Neurodegenerative Disease Treatment Drug PBT2 Breaks Intrinsic Polymyxin Resistance in Gram-Positive Bacteria
Abstract
1. Introduction
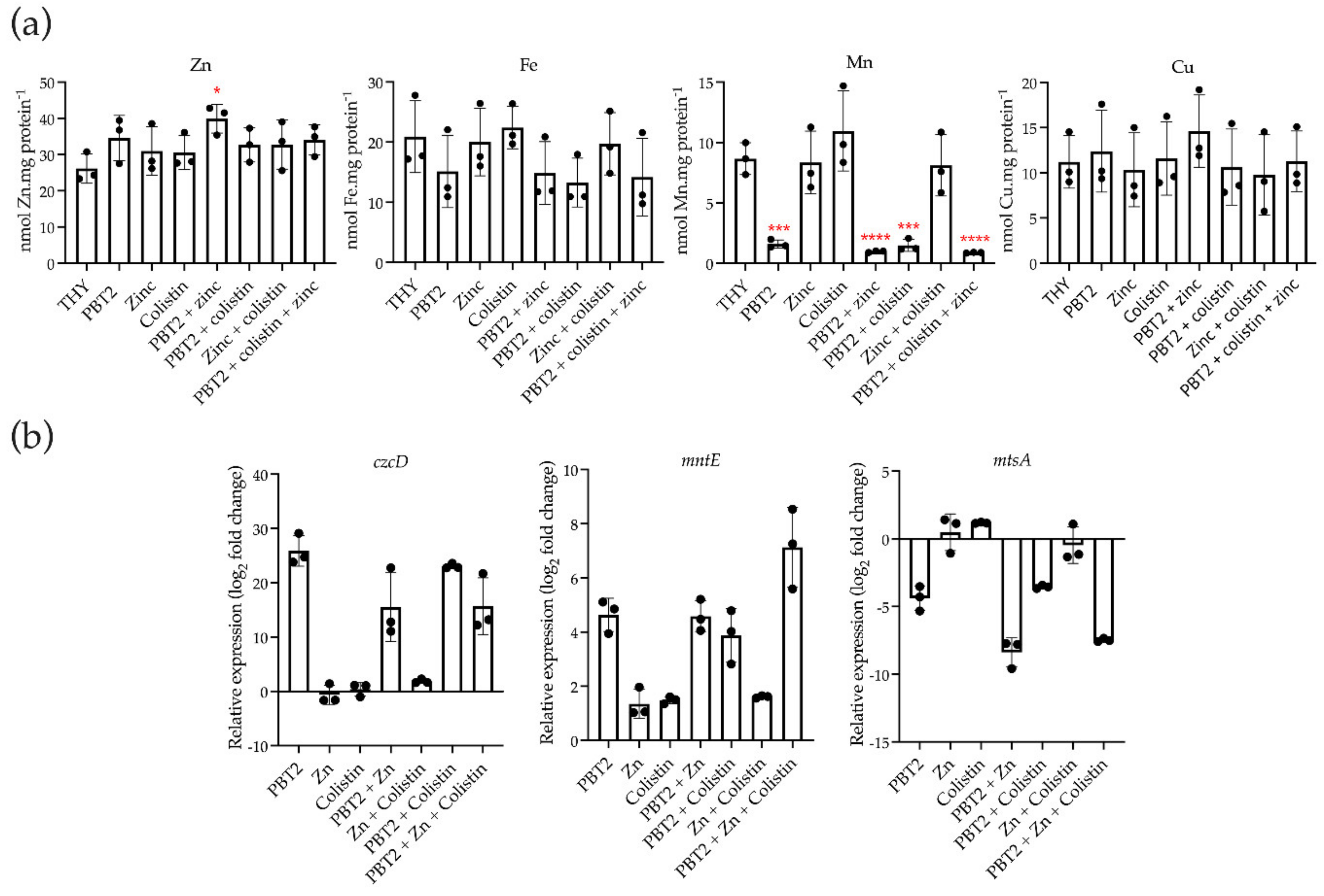
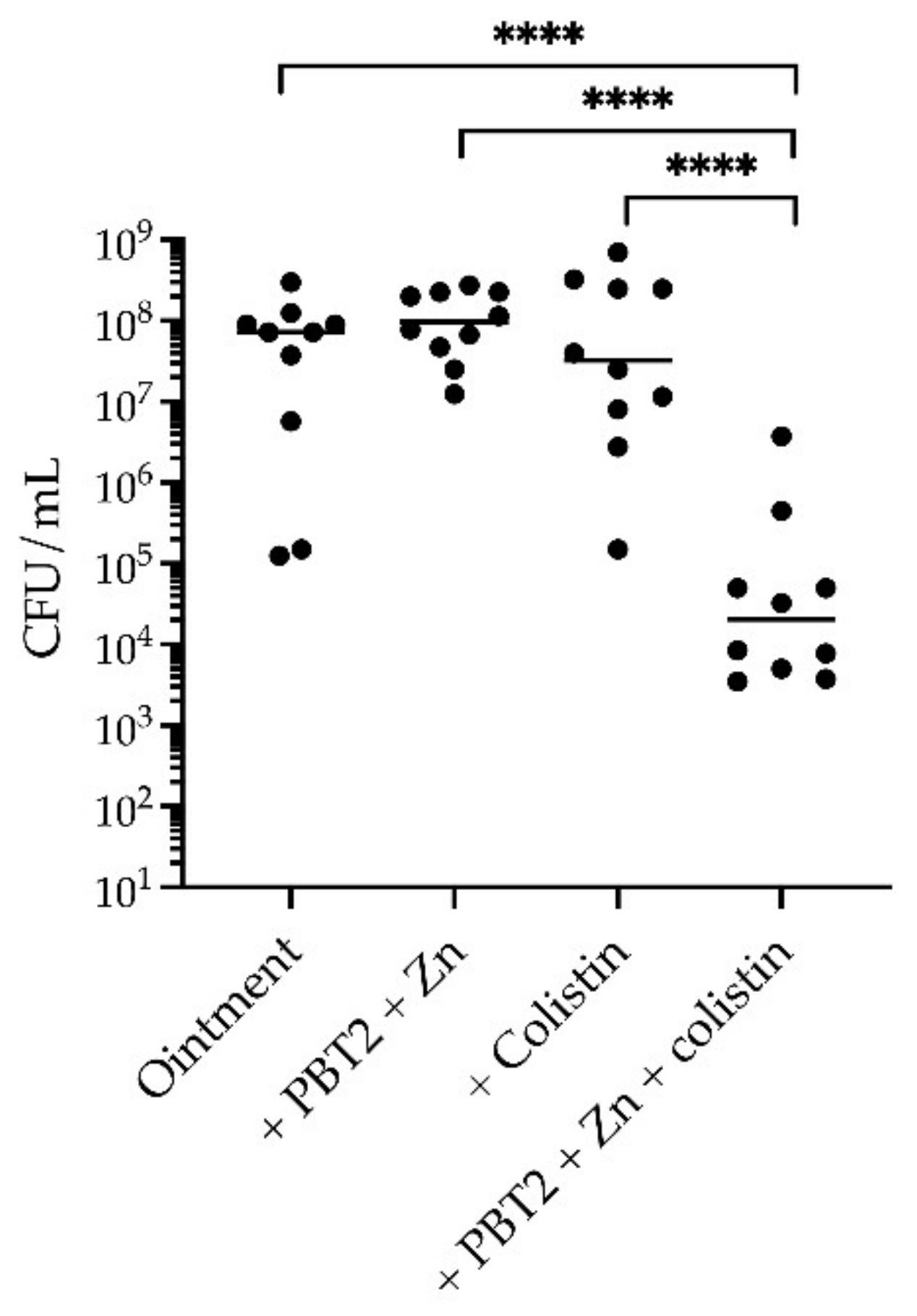
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Construction of 5448ΔmntE–ΔczcD Mutant
4.3. Bacterial Strains, Media, and Growth Conditions
4.4. Minimal Inhibitory Concentration (MIC) Determination
4.5. Bacterial Time-Kill Assays
4.6. Scanning Electron Microscopy (SEM)
4.7. Resistance Development Assays
4.8. Whole Genome Sequencing Analysis
4.9. Growth Analysis
4.10. Inductively Coupled Plasma Mass Spectrometry (ICP-MS)
4.11. RNA Isolation
4.12. Quantitative Real-Time PCR
4.13. Murine Wound Infection Model
4.14. Ethics
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Sabnis, A.; Hagart, K.L.H.; Klockner, A.; Becce, M.; Evans, L.E.; Furniss, R.C.D.; Mavridou, D.A.I.; Murphy, R.; Stevens, M.M.; Davies, J.C.; et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. eLife 2021, 10, e65836. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Cherny, R.A.; Ayton, S.; Finkelstein, D.I.; Bush, A.I.; McColl, G.; Massa, S.M. PBT2 reduces toxicity in a C. elegans model of polyQ aggregation and extends lifespan, reduces striatal atrophy and improves motor performance in the R6/2 mouse model of Huntington’s disease. J. Huntingt. Dis. 2012, 1, 211–219. [Google Scholar] [CrossRef]
- Lannfelt, L.; Blennow, K.; Zetterberg, H.; Batsman, S.; Ames, D.; Harrison, J.; Masters, C.L.; Targum, S.; Bush, A.I.; Murdoch, R.; et al. Safety, efficacy, and biomarker findings of PBT2 in targeting Abeta as a modifying therapy for Alzheimer’s disease: A phase IIa, double-blind, randomised, placebo-controlled trial. Lancet Neurol. 2008, 7, 779–786. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Bohlmann, L.; Conroy, T.; Jen, F.E.; Everest-Dass, A.; Hansford, K.A.; Bolisetti, R.; El-Deeb, I.M.; Forde, B.M.; Phan, M.D.; et al. Repurposing a neurodegenerative disease drug to treat Gram-negative antibiotic-resistant bacterial sepsis. Sci. Transl. Med. 2020, 12, eabb3791. [Google Scholar] [CrossRef]
- Walker, M.J.; Barnett, T.C.; McArthur, J.D.; Cole, J.N.; Gillen, C.M.; Henningham, A.; Sriprakash, K.S.; Sanderson-Smith, M.L.; Nizet, V. Disease manifestations and pathogenic mechanisms of group a Streptococcus. Clin. Microbiol. Rev. 2014, 27, 264–301. [Google Scholar] [CrossRef]
- Lebreton, F.; van Schaik, W.; McGuire, A.M.; Godfrey, P.; Griggs, A.; Mazumdar, V.; Corander, J.; Cheng, L.; Saif, S.; Young, S.; et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. Mbio 2013, 4, e00534-13. [Google Scholar] [CrossRef]
- Coombs, G.W.; Daley, D.A.; Thin, Y.; Pang, S.; Collignon, P.; Bradbury, S.; Gottlieb, T.; Robertson, G.; Branley, J.; Barbaro, D.; et al. Australian group on antimicrobial resistance (AGAR) Australian enterococcal sepsis outcome programme (AESOP) annual report 2016. Commun. Dis. Intell. 2018, 42. [Google Scholar] [CrossRef]
- O’Driscoll, T.; Crank, C.W. Vancomycin-resistant enterococcal infections: Epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 2015, 8, 217–230. [Google Scholar] [CrossRef]
- Cetinkaya, Y.; Falk, P.; Mayhall, C.G. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 2000, 13, 686–707. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.Y.; Perencevich, E.N.; Nair, R.; Nelson, R.E.; Samore, M.; Khader, K.; Chorazy, M.L.; Herwaldt, L.A.; Blevins, A.; Ward, M.A.; et al. Incidence and Outcomes Associated With Infections Caused by Vancomycin-Resistant Enterococci in the United States: Systematic Literature Review and Meta-Analysis. Infect. Control. Hosp. Epidemiol. 2017, 38, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, S.E.; Sakoulas, G.; Perencevich, E.N.; Schwaber, M.J.; Karchmer, A.W.; Carmeli, Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: A meta-analysis. Clin. Infect. Dis. 2003, 36, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Kaasch, A.J.; Barlow, G.; Edgeworth, J.D.; Fowler, V.G.; Hellmich, M.; Hopkins, S.; Kern, W.V.; Llewelyn, M.J.; Rieg, S.; Rodriguez-Bano, J.; et al. Staphylococcus aureus bloodstream infection: A pooled analysis of five prospective, observational studies. J. Infect. 2014, 69, 306–307. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Group A Streptococcal (GAS) Disease for Clinicians 2016. 2018. Available online: https://www.cdc.gov/groupastrep/diseases-hcp/index.html (accessed on 17 November 2021).
- Queensland Health. Invasive Group A Streptococcal Disease: Queensland Health Guidelines for Public Health Units. 2018. Available online: https://www.health.qld.gov.au/cdcg/index/igas (accessed on 17 November 2021).
- Angus, D.; Herd, C.; Stone, C.; Stout, J.; Wieler, M.; Rielmann, R.; Ritchie, C.W.; Dorsey, E.R.; Helles, K.; Kayson, E.; et al. Safety, tolerability, and efficacy of PBT2 in Huntington’s disease: A phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2015, 14, 39–47. [Google Scholar] [CrossRef]
- Villemagne, V.L.; Rowe, C.C.; Barnham, K.J.; Cherny, R.; Woodward, M.; Bozinosvski, S.; Salvado, O.; Bourgeat, P.; Perez, K.; Fowler, C.; et al. A randomized, exploratory molecular imaging study targeting amyloid beta with a novel 8-OH quinoline in Alzheimer’s disease: The PBT2-204 IMAGINE study. Alzheimers Dement. 2017, 3, 622–635. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. M100 Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, version 12.0. 2022. Available online: http://eucast.org (accessed on 8 January 2022).
- Aziz, R.K.; Kotb, M. Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerg. Infect. Dis. 2008, 14, 1511–1517. [Google Scholar] [CrossRef]
- Carrel, M.; Perencevich, E.N.; David, M.Z. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000–2013. Emerg. Infect. Dis. 2015, 21, 1973–1980. [Google Scholar] [CrossRef]
- Bohlmann, L.; de Oliveira, D.M.P.; El-Deeb, I.M.; Brazel, E.B.; Harbison-Price, N.; Ong, C.Y.; Rivera-Hernandez, T.; Ferguson, S.A.; Cork, A.J.; Phan, M.D.; et al. Chemical synergy between ionophore PBT2 and zinc reverses antibiotic resistance. MBio 2018, 9, e02391-18. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Feng, C.W.; Chiu, C.F.; Burne, R.A. cadDX operon of Streptococcus salivarius 57.I. Appl. Environ. Microbiol. 2008, 74, 1642–1645. [Google Scholar] [CrossRef]
- O’Brien, F.G.; Price, C.; Grubb, W.B.; Gustafson, J.E. Genetic characterization of the fusidic acid and cadmium resistance determinants of Staphylococcus aureus plasmid pUB101. J. Antimicrob. Chemother. 2002, 50, 313–321. [Google Scholar] [CrossRef]
- Turner, A.G.; Djoko, K.Y.; Ong, C.Y.; Barnett, T.C.; Walker, M.J.; McEwan, A.G. Group A Streptococcus co-ordinates manganese import and iron efflux in response to hydrogen peroxide stress. Biochem. J. 2019, 476, 595–611. [Google Scholar] [CrossRef]
- Bates, C.S.; Toukoki, C.; Neely, M.N.; Eichenbaum, Z. Characterization of MtsR, a new metal regulator in group A Streptococcus, involved in iron acquisition and virulence. Infect. Immun. 2005, 73, 5743–5753. [Google Scholar] [CrossRef]
- Turner, A.G.; Ong, C.L.; Gillen, C.M.; Davies, M.R.; West, N.P.; McEwan, A.G.; Walker, M.J. Manganese homeostasis in group A Streptococcus is critical for resistance to oxidative stress and virulence. MBio 2015, 6, e00278-15. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Phan, M.D.; Steiner, B.; Zhang, B.; Zuegg, J.; El-Deeb, I.M.; Li, G.; Keller, N.; Brouwer, S.; et al. Rescuing Tetracycline Class Antibiotics for the Treatment of Multidrug-Resistant Acinetobacter baumannii Pulmonary Infection. MBio 2022, 13, e0351721. [Google Scholar] [CrossRef] [PubMed]
- Harbison-Price, N.; Ferguson, S.A.; Heikal, A.; Taiaroa, G.; Hards, K.; Nakatani, Y.; Rennison, D.; Brimble, M.A.; El-Deeb, I.M.; Bohlmann, L.; et al. Multiple Bactericidal Mechanisms of the Zinc Ionophore PBT2. Msphere 2020, 5, e00157-20. [Google Scholar] [CrossRef] [PubMed]
- Brazel, E.B.; Tan, A.; Neville, S.L.; Iverson, A.R.; Udagedara, S.R.; Cunningham, B.A.; Sikanyika, M.; de Oliveira, D.M.P.; Keller, B.; Bohlmann, L.; et al. Dysregulation of Streptococcus pneumoniae zinc homeostasis breaks ampicillin resistance in a pneumonia infection model. Cell Rep. 2022, 38, 110202. [Google Scholar] [CrossRef]
- Trimble, M.J.; Mlynarcik, P.; Kolar, M.; Hancock, R.E.W. Polymyxin: Alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025288. [Google Scholar] [CrossRef]
- Gerlach, D.; Reichardt, W.; Vettermann, S. Extracellular superoxide dismutase from Streptococcus pyogenes type 12 strain is manganese-dependent. FEMS Microbiol. Lett. 1998, 160, 217–224. [Google Scholar] [CrossRef]
- Sobota, J.M.; Imlay, J.A. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. USA 2011, 108, 5402–5407. [Google Scholar] [CrossRef]
- Grifantini, R.; Toukoki, C.; Colaprico, A.; Gryllos, I. Peroxide stimulon and role of PerR in group A Streptococcus. J. Bacteriol. 2011, 193, 6539–6551. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.E.; Lisher, J.P.; Winkler, M.E.; Giedroc, D.P. Perturbation of manganese metabolism disrupts cell division in Streptococcus pneumoniae. Mol. Microbiol. 2017, 104, 334–348. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Gallardo-Godoy, A.; Swarbrick, J.D.; Blaskovich, M.A.T.; Elliott, A.G.; Han, M.L.; Thompson, P.E.; Roberts, K.D.; Huang, J.X.; Becker, B.; et al. Structure, function, and biosynthetic origin of octapeptin antibiotics active against extensively drug-Resistant Gram-negative bacteria. Cell Chem. Biol. 2018, 25, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Barnham, K.J.; Gautier, E.C.L.; Kok, G.B.; Krippner, G. 8-Hydroxy Quinoline Derivatives; (International patent classification no. PCT/WO2004/007461A1); World Intellectual Property Organization: Geneva, Switzerland, 2002. [Google Scholar]
- Ong, C.L.; Gillen, C.M.; Barnett, T.C.; Walker, M.J.; McEwan, A.G. An antimicrobial role for zinc in innate immune defense against group A Streptococcus. J. Infect. Dis. 2014, 209, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Sanderson-Smith, M.; De Oliveira, D.M.P.; Guglielmini, J.; McMillan, D.J.; Vu, T.; Holien, J.K.; Henningham, A.; Steer, A.C.; Bessen, D.E.; Dale, J.B.; et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J. Infect. Dis. 2014, 210, 1325–1338. [Google Scholar] [CrossRef]
- Towers, R.J.; Fagan, P.K.; Talay, S.R.; Currie, B.J.; Sriprakash, K.S.; Walker, M.J.; Chhatwal, G.S. Evolution of sfbI encoding streptococcal fibronectin-binding protein I: Horizontal genetic transfer and gene mosaic structure. J. Clin. Microbiol. 2003, 41, 5398–5406. [Google Scholar] [CrossRef]
- Davies, M.R.; McIntyre, L.; Mutreja, A.; Lacey, J.A.; Lees, J.A.; Towers, R.J.; Duchene, S.; Smeesters, P.R.; Frost, H.R.; Price, D.J.; et al. Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics. Nat. Genet. 2019, 51, 1035–1043. [Google Scholar] [CrossRef]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Eng. Anal. Multicell. Syst. Methods Protoc. 2014, 1151, 165–188. [Google Scholar] [CrossRef]



| Strain | Concentration (μM) | Colistin MIC (μg/mL) | ||||
|---|---|---|---|---|---|---|
| PBT2 | ZnSO4 | CA-MHB | +PBT2 | +ZnSO4 | +PBT2 + ZnSO4 | |
| Group A Streptococcus | ||||||
| 5448 | 7 | 50 | >128 | 32–64 | >128 | ≤0.125–0.25 |
| NS178 | 3.75 | 64 | >128 | 16 | >128 | 1 |
| NS415 | 3.25 | 64 | >128 | 32 | >128 | 0.25 |
| NS179 | 2.25 | 64 | >128 | 32 | >128 | 0.5 |
| NS730 | 3.25 | 64 | >128 | 16 | >128 | ≤0.125 |
| BL16 | 3.25 | 64 | >128 | 16 | >128 | ≤0.125 |
| NS365 | 3.25 | 64 | >128 | 16 | >128 | ≤0.125 |
| NS192 | 3.25 | 64 | >128 | 32 | >128 | 0.25 |
| NS731 | 3.75 | 64 | >128 | 32 | >128 | 1 |
| NS473 | 3.25 | 64 | >128 | 32 | >128 | 0.25 |
| S. aureus | ||||||
| USA300 (MRSA) | 8 | 50 | >128 | 32 | >128 | 2 |
| 25391-9848 | 2 | 60 | >128 | 64 | >128 | 2 |
| 18542-6683 | 2.5 | 60 | >128 | 64 | >128 | 2 |
| 19546-5182 | 2 | 60 | >128 | 32 | >128 | 0.5 |
| 13127-8512 | 2.5 | 60 | >128 | 64 | >128 | 1 |
| 27204-3593 | 3 | 60 | >128 | 32 | >128 | 0.5 |
| E. faecium | ||||||
| RBWH1 (VRE) | 1.75 | 128 | >128 | 16 | >128 | ≤0.125–0.25 |
| GP_044 (VRE) | 4 | 64 | >128 | 128 | >128 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Oliveira, D.M.P.; Keller, B.; Hayes, A.J.; Ong, C.-L.Y.; Harbison-Price, N.; El-Deeb, I.M.; Li, G.; Keller, N.; Bohlmann, L.; Brouwer, S.; et al. Neurodegenerative Disease Treatment Drug PBT2 Breaks Intrinsic Polymyxin Resistance in Gram-Positive Bacteria. Antibiotics 2022, 11, 449. https://doi.org/10.3390/antibiotics11040449
De Oliveira DMP, Keller B, Hayes AJ, Ong C-LY, Harbison-Price N, El-Deeb IM, Li G, Keller N, Bohlmann L, Brouwer S, et al. Neurodegenerative Disease Treatment Drug PBT2 Breaks Intrinsic Polymyxin Resistance in Gram-Positive Bacteria. Antibiotics. 2022; 11(4):449. https://doi.org/10.3390/antibiotics11040449
Chicago/Turabian StyleDe Oliveira, David M. P., Bernhard Keller, Andrew J. Hayes, Cheryl-Lynn Y. Ong, Nichaela Harbison-Price, Ibrahim M. El-Deeb, Gen Li, Nadia Keller, Lisa Bohlmann, Stephan Brouwer, and et al. 2022. "Neurodegenerative Disease Treatment Drug PBT2 Breaks Intrinsic Polymyxin Resistance in Gram-Positive Bacteria" Antibiotics 11, no. 4: 449. https://doi.org/10.3390/antibiotics11040449
APA StyleDe Oliveira, D. M. P., Keller, B., Hayes, A. J., Ong, C.-L. Y., Harbison-Price, N., El-Deeb, I. M., Li, G., Keller, N., Bohlmann, L., Brouwer, S., Turner, A. G., Cork, A. J., Jones, T. R., Paterson, D. L., McEwan, A. G., Davies, M. R., McDevitt, C. A., Itzstein, M. v., & Walker, M. J. (2022). Neurodegenerative Disease Treatment Drug PBT2 Breaks Intrinsic Polymyxin Resistance in Gram-Positive Bacteria. Antibiotics, 11(4), 449. https://doi.org/10.3390/antibiotics11040449








