumab (MEDI4893; AstraZeneca, outlicensed to Aridis Pharmaceuticals) specifically binds to and neutralizes the pore-forming alpha-toxin of S. aureus, a highly conserved key virulence factor which is expressed in a vast majority of clinical respiratory S. aureus isolates [228]. Suvratoxumab was shown to confer potent protection in animal models of lethal S. aureus pneumonia, and no severe adverse events were reported upon administration in healthy human adults in a phase 1 placebo-controlled trial [207,229,230]. Efficacy and safety of Suvratoxumab for the prophylaxis of S. aureus VAP in mechanically ventilated ICU patients colonized with S. aureus were further assessed in a phase 2 trial. While the study failed to meet its primary endpoint of a 50% reduction in S. aureus pneumonia in the Suvratoxumab versus the placebo control group, subgroup analyses of patients younger than 65 years showed significant reduction (47%) in the Suvratoxumab-treated group and an associated reduction in the duration of hospitalization and ICU stay [231,232]. A planned phase 3 study will further assess the efficacy of Suvratoxumab in VAP prevention in under 65 year old patients. Another human monoclonal antibody targeting S. aureus alpha-toxin, Tosatoxumab (AR-301, Salvecin; Aridis Pharmaceuticals), is currently in phase 3 clinical development for adjunctive therapeutic treatment of S. aureus VAP in combination with standard of care antibiotics [233]. In addition, several antibacterial human mAbs for the potential treatment of respiratory disease are in pre-clinical and early clinical development including AR-401 (Aridis Pharmaceuticals) and ASN-5 (Arsanis, outlicensed to BB200) targeting A. baumannii and K. pneumoniae, respectively [208].
3.2.2. Bacteriophages and Phage Lysins
Bacteriophages are viruses that specifically infect bacteria by attaching to bacterial target cells, penetrating the bacterial cell membrane and injecting their genetic material into the host cytoplasm. Obligately lytic phages, which are predominantly used for phage-based therapeutics, then hijack the host’s transcriptional and translational machinery and replicate intracellularly. The newly-assembled virions mature and, after reaching a critical mass, phage-derived lytic enzymes (lysins), dissolve the bacterial cell wall to release the phage progeny into the environment [234]. Bacteriophages have been used as early as 1919 to treat bacterial dysentery but have been widely disregarded in Western medicine upon the discovery of antibiotics in the 1940s [234]. Meanwhile, phage-based therapeutics persisted in Eastern Europe and the former Soviet Union to treat, among others, respiratory infections with S. aureus, P. aeruginosa, K. pneumoniae and E. coli [234,235]. With the increase in drug-resistant bacterial isolates and the search for alternative intervention strategies, phage therapy has received renewed clinical interest in Western countries to treat respiratory infections with MDR bacterial pathogens. A clear advantage of using phages over antibiotics is their defined bacterial host range which allows them to selectively eradicate target bacteria without adversely affecting the host microbiota. In addition, their ability to penetrate and disrupt bacterial biofilms, as well as their compatibility with antibiotics and low immunogenicity renders them an attractive alternative to traditional antibiotics. Furthermore, phages have been shown to restore antibiotic-sensitivity to MDR bacteria and features of natural phages such as breadth of strain coverage and resistance development can be modified by genetic engineering to generate highly versatile synthetic phages [236]. Furthermore, nonlytic genetically engineered bacteriophages as a vehicle to selectively deliver genetic material encoding bactericidal proteins are currently in pre-clinical development by Phico Therapeutics to target P. aeruginosa-mediated VAP (SASPject PT3.9), S. aureus (SASPject PT1.2), K. pneumonia (SASPject PT4) and E. coli (SASPject PT5). Phage formulations exhibited potent efficacy in the treatment of P. aeruginosa, K. pneumoniae, A. baumannii or E. coli and in mouse models of lung infection and are currently being evaluated in the clinic for the treatment of human respiratory infections [237,238,239,240,241,242,243,244,245,246]. AP-PA01 (Armata Pharmaceuticals), a cocktail of four obligately lytic bacteriophages targeting P. aeruginosa respiratory infections, was used under the FDA expanded access program to successfully treat a CF patient suffering from an MDR P. aeruginosa infection as well as a patient with ventilator-associated pneumonia and empyema [247,248]. A second-generation phage cocktail, AP-PA02 (Armata Pharmaceuticals), with improved host range and increased potency, is currently being evaluated for safety, tolerability and preliminary efficacy in inhaled form in a Phase 1b/2a study (SWARM-Pa) in individuals with chronic P. aeruginosa lung infections and CF [249].
Besides using whole bacteriophage preparations, the use of phage-derived endolysins or engineered lysins was suggested to combat bacterial infections. Lysins exhibit several advantages over antibiotics and whole phage preparations, as resistance is unlikely to develop due to the conserved nature of their cell wall targets. Purified phage-derived and bioengineered chimeric endolysins show potent in vitro bactericidal activity against Gram-positive as well as Gram-negative respiratory pathogens including planktonic cultures and biofilms of S. pneumoniae, S. aureus, P. aeruginosa and K. pneumoniae [250]. Importantly, administration of phage endolysins drastically reduced S. pneumoniae titers in a mouse model of nasopharyngeal colonization and protected mice from fatal pneumococcal pneumonia or P. aeruginosa lung infection, respectively, underlining their potential to prevent and treat respiratory bacterial infections [177,251,252].
4. Conclusions and Future Outlook
Multi-drug resistant bacterial infections are on the rise and it is predicted that 10 million people will succumb to untreatable infections annually by 2050, estimated to surpass cancer and cardiovascular diseases combined [4]. In 2019 alone, 1.27 million deaths worldwide were directly attributable to infections with MDR pathogens, while 4.95 million deaths were associated with infections [253]. With 400,000 directly attributable deaths globally, lower respiratory infections are the leading cause of death among all MDR infections and are often associated with the priority pathogens S. aureus, K. pneumoniae, S. pneumoniae, A. baumannii and P. aeruginosa. Meanwhile, hundreds of millions of individuals globally suffer from chronic lung disease and respiratory viral infections that are becoming increasingly difficult to manage due to the emergence of novel viral variants. Hence, there is a critical need for novel therapeutic and preventive strategies, and due to the key role of lung and gut dysbiosis in lower airway disease, modulation of the microbiome has emerged as one potential intervention avenue.
Dietary supplementation with prebiotics, probiotics or postbiotics to deliberately alter the gut-lung axis would be a relatively simple approach for managing lower airway disease. Nevertheless, apart from a clear correlation of increased dietary fiber intake with lung health, there is limited evidence that intake of probiotics or postbiotics results in clinical improvement of lower respiratory conditions. This may at least in part be due to the high interindividual variability in response to colonization with probiotic strains where some individuals were shown to be more permissive, while others are resistant and host pathways were differentially affected upon colonization [254,255,256]. In addition, the effects of probiotics are generally transient, even in permissive individuals and only observed during or shortly after consumption [254,257,258,259,260,261]. The use of microbiome-specific tailored probiotics or purified microbial products and metabolites, such as SCFAs, could potentially overcome these issues. However, while SCFAs and SCFA receptor agonists were successfully used to treat and prevent lower respiratory infections and allergic airway inflammation in animal models, no efficacy was observed in the clinic so far [65,76,78,83,86,98,100,117,156,158,161].
Future research to better understand the complex host-microbiome crosstalk as well as interactions between individual members of the microbiota are needed and will aid in developing personalized strategies to modulate the microbiota in individual patients in order to achieve the highest possible treatment outcome. Promising results have been obtained from using antibacterial antibodies or bacteriophages in the prevention and treatment of nosocomial respiratory tract infections with opportunistic priority pathogens in at-risk patient populations. Further clinical evaluation is currently underway, and with innovations in phage and mAb technologies such as the generation of multivalent molecules with different modes of action and potential combined approaches of several mAbs to target multiple respiratory pathogens, they represent a viable and urgently needed alternative to current antibiotics [208,217].
Author Contributions
B.C.M. wrote the article and A.D. contributed to writing and editing. All authors have read and agreed to the published version of the manuscript.
Funding
The funding required for the preparation of this manuscript was provided by AstraZeneca.
Conflicts of Interest
B.C.M. and A.D. are employees of AstraZeneca and may hold stock options.
References
- National Center for Farmworker Health—Statistics. Monthly Provisional Counts of Deaths by Select Causes, 2020–2021. Available online: https://data.cdc.gov/NCHS/Monthly-Provisional-Counts-of-Deaths-by-Select-Cau/9dzk-mvmi (accessed on 8 December 2021).
- GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Syamlal, G.; Bhattacharya, A.; Dodd, K.E. Medical Expenditures Attributed to Asthma and Chronic Obstructive Pulmonary Disease Among Workers—United States, 2011–2015. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 809–814. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. In The Review on Antimicrobial Resistance. 2014. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 10 January 2022).
- Centers for Disease Control and Prevention, U.S. Department of Health and Human Services. Antibiotics Resistance Threats in the United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013.
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Mackay, C.R. Diet, gut microbiota and immune responses. Nat. Immunol. 2011, 12, 5–9. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Blander, J.M.; Longman, R.S.; Iliev, I.D.; Sonnenberg, G.F.; Artis, D. Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 2017, 18, 851–860. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Cook, M.D.; Allen, J.M.; Pence, B.D.; Wallig, M.A.; Gaskins, H.R.; White, B.A.; Woods, J.A. Exercise and gut immune function: Evidence of alterations in colon immune cell homeostasis and microbiome characteristics with exercise training. Immunol. Cell Biol. 2016, 94, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Benedict, C.; Vogel, H.; Jonas, W.; Woting, A.; Blaut, M.; Schurmann, A.; Cedernaes, J. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol. Metab. 2016, 5, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Karl, J.P.; Margolis, L.M.; Madslien, E.H.; Murphy, N.E.; Castellani, J.W.; Gundersen, Y.; Hoke, A.V.; Levangie, M.W.; Kumar, R.; Chakraborty, N.; et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G559–G571. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4554–4561. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Gevers, D.; Knight, R.; Petrosino, J.F.; Huang, K.; McGuire, A.L.; Birren, B.W.; Nelson, K.E.; White, O.; Methe, B.A.; Huttenhower, C. The Human Microbiome Project: A community resource for the healthy human microbiome. PLoS Biol. 2012, 10, e1001377. [Google Scholar] [CrossRef]
- HMP Integrative. The Integrative Human Microbiome Project: Dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host. Microbe. 2014, 16, 276–289. [Google Scholar] [CrossRef]
- HMP Integrative. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.Y.; Zhang, Z.; Feng, Y. The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 11, 301. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut-lung axis. Mucosal. Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Quinn, R.A.; Debelius, J.; Xu, Z.Z.; Morton, J.; Garg, N.; Jansson, J.K.; Dorrestein, P.C.; Knight, R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 2016, 535, 94–103. [Google Scholar] [CrossRef]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, F.; Sun, R.; Gao, X.; Wei, H.; Li, L.J.; Tian, Z. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat. Commun. 2013, 4, 2106. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.L.; Sequeira, R.P.; Clarke, T.B. The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 2017, 8, 1512. [Google Scholar] [CrossRef] [PubMed]
- Man, W.H.; de Steenhuijsen Piters, W.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Budden, K.F.; Shukla, S.D.; Rehman, S.F.; Bowerman, K.L.; Keely, S.; Hugenholtz, P.; Armstrong-James, D.P.H.; Adcock, I.M.; Chotirmall, S.H.; Chung, K.F.; et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir. Med. 2019, 7, 907–920. [Google Scholar] [CrossRef]
- Mathieu, E.; Escribano-Vazquez, U.; Descamps, D.; Cherbuy, C.; Langella, P.; Riffault, S.; Remot, A.; Thomas, M. Paradigms of Lung Microbiota Functions in Health and Disease, Particularly, in Asthma. Front. Physiol. 2018, 9, 1168. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal. Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Segal, L.N.; Alekseyenko, A.V.; Clemente, J.C.; Kulkarni, R.; Wu, B.; Gao, Z.; Chen, H.; Berger, K.I.; Goldring, R.M.; Rom, W.N.; et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013, 1, 19. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Beck, J.M.; Huffnagle, G.B.; Curtis, J.L. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann. Am. Thorac. Soc. 2015, 12, 821–830. [Google Scholar] [CrossRef]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, e00037. [Google Scholar] [CrossRef]
- Marsland, B.J.; Gollwitzer, E.S. Host-microorganism interactions in lung diseases. Nat. Rev. Immunol. 2014, 14, 827–835. [Google Scholar] [CrossRef]
- Eklof, J.; Sorensen, R.; Ingebrigtsen, T.S.; Sivapalan, P.; Achir, I.; Boel, J.B.; Bangsborg, J.; Ostergaard, C.; Dessau, R.B.; Jensen, U.S.; et al. Pseudomonas aeruginosa and risk of death and exacerbations in patients with chronic obstructive pulmonary disease: An observational cohort study of 22 053 patients. Clin. Microbiol. Infect. 2020, 26, 227–234. [Google Scholar] [CrossRef]
- Engler, K.; Muhlemann, K.; Garzoni, C.; Pfahler, H.; Geiser, T.; von Garnier, C. Colonisation with Pseudomonas aeruginosa and antibiotic resistance patterns in COPD patients. Swiss Med. Wkly. 2012, 142, w13509. [Google Scholar] [CrossRef]
- Flynn, K.M.; Dowell, G.; Johnson, T.M.; Koestler, B.J.; Waters, C.M.; Cooper, V.S. Evolution of Ecological Diversity in Biofilms of Pseudomonas aeruginosa by Altered Cyclic Diguanylate Signaling. J. Bacteriol. 2016, 198, 2608–2618. [Google Scholar] [CrossRef]
- Gallego, M.; Pomares, X.; Espasa, M.; Castaner, E.; Sole, M.; Suarez, D.; Monso, E.; Monton, C. Pseudomonas aeruginosa isolates in severe chronic obstructive pulmonary disease: Characterization and risk factors. BMC Pulm. Med. 2014, 14, 103. [Google Scholar] [CrossRef]
- Jacobs, D.M.; Ochs-Balcom, H.M.; Noyes, K.; Zhao, J.; Leung, W.Y.; Pu, C.Y.; Murphy, T.F.; Sethi, S. Impact of Pseudomonas aeruginosa Isolation on Mortality and Outcomes in an Outpatient Chronic Obstructive Pulmonary Disease Cohort. Open. Forum. Infect. Dis. 2020, 7, ofz546. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.F.; Brauer, A.L.; Eschberger, K.; Lobbins, P.; Grove, L.; Cai, X.; Sethi, S. Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2008, 177, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Belchamber, K.B.R.; Singh, R.; Batista, C.M.; Whyte, M.K.; Dockrell, D.H.; Kilty, I.; Robinson, M.J.; Wedzicha, J.A.; Barnes, P.J.; Donnelly, L.E.; et al. Defective bacterial phagocytosis is associated with dysfunctional mitochondria in COPD macrophages. Eur. Respir. J. 2019, 54, 1802244. [Google Scholar] [CrossRef] [PubMed]
- Berenson, C.S.; Kruzel, R.L.; Wrona, C.T.; Mammen, M.J.; Sethi, S. Impaired Innate COPD Alveolar Macrophage Responses and Toll-Like Receptor-9 Polymorphisms. PLoS ONE 2015, 10, e0134209. [Google Scholar] [CrossRef]
- Bewley, M.A.; Belchamber, K.B.; Chana, K.K.; Budd, R.C.; Donaldson, G.; Wedzicha, J.A.; Brightling, C.E.; Kilty, I.; Donnelly, L.E.; Barnes, P.J.; et al. Differential Effects of p38, MAPK, PI3K or Rho Kinase Inhibitors on Bacterial Phagocytosis and Efferocytosis by Macrophages in COPD. PLoS ONE 2016, 11, e0163139. [Google Scholar] [CrossRef]
- Singh, R.; Belchamber, K.B.R.; Fenwick, P.S.; Chana, K.; Donaldson, G.; Wedzicha, J.A.; Barnes, P.J.; Donnelly, L.E.; Consortium, C. Defective monocyte-derived macrophage phagocytosis is associated with exacerbation frequency in COPD. Respir. Res. 2021, 22, 113. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.E.; Finney-Hayward, T.K.; Quint, J.K.; Thomas, C.M.; Tudhope, S.J.; Wedzicha, J.A.; Barnes, P.J.; Donnelly, L.E. Defective macrophage phagocytosis of bacteria in COPD. Eur. Respir. J. 2010, 35, 1039–1047. [Google Scholar] [CrossRef]
- Fitzpatrick, A.M.; Holguin, F.; Teague, W.G.; Brown, L.A. Alveolar macrophage phagocytosis is impaired in children with poorly controlled asthma. J. Allergy Clin. Immunol. 2008, 121, 1372–1378.e3. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, Q.; Thomas, C.M.; Chana, K.K.; Gibeon, D.; Barnes, P.J.; Chung, K.F.; Bhavsar, P.K.; Donnelly, L.E. Impaired macrophage phagocytosis of bacteria in severe asthma. Respir. Res. 2014, 15, 72. [Google Scholar] [CrossRef]
- Van de Weert-van Leeuwen, P.B.; Van Meegen, M.A.; Speirs, J.J.; Pals, D.J.; Rooijakkers, S.H.; Van der Ent, C.K.; Terheggen-Lagro, S.W.; Arets, H.G.; Beekman, J.M. Optimal complement-mediated phagocytosis of Pseudomonas aeruginosa by monocytes is cystic fibrosis transmembrane conductance regulator-dependent. Am. J. Respir. Cell Mol. Biol. 2013, 49, 463–470. [Google Scholar] [CrossRef]
- Allden, S.J.; Ogger, P.P.; Ghai, P.; McErlean, P.; Hewitt, R.; Toshner, R.; Walker, S.A.; Saunders, P.; Kingston, S.; Molyneaux, P.L.; et al. The Transferrin Receptor CD71 Delineates Functionally Distinct Airway Macrophage Subsets during Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 200, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Sencio, V.; Barthelemy, A.; Tavares, L.P.; Machado, M.G.; Soulard, D.; Cuinat, C.; Queiroz-Junior, C.M.; Noordine, M.L.; Salome-Desnoulez, S.; Deryuter, L.; et al. Gut Dysbiosis during Influenza Contributes to Pulmonary Pneumococcal Superinfection through Altered Short-Chain Fatty Acid Production. Cell Rep. 2020, 30, 2934–2947.e6. [Google Scholar] [CrossRef] [PubMed]
- Hang, D.T.T.; Choi, E.J.; Song, J.Y.; Kim, S.E.; Kwak, J.; Shin, Y.K. Differential effect of prior influenza infection on alveolar macrophage phagocytosis of Staphylococcus aureus and Escherichia coli: Involvement of interferon-gamma production. Microbiol. Immunol. 2011, 55, 751–759. [Google Scholar] [CrossRef]
- Sun, K.; Metzger, D.W. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat. Med. 2008, 14, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Nelson, C.E.; Brodie, E.L.; Desantis, T.Z.; Baek, M.S.; Liu, J.; Woyke, T.; Allgaier, M.; Bristow, J.; Wiener-Kronish, J.P.; et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 2011, 127, 372–381.e3. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Mac Aogain, M.; Chotirmall, S.H. The current understanding and future directions for sputum microbiome profiling in chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 2021, 28, 121–133. [Google Scholar] [CrossRef]
- Patel, I.S.; Seemungal, T.A.; Wilks, M.; Lloyd-Owen, S.J.; Donaldson, G.C.; Wedzicha, J.A. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax 2002, 57, 759–764. [Google Scholar] [CrossRef]
- Abt, M.C.; Osborne, L.C.; Monticelli, L.A.; Doering, T.A.; Alenghat, T.; Sonnenberg, G.F.; Paley, M.A.; Antenus, M.; Williams, K.L.; Erikson, J.; et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012, 37, 158–170. [Google Scholar] [CrossRef]
- Bradley, K.C.; Finsterbusch, K.; Schnepf, D.; Crotta, S.; Llorian, M.; Davidson, S.; Fuchs, S.Y.; Staeheli, P.; Wack, A. Microbiota-Driven Tonic Interferon Signals in Lung Stromal Cells Protect from Influenza Virus Infection. Cell Rep. 2019, 28, 245–256.e4. [Google Scholar] [CrossRef]
- Clarke, T.B.; Davis, K.M.; Lysenko, E.S.; Zhou, A.Y.; Yu, Y.; Weiser, J.N. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010, 16, 228–231. [Google Scholar] [CrossRef]
- Clarke, T.B. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect. Immun. 2014, 82, 4596–4606. [Google Scholar] [CrossRef] [PubMed]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [PubMed]
- Cait, A.; Hughes, M.R.; Antignano, F.; Cait, J.; Dimitriu, P.A.; Maas, K.R.; Reynolds, L.A.; Hacker, L.; Mohr, J.; Finlay, B.B.; et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal. Immunol. 2018, 11, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Steed, A.L.; Christophi, G.P.; Kaiko, G.E.; Sun, L.; Goodwin, V.M.; Jain, U.; Esaulova, E.; Artyomov, M.N.; Morales, D.J.; Holtzman, M.J.; et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 2017, 357, 498–502. [Google Scholar] [CrossRef]
- Huang, Y.; Mao, K.; Chen, X.; Sun, M.A.; Kawabe, T.; Li, W.; Usher, N.; Zhu, J.; Urban, J.F., Jr.; Paul, W.E.; et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 2018, 359, 114–119. [Google Scholar] [CrossRef]
- Gray, J.; Oehrle, K.; Worthen, G.; Alenghat, T.; Whitsett, J.; Deshmukh, H. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci. Transl. Med. 2017, 9, eaaf9412. [Google Scholar] [CrossRef]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef]
- Antunes, K.H.; Fachi, J.L.; de Paula, R.; da Silva, E.F.; Pral, L.P.; Dos Santos, A.A.; Dias, G.B.M.; Vargas, J.E.; Puga, R.; Mayer, F.Q.; et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 2019, 10, 3273. [Google Scholar] [CrossRef]
- Yazar, A.; Atis, S.; Konca, K.; Pata, C.; Akbay, E.; Calikoglu, M.; Hafta, A. Respiratory symptoms and pulmonary functional changes in patients with irritable bowel syndrome. Am. J. Gastroenterol. 2001, 96, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Fagundes, C.T.; Amaral, F.A.; Vieira, A.T.; Soares, A.C.; Pinho, V.; Nicoli, J.R.; Vieira, L.Q.; Teixeira, M.M.; Souza, D.G. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J. Immunol. 2012, 188, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.; Wang, B.; Shafiei Jahani, P.; Hurrell, B.P.; Banie, H.; Aleman Muench, G.R.; Maazi, H.; Helou, D.G.; Howard, E.; Galle-Treger, L.; et al. Dietary Fiber-Induced Microbial Short Chain Fatty Acids Suppress ILC2-Dependent Airway Inflammation. Front. Immunol. 2019, 10, 2051. [Google Scholar] [CrossRef]
- Mateer, S.W.; Maltby, S.; Marks, E.; Foster, P.S.; Horvat, J.C.; Hansbro, P.M.; Keely, S. Potential mechanisms regulating pulmonary pathology in inflammatory bowel disease. J. Leukoc. Biol. 2015, 98, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Black, H.; Mendoza, M.; Murin, S. Thoracic manifestations of inflammatory bowel disease. Chest 2007, 131, 524–532. [Google Scholar] [CrossRef]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Bjorksten, B.; Engstrand, L.; Jenmalm, M.C. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 2014, 44, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.L.; Gold, M.J.; Hartmann, M.; Willing, B.P.; Thorson, L.; Wlodarska, M.; Gill, N.; Blanchet, M.R.; Mohn, W.W.; McNagny, K.M.; et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012, 13, 440–447. [Google Scholar] [CrossRef]
- Slob, E.M.A.; Brew, B.K.; Vijverberg, S.J.H.; Kats, C.; Longo, C.; Pijnenburg, M.W.; van Beijsterveldt, T.; Dolan, C.V.; Bartels, M.; Magnusson, P.; et al. Early-life antibiotic use and risk of asthma and eczema: Results of a discordant twin study. Eur. Respir. J. 2020, 55, 1902021. [Google Scholar] [CrossRef]
- Ahmadizar, F.; Vijverberg, S.J.H.; Arets, H.G.M.; de Boer, A.; Turner, S.; Devereux, G.; Arabkhazaeli, A.; Soares, P.; Mukhopadhyay, S.; Garssen, J.; et al. Early life antibiotic use and the risk of asthma and asthma exacerbations in children. Pediatr. Allergy Immunol. 2017, 28, 430–437. [Google Scholar] [CrossRef]
- Bisgaard, H.; Li, N.; Bonnelykke, K.; Chawes, B.L.; Skov, T.; Paludan-Muller, G.; Stokholm, J.; Smith, B.; Krogfelt, K.A. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 2011, 128, 646–652.e5. [Google Scholar] [CrossRef]
- Marra, F.; Marra, C.A.; Richardson, K.; Lynd, L.D.; Kozyrskyj, A.; Patrick, D.M.; Bowie, W.R.; Fitzgerald, J.M. Antibiotic use in children is associated with increased risk of asthma. Pediatrics 2009, 123, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Metsala, J.; Lundqvist, A.; Virta, L.J.; Kaila, M.; Gissler, M.; Virtanen, S.M. Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin. Exp. Allergy 2015, 45, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Jiang, W.; Zou, B.; Feng, J.; Cheng, X.; Gu, J.; Chu, T.; Niu, C.; He, R.; Chu, Y.; et al. LPS inactivation by a host lipase allows lung epithelial cell sensitization for allergic asthma. J. Exp. Med. 2018, 215, 2397–2412. [Google Scholar] [CrossRef] [PubMed]
- Theiler, A.; Barnthaler, T.; Platzer, W.; Richtig, G.; Peinhaupt, M.; Rittchen, S.; Kargl, J.; Ulven, T.; Marsh, L.M.; Marsche, G.; et al. Butyrate ameliorates allergic airway inflammation by limiting eosinophil trafficking and survival. J. Allergy Clin. Immunol. 2019, 144, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Thio, C.L.; Chi, P.Y.; Lai, A.C.; Chang, Y.J. Regulation of type 2 innate lymphoid cell-dependent airway hyperreactivity by butyrate. J. Allergy Clin. Immunol. 2018, 142, 1867–1883.e12. [Google Scholar] [CrossRef]
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.; Stanley, D.; Macia, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Jeffery, I.B. Gut microbiota and aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef]
- Chiu, Y.C.; Lee, S.W.; Liu, C.W.; Lan, T.Y.; Wu, L.S. Relationship between gut microbiota and lung function decline in patients with chronic obstructive pulmonary disease: A 1-year follow-up study. Respir. Res. 2022, 23, 10. [Google Scholar] [CrossRef]
- Biedermann, L.; Zeitz, J.; Mwinyi, J.; Sutter-Minder, E.; Rehman, A.; Ott, S.J.; Steurer-Stey, C.; Frei, A.; Frei, P.; Scharl, M.; et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS ONE 2013, 8, e59260. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, A.; Frazer, Z.A.; Hansbro, P.M.; Yang, I.A. COPD and the gut-lung axis: The therapeutic potential of fibre. J. Thorac. Dis. 2019, 11, S2173–S2180. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.O.; Lee, S.H.; Choi, J.J.; Kim, D.H.; Choi, J.M.; Kang, M.J.; Oh, Y.M.; Park, Y.J.; Shin, Y.; Lee, S.W. Fecal microbial transplantation and a high fiber diet attenuates emphysema development by suppressing inflammation and apoptosis. Exp. Mol. Med. 2020, 52, 1128–1139. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Dai, Z.; Wang, Z.; Deng, Z.; Zhang, J.; Pu, J.; Cao, W.; Pan, T.; Zhou, Y.; Yang, Z.; et al. Gut microbiota dysbiosis contributes to the development of chronic obstructive pulmonary disease. Respir. Res. 2021, 22, 274. [Google Scholar] [CrossRef]
- Rutten, E.P.A.; Lenaerts, K.; Buurman, W.A.; Wouters, E.F.M. Disturbed intestinal integrity in patients with COPD: Effects of activities of daily living. Chest 2014, 145, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Keely, S.; Talley, N.J.; Hansbro, P.M. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal. Immunol. 2012, 5, 7–18. [Google Scholar] [CrossRef]
- Ekbom, A.; Brandt, L.; Granath, F.; Lofdahl, C.G.; Egesten, A. Increased risk of both ulcerative colitis and Crohn’s disease in a population suffering from COPD. Lung 2008, 186, 167–172. [Google Scholar] [CrossRef]
- Bowerman, K.L.; Rehman, S.F.; Vaughan, A.; Lachner, N.; Budden, K.F.; Kim, R.Y.; Wood, D.L.A.; Gellatly, S.L.; Shukla, S.D.; Wood, L.G.; et al. Disease-associated gut microbiome and metabolome changes in patients with chronic obstructive pulmonary disease. Nat. Commun. 2020, 11, 5886. [Google Scholar] [CrossRef]
- Walker, C.L.; Perin, J.; Katz, J.; Tielsch, J.M.; Black, R.E. Diarrhea as a risk factor for acute lower respiratory tract infections among young children in low income settings. J. Glob. Health 2013, 3, 010402. [Google Scholar] [CrossRef][Green Version]
- Schuijt, T.J.; Lankelma, J.M.; Scicluna, B.P.; de Sousae Melo, F.; Roelofs, J.J.; de Boer, J.D.; Hoogendijk, A.J.; de Beer, R.; de Vos, A.; Belzer, C.; et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016, 65, 575–583. [Google Scholar] [CrossRef]
- Robak, O.H.; Heimesaat, M.M.; Kruglov, A.A.; Prepens, S.; Ninnemann, J.; Gutbier, B.; Reppe, K.; Hochrein, H.; Suter, M.; Kirschning, C.J.; et al. Antibiotic treatment-induced secondary IgA deficiency enhances susceptibility to Pseudomonas aeruginosa pneumonia. J. Clin. Investig. 2018, 128, 3535–3545. [Google Scholar] [CrossRef] [PubMed]
- Dessein, R.; Bauduin, M.; Grandjean, T.; Le Guern, R.; Figeac, M.; Beury, D.; Faure, K.; Faveeuw, C.; Guery, B.; Gosset, P.; et al. Antibiotic-related gut dysbiosis induces lung immunodepression and worsens lung infection in mice. Crit. Care 2020, 24, 611. [Google Scholar] [CrossRef] [PubMed]
- Gauguet, S.; D’Ortona, S.; Ahnger-Pier, K.; Duan, B.; Surana, N.K.; Lu, R.; Cywes-Bentley, C.; Gadjeva, M.; Shan, Q.; Priebe, G.P.; et al. Intestinal Microbiota of Mice Influences Resistance to Staphylococcus aureus Pneumonia. Infect. Immun. 2015, 83, 4003–4014. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Pattaroni, C.; Lopez-Mejia, I.C.; Riva, E.; Pernot, J.; Ubags, N.; Fajas, L.; Nicod, L.P.; Marsland, B.J. Dietary Fiber Confers Protection against Flu by Shaping Ly6c(−) Patrolling Monocyte Hematopoiesis and CD8(+) T Cell Metabolism. Immunity 2018, 48, 992–1005.e8. [Google Scholar] [CrossRef] [PubMed]
- Dilantika, C.; Sedyaningsih, E.R.; Kasper, M.R.; Agtini, M.; Listiyaningsih, E.; Uyeki, T.M.; Burgess, T.H.; Blair, P.J.; Putnam, S.D. Influenza virus infection among pediatric patients reporting diarrhea and influenza-like illness. BMC Infect. Dis. 2010, 10, 3. [Google Scholar] [CrossRef]
- Qin, N.; Zheng, B.; Yao, J.; Guo, L.; Zuo, J.; Wu, L.; Zhou, J.; Liu, L.; Guo, J.; Ni, S.; et al. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci. Rep. 2015, 5, 14771. [Google Scholar] [CrossRef]
- Bartley, J.M.; Zhou, X.; Kuchel, G.A.; Weinstock, G.M.; Haynes, L. Impact of Age, Caloric Restriction, and Influenza Infection on Mouse Gut Microbiome: An Exploratory Study of the Role of Age-Related Microbiome Changes on Influenza Responses. Front. Immunol. 2017, 8, 1164. [Google Scholar] [CrossRef]
- Deriu, E.; Boxx, G.M.; He, X.; Pan, C.; Benavidez, S.D.; Cen, L.; Rozengurt, N.; Shi, W.; Cheng, G. Influenza Virus Affects Intestinal Microbiota and Secondary Salmonella Infection in the Gut through Type I Interferons. PLoS Pathog. 2016, 12, e1005572. [Google Scholar] [CrossRef]
- Groves, H.T.; Cuthbertson, L.; James, P.; Moffatt, M.F.; Cox, M.J.; Tregoning, J.S. Respiratory Disease following Viral Lung Infection Alters the Murine Gut Microbiota. Front. Immunol. 2018, 9, 182. [Google Scholar] [CrossRef]
- Wang, J.; Li, F.; Wei, H.; Lian, Z.X.; Sun, R.; Tian, Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 2014, 211, 2397–2410. [Google Scholar] [CrossRef]
- Yildiz, S.; Mazel-Sanchez, B.; Kandasamy, M.; Manicassamy, B.; Schmolke, M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome 2018, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, J.; Feng, J.W.; Hu, X.T.; Wang, T.; Gong, W.X.; Huang, K.; Guo, Y.X.; Zou, Z.; Lin, X.; et al. Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection. Genome. Biol. 2020, 21, 99. [Google Scholar] [CrossRef] [PubMed]
- Harding, J.N.; Siefker, D.; Vu, L.; You, D.; DeVincenzo, J.; Pierre, J.F.; Cormier, S.A. Altered gut microbiota in infants is associated with respiratory syncytial virus disease severity. BMC Microbiol. 2020, 20, 140. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Young, R.P.; Hopkins, R.J.; Hanson, C. Connecting Dietary Fiber Directly with Good Lung Health. Ann. Am. Thorac. Soc. 2016, 13, 1869–1870. [Google Scholar] [CrossRef]
- Young, R.P.; Hopkins, R.J.; Marsland, B. The Gut-Liver-Lung Axis. Modulation of the Innate Immune Response and Its Possible Role in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2016, 54, 161–169. [Google Scholar] [CrossRef]
- Hanson, C.; Lyden, E.; Rennard, S.; Mannino, D.M.; Rutten, E.P.; Hopkins, R.; Young, R. The Relationship between Dietary Fiber Intake and Lung Function in the National Health and Nutrition Examination Surveys. Ann. Am. Thorac. Soc. 2016, 13, 643–650. [Google Scholar] [CrossRef]
- Wypych, T.P.; Marsland, B.J.; Ubags, N.D.J. The Impact of Diet on Immunity and Respiratory Diseases. Ann. Am. Thorac. Soc. 2017, 14, S339–S347. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Root, M.M.; Houser, S.M.; Anderson, J.J.; Dawson, H.R. Healthy Eating Index 2005 and selected macronutrients are correlated with improved lung function in humans. Nutr. Res. 2014, 34, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.C.; Norat, T.; Murphy, N.; Olsen, A.; Tjonneland, A.; Overvad, K.; Boutron-Ruault, M.C.; Perquier, F.; Dartois, L.; Kaaks, R.; et al. Fiber intake and total and cause-specific mortality in the European Prospective Investigation into Cancer and Nutrition cohort. Am. J. Clin. Nutr. 2012, 96, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Subar, A.F.; Hollenbeck, A.; Schatzkin, A. Dietary fiber intake and mortality in the NIH-AARP diet and health study. Arch. Intern. Med. 2011, 171, 1061–1068. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [PubMed]
- Le Poul, E.; Loison, C.; Struyf, S.; Springael, J.Y.; Lannoy, V.; Decobecq, M.E.; Brezillon, S.; Dupriez, V.; Vassart, G.; Van Damme, J.; et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 2003, 278, 25481–25489. [Google Scholar] [CrossRef] [PubMed]
- Taggart, A.K.; Kero, J.; Gan, X.; Cai, T.Q.; Cheng, K.; Ippolito, M.; Ren, N.; Kaplan, R.; Wu, K.; Wu, T.J.; et al. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J. Biol. Chem. 2005, 280, 26649–26652. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Berthon, B.S.; Macdonald-Wicks, L.K.; Gibson, P.G.; Wood, L.G. Investigation of the association between dietary intake, disease severity and airway inflammation in asthma. Respirology 2013, 18, 447–454. [Google Scholar] [CrossRef]
- Halnes, I.; Baines, K.J.; Berthon, B.S.; MacDonald-Wicks, L.K.; Gibson, P.G.; Wood, L.G. Soluble Fibre Meal Challenge Reduces Airway Inflammation and Expression of GPR43 and GPR41 in Asthma. Nutrients 2017, 9, 57. [Google Scholar] [CrossRef]
- Kan, H.; Stevens, J.; Heiss, G.; Rose, K.M.; London, S.J. Dietary fiber, lung function, and chronic obstructive pulmonary disease in the atherosclerosis risk in communities study. Am. J. Epidemiol. 2008, 167, 570–578. [Google Scholar] [CrossRef]
- Varraso, R.; Willett, W.C.; Camargo, C.A., Jr. Prospective study of dietary fiber and risk of chronic obstructive pulmonary disease among US women and men. Am. J. Epidemiol. 2010, 171, 776–784. [Google Scholar] [CrossRef]
- Jang, Y.O.; Kim, O.H.; Kim, S.J.; Lee, S.H.; Yun, S.; Lim, S.E.; Yoo, H.J.; Shin, Y.; Lee, S.W. High-fiber diets attenuate emphysema development via modulation of gut microbiota and metabolism. Sci. Rep. 2021, 11, 7008. [Google Scholar] [CrossRef]
- Szmidt, M.K.; Kaluza, J.; Harris, H.R.; Linden, A.; Wolk, A. Long-term dietary fiber intake and risk of chronic obstructive pulmonary disease: A prospective cohort study of women. Eur. J. Nutr. 2020, 59, 1869–1879. [Google Scholar] [CrossRef] [PubMed]
- Kaluza, J.; Harris, H.; Wallin, A.; Linden, A.; Wolk, A. Dietary Fiber Intake and Risk of Chronic Obstructive Pulmonary Disease: A Prospective Cohort Study of Men. Epidemiology 2018, 29, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Ichinohe, T. High ambient temperature dampens adaptive immune responses to influenza A virus infection. Proc. Natl. Acad. Sci. USA 2019, 116, 3118–3125. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Raundhal, M.; Chen, B.B.; Morse, C.; Tyurina, Y.Y.; Khare, A.; Oriss, T.B.; Huff, R.; Lee, J.S.; St Croix, C.M.; et al. The mito-DAMP cardiolipin blocks IL-10 production causing persistent inflammation during bacterial pneumonia. Nat. Commun. 2017, 8, 13944. [Google Scholar] [CrossRef] [PubMed]
- Galvao, I.; Tavares, L.P.; Correa, R.O.; Fachi, J.L.; Rocha, V.M.; Rungue, M.; Garcia, C.C.; Cassali, G.; Ferreira, C.M.; Martins, F.S.; et al. The Metabolic Sensor GPR43 Receptor Plays a Role in the Control of Klebsiella pneumoniae Infection in the Lung. Front. Immunol. 2018, 9, 142. [Google Scholar] [CrossRef]
- Fiber Metabolism in Chronic Obstructive Pulmonary Disease. Available online: https://ClinicalTrials.gov/show/NCT04459156 (accessed on 19 January 2022).
- The Effects of Prebiotics on Gut Bacterial Parameters, Immune Function & Exercise-Induced Airway Inflammation. Available online: https://ClinicalTrials.gov/show/NCT02872675 (accessed on 15 February 2022).
- Machado, M.G.; Patente, T.A.; Rouille, Y.; Heumel, S.; Melo, E.M.; Deruyter, L.; Pourcet, B.; Sencio, V.; Teixeira, M.M.; Trottein, F. Acetate Improves the Killing of Streptococcus pneumoniae by Alveolar Macrophages via NLRP3 Inflammasome and Glycolysis-HIF-1alpha Axis. Front. Immunol. 2022, 13, 773261. [Google Scholar] [CrossRef]
- Tan, P.; Li, X.; Shen, J.; Feng, Q. Fecal Microbiota Transplantation for the Treatment of Inflammatory Bowel Disease: An Update. Front. Pharmacol. 2020, 11, 574533. [Google Scholar] [CrossRef]
- The Impact of Fecal Microbiota Transplantation as an Immunomodulation on the Risk Reduction of COVID-19 Disease Progression With Escalating Cytokine Storm and Inflammatory Parameters. Available online: https://ClinicalTrials.gov/show/NCT04824222 (accessed on 18 January 2022).
- DeFilipp, Z.; Bloom, P.P.; Torres Soto, M.; Mansour, M.K.; Sater, M.R.A.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.B.; Hohmann, E.L. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Freedman, S.B.; Schnadower, D.; Tarr, P.I. The Probiotic Conundrum: Regulatory Confusion, Conflicting Studies, and Safety Concerns. JAMA 2020, 323, 823–824. [Google Scholar] [CrossRef]
- Clarke, T.C.; Black, L.I.; Stussman, B.J.; Barnes, P.M.; Nahin, R.L. Trends in the use of complementary health approaches among adults: United States, 2002–2012. Natl. Health Stat. Rep. 2015, 79, 1–16. [Google Scholar]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. Biomed. Res. Int. 2018, 2018, 9478630. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Boushey, H.A. The microbiome and development of allergic disease. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Feleszko, W.; Jaworska, J.; Rha, R.D.; Steinhausen, S.; Avagyan, A.; Jaudszus, A.; Ahrens, B.; Groneberg, D.A.; Wahn, U.; Hamelmann, E. Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin. Exp. Allergy 2007, 37, 498–505. [Google Scholar] [CrossRef]
- Forsythe, P.; Inman, M.D.; Bienenstock, J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am. J. Respir. Crit. Care Med. 2007, 175, 561–569. [Google Scholar] [CrossRef]
- Hougee, S.; Vriesema, A.J.; Wijering, S.C.; Knippels, L.M.; Folkerts, G.; Nijkamp, F.P.; Knol, J.; Garssen, J. Oral treatment with probiotics reduces allergic symptoms in ovalbumin-sensitized mice: A bacterial strain comparative study. Int. Arch. Allergy Immunol. 2010, 151, 107–117. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Demoor, T.; Rauch, M.; Faruqi, A.A.; Jang, S.; Johnson, C.C.; Boushey, H.A.; Zoratti, E.; Ownby, D.; Lukacs, N.W.; et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 805–810. [Google Scholar] [CrossRef]
- Wu, C.T.; Chen, P.J.; Lee, Y.T.; Ko, J.L.; Lue, K.H. Effects of immunomodulatory supplementation with Lactobacillus rhamnosus on airway inflammation in a mouse asthma model. J. Microbiol. Immunol. Infect. 2016, 49, 625–635. [Google Scholar] [CrossRef]
- Zhang, B.; An, J.; Shimada, T.; Liu, S.; Maeyama, K. Oral administration of Enterococcus faecalis FK-23 suppresses Th17 cell development and attenuates allergic airway responses in mice. Int. J. Mol. Med. 2012, 30, 248–254. [Google Scholar] [CrossRef]
- Carvalho, J.L.; Miranda, M.; Fialho, A.K.; Castro-Faria-Neto, H.; Anatriello, E.; Keller, A.C.; Aimbire, F. Oral feeding with probiotic Lactobacillus rhamnosus attenuates cigarette smoke-induced COPD in C57Bl/6 mice: Relevance to inflammatory markers in human bronchial epithelial cells. PLoS ONE 2020, 15, e0225560. [Google Scholar] [CrossRef]
- Lai, H.C.; Lin, T.L.; Chen, T.W.; Kuo, Y.L.; Chang, C.J.; Wu, T.R.; Shu, C.C.; Tsai, Y.H.; Swift, S.; Lu, C.C. Gut microbiota modulates COPD pathogenesis: Role of anti-inflammatory Parabacteroides goldsteinii lipopolysaccharide. Gut 2021, 71, 209–321. [Google Scholar] [CrossRef] [PubMed]
- Zelaya, H.; Alvarez, S.; Kitazawa, H.; Villena, J. Respiratory Antiviral Immunity and Immunobiotics: Beneficial Effects on Inflammation-Coagulation Interaction during Influenza Virus Infection. Front. Immunol. 2016, 7, 633. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, W.; Lucey, K.; Jang, S.; Fujimura, K.E.; Rasky, A.; Ting, H.A.; Petersen, J.; Johnson, C.C.; Boushey, H.A.; Zoratti, E.; et al. Lactobacillus johnsonii supplementation attenuates respiratory viral infection via metabolic reprogramming and immune cell modulation. Mucosal. Immunol. 2017, 10, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Rice, T.A.; Brenner, T.A.; Percopo, C.M.; Ma, M.; Keicher, J.D.; Domachowske, J.B.; Rosenberg, H.F. Signaling via pattern recognition receptors NOD2 and TLR2 contributes to immunomodulatory control of lethal pneumovirus infection. Antiviral. Res. 2016, 132, 131–140. [Google Scholar] [CrossRef][Green Version]
- Alvarez, S.; Herrero, C.; Bru, E.; Perdigon, G. Effect of Lactobacillus casei and yogurt administration on prevention of Pseudomonas aeruginosa infection in young mice. J. Food Prot. 2001, 64, 1768–1774. [Google Scholar] [CrossRef]
- Villena, J.; Oliveira, M.L.; Ferreira, P.C.; Salva, S.; Alvarez, S. Lactic acid bacteria in the prevention of pneumococcal respiratory infection: Future opportunities and challenges. Int. Immunopharmacol. 2011, 11, 1633–1645. [Google Scholar] [CrossRef]
- Vieira, A.T.; Rocha, V.M.; Tavares, L.; Garcia, C.C.; Teixeira, M.M.; Oliveira, S.C.; Cassali, G.D.; Gamba, C.; Martins, F.S.; Nicoli, J.R. Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 5(1A). Microbes. Infect. 2016, 18, 180–189. [Google Scholar] [CrossRef]
- Hao, Q.; Dong, B.R.; Wu, T. Probiotics for preventing acute upper respiratory tract infections. Cochrane. Database Syst. Rev. 2015, 7, CD006895. [Google Scholar] [CrossRef]
- Morrow, L.E.; Kollef, M.H.; Casale, T.B. Probiotic prophylaxis of ventilator-associated pneumonia: A blinded, randomized, controlled trial. Am. J. Respir. Crit. Care Med. 2010, 182, 1058–1064. [Google Scholar] [CrossRef]
- d’Ettorre, G.; Ceccarelli, G.; Marazzato, M.; Campagna, G.; Pinacchio, C.; Alessandri, F.; Ruberto, F.; Rossi, G.; Celani, L.; Scagnolari, C.; et al. Challenges in the Management of SARS-CoV2 Infection: The Role of Oral Bacteriotherapy as Complementary Therapeutic Strategy to Avoid the Progression of COVID-19. Front. Med. 2020, 7, 389. [Google Scholar] [CrossRef]
- Dashiff, A.; Junka, R.A.; Libera, M.; Kadouri, D.E. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J. Appl. Microbiol. 2011, 110, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ye, J.; Hou, Y.; Chen, H.; Cao, J.; Zhou, T. Predation Efficacy of Bdellovibrio bacteriovorus on Multidrug-Resistant Clinical Pathogens and Their Corresponding Biofilms. Jpn. J. Infect. Dis. 2017, 70, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Kadouri, D.E.; To, K.; Shanks, R.M.; Doi, Y. Predatory bacteria: A potential ally against multidrug-resistant Gram-negative pathogens. PLoS ONE 2013, 8, e63397. [Google Scholar] [CrossRef] [PubMed]
- Iebba, V.; Totino, V.; Santangelo, F.; Gagliardi, A.; Ciotoli, L.; Virga, A.; Ambrosi, C.; Pompili, M.; De Biase, R.V.; Selan, L.; et al. Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus Cystic fibrosis isolates. Front. Microbiol. 2014, 5, 280. [Google Scholar] [CrossRef]
- Shatzkes, K.; Singleton, E.; Tang, C.; Zuena, M.; Shukla, S.; Gupta, S.; Dharani, S.; Rinaggio, J.; Kadouri, D.E.; Connell, N.D. Examining the efficacy of intravenous administration of predatory bacteria in rats. Sci. Rep. 2017, 7, 1864. [Google Scholar] [CrossRef]
- Shatzkes, K.; Chae, R.; Tang, C.; Ramirez, G.C.; Mukherjee, S.; Tsenova, L.; Connell, N.D.; Kadouri, D.E. Examining the safety of respiratory and intravenous inoculation of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus in a mouse model. Sci. Rep. 2015, 5, 12899. [Google Scholar] [CrossRef]
- Shatzkes, K.; Singleton, E.; Tang, C.; Zuena, M.; Shukla, S.; Gupta, S.; Dharani, S.; Onyile, O.; Rinaggio, J.; Connell, N.D.; et al. Predatory Bacteria Attenuate Klebsiella pneumoniae Burden in Rat Lungs. mBio 2016, 7, e01847-16. [Google Scholar] [CrossRef]
- Schwudke, D.; Linscheid, M.; Strauch, E.; Appel, B.; Zahringer, U.; Moll, H.; Muller, M.; Brecker, L.; Gronow, S.; Lindner, B. The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid A containing alpha-D-Mannoses that replace phosphate residues: Similarities and differences between the lipid As and the lipopolysaccharides of the wild type strain B. bacteriovorus HD100 and its host-independent derivative HI100. J. Biol. Chem. 2003, 278, 27502–27512. [Google Scholar] [CrossRef]
- Dwidar, M.; Monnappa, A.K.; Mitchell, R.J. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 2012, 45, 71–78. [Google Scholar] [CrossRef]
- Iebba, V.; Santangelo, F.; Totino, V.; Nicoletti, M.; Gagliardi, A.; De Biase, R.V.; Cucchiara, S.; Nencioni, L.; Conte, M.P.; Schippa, S. Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS ONE 2013, 8, e61608. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- Braido, F.; Melioli, G.; Candoli, P.; Cavalot, A.; Di Gioacchino, M.; Ferrero, V.; Incorvaia, C.; Mereu, C.; Ridolo, E.; Rolla, G.; et al. The bacterial lysate Lantigen B reduces the number of acute episodes in patients with recurrent infections of the respiratory tract: The results of a double blind, placebo controlled, multicenter clinical trial. Immunol. Lett. 2014, 162, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Braido, F.; Melioli, G.; Cazzola, M.; Fabbri, L.; Blasi, F.; Moretta, L.; Canonica, G.W.; Group, A.S. Sub-lingual administration of a polyvalent mechanical bacterial lysate (PMBL) in patients with moderate, severe, or very severe chronic obstructive pulmonary disease (COPD) according to the GOLD spirometric classification: A multicentre, double-blind, randomised, controlled, phase IV study (AIACE study: Advanced Immunological Approach in COPD Exacerbation). Pulm. Pharmacol. Ther. 2015, 33, 75–80. [Google Scholar] [CrossRef]
- Braido, F.; Tarantini, F.; Ghiglione, V.; Melioli, G.; Canonica, G.W. Bacterial lysate in the prevention of acute exacerbation of COPD and in respiratory recurrent infections. Int. J. Chron. Obstruct. Pulmon. Dis. 2007, 2, 335–345. [Google Scholar] [PubMed]
- Cazzola, M.; Anapurapu, S.; Page, C.P. Polyvalent mechanical bacterial lysate for the prevention of recurrent respiratory infections: A meta-analysis. Pulm. Pharmacol. Ther. 2012, 25, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Cazzola, M.; Noschese, P.; Di Perna, F. Value of adding a polyvalent mechanical bacterial lysate to therapy of COPD patients under regular treatment with salmeterol/fluticasone. Ther. Adv. Respir. Dis. 2009, 3, 59–63. [Google Scholar] [CrossRef]
- Kearney, S.C.; Dziekiewicz, M.; Feleszko, W. Immunoregulatory and immunostimulatory responses of bacterial lysates in respiratory infections and asthma. Ann. Allergy Asthma. Immunol. 2015, 114, 364–369. [Google Scholar] [CrossRef]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.; Harper, D.; et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef]
- Jones-Nelson, O.; Tovchigrechko, A.; Glover, M.S.; Fernandes, F.; Rangaswamy, U.; Liu, H.; Tabor, D.E.; Boyd, J.; Warrener, P.; Martinez, J.; et al. Antibacterial Monoclonal Antibodies Do Not Disrupt the Intestinal Microbiome or Its Function. Antimicrob. Agents. Chemother. 2020, 64, e02347-19. [Google Scholar] [CrossRef]
- DiGiandomenico, A.; Keller, A.E.; Gao, C.; Rainey, G.J.; Warrener, P.; Camara, M.M.; Bonnell, J.; Fleming, R.; Bezabeh, B.; Dimasi, N.; et al. A multifunctional bispecific antibody protects against Pseudomonas aeruginosa. Sci. Transl. Med. 2014, 6, 262ra155. [Google Scholar] [CrossRef]
- Yu, X.Q.; Robbie, G.J.; Wu, Y.; Esser, M.T.; Jensen, K.; Schwartz, H.I.; Bellamy, T.; Hernandez-Illas, M.; Jafri, H.S. Safety, Tolerability, and Pharmacokinetics of MEDI4893, an Investigational, Extended-Half-Life, Anti-Staphylococcus aureus Alpha-Toxin Human Monoclonal Antibody, in Healthy Adults. Antimicrob. Agents. Chemother. 2017, 61, 1. [Google Scholar] [CrossRef]
- Zurawski, D.V.; McLendon, M.K. Monoclonal Antibodies as an Antibacterial Approach Against Bacterial Pathogens. Antibiotics 2020, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- Robbie, G.J.; Criste, R.; Dall’acqua, W.F.; Jensen, K.; Patel, N.K.; Losonsky, G.A.; Griffin, M.P. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrob. Agents. Chemother. 2013, 57, 6147–6153. [Google Scholar] [CrossRef]
- Wilcox, M.H.; Gerding, D.N.; Poxton, I.R.; Kelly, C.; Nathan, R.; Birch, T.; Cornely, O.A.; Rahav, G.; Bouza, E.; Lee, C.; et al. Bezlotoxumab for Prevention of Recurrent Clostridium difficile Infection. N. Engl. J. Med. 2017, 376, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlen, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin. Infect. Dis. 2010, 51 (Suppl. 1), S81–S87. [Google Scholar] [CrossRef] [PubMed]
- Koulenti, D.; Tsigou, E.; Rello, J. Nosocomial pneumonia in 27 ICUs in Europe: Perspectives from the EU-VAP/CAP study. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1999–2006. [Google Scholar] [CrossRef] [PubMed]
- Kollef, M.H.; Chastre, J.; Fagon, J.Y.; Francois, B.; Niederman, M.S.; Rello, J.; Torres, A.; Vincent, J.L.; Wunderink, R.G.; Go, K.W.; et al. Global prospective epidemiologic and surveillance study of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit. Care Med. 2014, 42, 2178–2187. [Google Scholar] [CrossRef]
- Paling, F.P.; Hazard, D.; Bonten, M.J.M.; Goossens, H.; Jafri, H.S.; Malhotra-Kumar, S.; Sifakis, F.; Weber, S.; Kluytmans, J.; for the ASPIRE-ICU Study Team. Association of Staphylococcus aureus Colonization and Pneumonia in the Intensive Care Unit. JAMA Netw. Open. 2020, 3, e2012741. [Google Scholar] [CrossRef]
- Bonten, M.J.; Kollef, M.H.; Hall, J.B. Risk factors for ventilator-associated pneumonia: From epidemiology to patient management. Clin. Infect. Dis. 2004, 38, 1141–1149. [Google Scholar] [CrossRef]
- DiGiandomenico, A.; Sellman, B.R. Antibacterial monoclonal antibodies: The next generation? Curr. Opin. Microbiol. 2015, 27, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Tabor, D.E.; Oganesyan, V.; Keller, A.E.; Yu, L.; McLaughlin, R.E.; Song, E.; Warrener, P.; Rosenthal, K.; Esser, M.; Qi, Y.; et al. Pseudomonas aeruginosa PcrV and Psl, the Molecular Targets of Bispecific Antibody MEDI3902, Are Conserved Among Diverse Global Clinical Isolates. J. Infect. Dis. 2018, 218, 1983–1994. [Google Scholar] [CrossRef] [PubMed]
- Thanabalasuriar, A.; Surewaard, B.G.; Willson, M.E.; Neupane, A.S.; Stover, C.K.; Warrener, P.; Wilson, G.; Keller, A.E.; Sellman, B.R.; DiGiandomenico, A.; et al. Bispecific antibody targets multiple Pseudomonas aeruginosa evasion mechanisms in the lung vasculature. J. Clin. Investig. 2017, 127, 2249–2261. [Google Scholar] [CrossRef] [PubMed]
- Le, H.N.; Quetz, J.S.; Tran, V.G.; Le, V.T.M.; Aguiar-Alves, F.; Pinheiro, M.G.; Cheng, L.; Yu, L.; Sellman, B.R.; Stover, C.K.; et al. MEDI3902 Correlates of Protection against Severe Pseudomonas aeruginosa Pneumonia in a Rabbit Acute Pneumonia Model. Antimicrob. Agents. Chemother. 2018, 62, e02565-17. [Google Scholar] [CrossRef] [PubMed]
- Le, H.N.; Tran, V.G.; Vu, T.T.T.; Gras, E.; Le, V.T.M.; Pinheiro, M.G.; Aguiar-Alves, F.; Schneider-Smith, E.; Carter, H.C.; Sellman, B.R.; et al. Treatment Efficacy of MEDI3902 in Pseudomonas aeruginosa Bloodstream Infection and Acute Pneumonia Rabbit Models. Antimicrob. Agents. Chemother. 2019, 63, e22710-19. [Google Scholar] [CrossRef] [PubMed]
- Aguiar-Alves, F.; Le, H.N.; Tran, V.G.; Gras, E.; Vu, T.; Dong, O.; Quetz, J.S.; Cheng, L.I.; Yu, L.; Sellman, B.R.; et al. Anti-virulence Bispecific Monoclonal Antibody Mediated Protection Against Pseudomonas aeruginosa Ventilator-Associated Pneumonia in a Rabbit Model. Antimicrob. Agents. Chemother. 2021, 66, e02022-21. [Google Scholar] [CrossRef]
- Chastre, J.; François, B.; Bourgeois, M.; Komnos, A.; Ferrer, R.; Rahav, G.; De Schryver, N.; Lepape, A.; Koksal, I.; Luyt, C.-E.; et al. Efficacy, Pharmacokinetics (PK), and Safety Profile of MEDI3902, an Anti-Pseudomonas aeruginosa Bispecific Human Monoclonal Antibody in Mechanically Ventilated Intensive Care Unit Patients; Results of the Phase 2 EVADE Study Conducted by the Public-Private COMBACTE-MAGNET Consortium in the Innovative Medicines Initiative (IMI) Program. Open. Forum. Infect. Dis. 2020, 7, S377–S378. [Google Scholar] [CrossRef]
- Lu, Q.; Eggimann, P.; Luyt, C.E.; Wolff, M.; Tamm, M.; Francois, B.; Mercier, E.; Garbino, J.; Laterre, P.F.; Koch, H.; et al. Pseudomonas aeruginosa serotypes in nosocomial pneumonia: Prevalence and clinical outcomes. Crit. Care. 2014, 18, R17. [Google Scholar] [CrossRef]
- Horn, M.P.; Zuercher, A.W.; Imboden, M.A.; Rudolf, M.P.; Lazar, H.; Wu, H.; Hoiby, N.; Fas, S.C.; Lang, A.B. Preclinical in vitro and in vivo characterization of the fully human monoclonal IgM antibody KBPA101 specific for Pseudomonas aeruginosa serotype IATS-O11. Antimicrob. Agents. Chemother. 2010, 54, 2338–2344. [Google Scholar] [CrossRef]
- Secher, T.; Fauconnier, L.; Szade, A.; Rutschi, O.; Fas, S.C.; Ryffel, B.; Rudolf, M.P. Anti-Pseudomonas aeruginosa serotype O11 LPS immunoglobulin M monoclonal antibody panobacumab (KBPA101) confers protection in a murine model of acute lung infection. J. Antimicrob. Chemother. 2011, 66, 1100–1109. [Google Scholar] [CrossRef]
- Que, Y.A.; Lazar, H.; Wolff, M.; Francois, B.; Laterre, P.F.; Mercier, E.; Garbino, J.; Pagani, J.L.; Revelly, J.P.; Mus, E.; et al. Assessment of panobacumab as adjunctive immunotherapy for the treatment of nosocomial Pseudomonas aeruginosa pneumonia. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1861–1867. [Google Scholar] [CrossRef] [PubMed]
- Tabor, D.E.; Yu, L.; Mok, H.; Tkaczyk, C.; Sellman, B.R.; Wu, Y.; Oganesyan, V.; Slidel, T.; Jafri, H.; McCarthy, M.; et al. Staphylococcus aureus Alpha-Toxin Is Conserved among Diverse Hospital Respiratory Isolates Collected from a Global Surveillance Study and Is Neutralized by Monoclonal Antibody MEDI4893. Antimicrob. Agents. Chemother. 2016, 60, 5312–5321. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Hilliard, J.J.; Shi, Y.; Tkaczyk, C.; Cheng, L.I.; Yu, X.; Datta, V.; Ren, S.; Feng, H.; Zinsou, R.; et al. Assessment of an anti-alpha-toxin monoclonal antibody for prevention and treatment of Staphylococcus aureus-induced pneumonia. Antimicrob. Agents. Chemother. 2014, 58, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Oganesyan, V.; Peng, L.; Damschroder, M.M.; Cheng, L.; Sadowska, A.; Tkaczyk, C.; Sellman, B.R.; Wu, H.; Dall’Acqua, W.F. Mechanisms of neutralization of a human anti-alpha-toxin antibody. J. Biol. Chem. 2014, 289, 29874–29880. [Google Scholar] [CrossRef]
- Francois, B.; Jafri, H.S.; Chastre, J.; Sanchez-Garcia, M.; Eggimann, P.; Dequin, P.F.; Huberlant, V.; Vina Soria, L.; Boulain, T.; Bretonniere, C.; et al. Efficacy and safety of suvratoxumab for prevention of Staphylococcus aureus ventilator-associated pneumonia (SAATELLITE): A multicentre, randomised, double-blind, placebo-controlled, parallel-group, phase 2 pilot trial. Lancet Infect. Dis. 2021, 21, 1313–1323. [Google Scholar] [CrossRef]
- Kollef, M.H.; Betthauser, K.D. Monoclonal antibodies as antibacterial therapies: Thinking outside of the box. Lancet Infect. Dis. 2021, 21, 1201–1202. [Google Scholar] [CrossRef]
- Adjunctive Therapy to Antibiotics in the Treatment of S. Aureus Ventilator-Associated Pneumonia With AR-301. Available online: https://ClinicalTrials.gov/show/NCT03816956 (accessed on 11 January 2022).
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G., Jr. Bacteriophage therapy. Antimicrob. Agents. Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Debarbieux, L.; Leduc, D.; Maura, D.; Morello, E.; Criscuolo, A.; Grossi, O.; Balloy, V.; Touqui, L. Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J. Infect. Dis. 2010, 201, 1096–1104. [Google Scholar] [CrossRef]
- Pabary, R.; Singh, C.; Morales, S.; Bush, A.; Alshafi, K.; Bilton, D.; Alton, E.W.; Smithyman, A.; Davies, J.C. Antipseudomonal Bacteriophage Reduces Infective Burden and Inflammatory Response in Murine Lung. Antimicrob. Agents. Chemother. 2016, 60, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell Host. Microbe. 2017, 22, 38–47.e4. [Google Scholar] [CrossRef] [PubMed]
- Forti, F.; Roach, D.R.; Cafora, M.; Pasini, M.E.; Horner, D.S.; Fiscarelli, E.V.; Rossitto, M.; Cariani, L.; Briani, F.; Debarbieux, L.; et al. Design of a Broad-Range Bacteriophage Cocktail That Reduces Pseudomonas aeruginosa Biofilms and Treats Acute Infections in Two Animal Models. Antimicrob. Agents. Chemother. 2018, 62, e02573-17. [Google Scholar] [CrossRef] [PubMed]
- Chegini, Z.; Khoshbayan, A.; Taati Moghadam, M.; Farahani, I.; Jazireian, P.; Shariati, A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Ryu, C.M.; Lee, J.Y.; Park, J.H.; Yong, D.; Lee, K. In Vivo Application of Bacteriophage as a Potential Therapeutic Agent To Control OXA-66-Like Carbapenemase-Producing Acinetobacter baumannii Strains Belonging to Sequence Type 357. Appl. Environ. Microbiol. 2016, 82, 4200–4208. [Google Scholar] [CrossRef]
- Hua, Y.; Luo, T.; Yang, Y.; Dong, D.; Wang, R.; Wang, Y.; Xu, M.; Guo, X.; Hu, F.; He, P. Phage Therapy as a Promising New Treatment for Lung Infection Caused by Carbapenem-Resistant Acinetobacter baumannii in Mice. Front. Microbiol. 2017, 8, 2659. [Google Scholar] [CrossRef]
- Singla, S.; Harjai, K.; Katare, O.P.; Chhibber, S. Bacteriophage-loaded nanostructured lipid carrier: Improved pharmacokinetics mediates effective resolution of Klebsiella pneumoniae-induced lobar pneumonia. J. Infect. Dis. 2015, 212, 325–334. [Google Scholar] [CrossRef]
- Dufour, N.; Debarbieux, L.; Fromentin, M.; Ricard, J.D. Treatment of Highly Virulent Extraintestinal Pathogenic Escherichia coli Pneumonia With Bacteriophages. Crit. Care Med. 2015, 43, e190–e198. [Google Scholar] [CrossRef]
- Furfaro, L.L.; Payne, M.S.; Chang, B.J. Bacteriophage Therapy: Clinical Trials and Regulatory Hurdles. Front. Cell Infect. Microbiol. 2018, 8, 376. [Google Scholar] [CrossRef]
- Law, N.; Logan, C.; Yung, G.; Furr, C.L.; Lehman, S.M.; Morales, S.; Rosas, F.; Gaidamaka, A.; Bilinsky, I.; Grint, P.; et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 2019, 47, 665–668. [Google Scholar] [CrossRef]
- Maddocks, S.; Fabijan, A.P.; Ho, J.; Lin, R.C.Y.; Ben Zakour, N.L.; Dugan, C.; Kliman, I.; Branston, S.; Morales, S.; Iredell, J.R. Bacteriophage Therapy of Ventilator-associated Pneumonia and Empyema Caused by Pseudomonas aeruginosa. Am. J. Respir. Crit. Care Med. 2019, 200, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Ph 1/2 Study Evaluating Safety and Tolerability of Inhaled AP-PA02 in Subjects with Chronic Pseudomonas Aeruginosa Lung Infections and Cystic Fibrosis. Available online: https://ClinicalTrials.gov/show/NCT04596319 (accessed on 10 January 2022).
- Vazquez, R.; Garcia, E.; Garcia, P. Phage Lysins for Fighting Bacterial Respiratory Infections: A New Generation of Antimicrobials. Front. Immunol. 2018, 9, 2252. [Google Scholar] [CrossRef] [PubMed]
- Witzenrath, M.; Schmeck, B.; Doehn, J.M.; Tschernig, T.; Zahlten, J.; Loeffler, J.M.; Zemlin, M.; Muller, H.; Gutbier, B.; Schutte, H.; et al. Systemic use of the endolysin Cpl-1 rescues mice with fatal pneumococcal pneumonia. Crit. Care Med. 2009, 37, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Raz, A.; Serrano, A.; Hernandez, A.; Euler, C.W.; Fischetti, V.A. Isolation of Phage Lysins That Effectively Kill Pseudomonas aeruginosa in Mouse Models of Lung and Skin Infection. Antimicrob. Agents. Chemother. 2019, 63, e00024-19. [Google Scholar] [CrossRef]
- Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef]
- Zhang, C.; Derrien, M.; Levenez, F.; Brazeilles, R.; Ballal, S.A.; Kim, J.; Degivry, M.C.; Quere, G.; Garault, P.; van Hylckama Vlieg, J.E.; et al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. 2016, 10, 2235–2245. [Google Scholar] [CrossRef]
- Senok, A.C.; Ismaeel, A.Y.; Botta, G.A. Probiotics: Facts and myths. Clin. Microbiol. Infect. 2005, 11, 958–966. [Google Scholar] [CrossRef]
- Sierra, S.; Lara-Villoslada, F.; Sempere, L.; Olivares, M.; Boza, J.; Xaus, J. Intestinal and immunological effects of daily oral administration of Lactobacillus salivarius CECT5713 to healthy adults. Anaerobe 2010, 16, 195–200. [Google Scholar] [CrossRef]
- Lahti, L.; Salonen, A.; Kekkonen, R.A.; Salojarvi, J.; Jalanka-Tuovinen, J.; Palva, A.; Oresic, M.; de Vos, W.M. Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. PeerJ 2013, 1, e32. [Google Scholar] [CrossRef]
- Wang, C.; Nagata, S.; Asahara, T.; Yuki, N.; Matsuda, K.; Tsuji, H.; Takahashi, T.; Nomoto, K.; Yamashiro, Y. Intestinal Microbiota Profiles of Healthy Pre-School and School-Age Children and Effects of Probiotic Supplementation. Ann. Nutr. Metab. 2015, 67, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Rochet, V.; Rigottier-Gois, L.; Sutren, M.; Krementscki, M.N.; Andrieux, C.; Furet, J.P.; Tailliez, P.; Levenez, F.; Mogenet, A.; Bresson, J.L.; et al. Effects of orally administered Lactobacillus casei DN-114 001 on the composition or activities of the dominant faecal microbiota in healthy humans. Br. J. Nutr. 2006, 95, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Goossens, D.; Jonkers, D.; Russel, M.; Stobberingh, E.; Van Den Bogaard, A.; Stockbrügger, R. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: A placebo-controlled study on the onset and duration of effects. Aliment. Pharmacol. Ther. 2003, 18, 495–505. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).