Escherichia coli Isolated from Organic Laying Hens Reveal a High Level of Antimicrobial Resistance despite No Antimicrobial Treatments
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Bacteriology
4.3. Antimicrobial Susceptibility Test
4.4. Antimicrobial Resistance Profiles
4.5. Analyses of Data
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Boeckel, T.P.; Brower, C.; Gilber, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Layminarayan, R. Global trends in antimicrobial use in food animals. Proc. Nat. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, H.; Cheng, G.; Iqbal, Z.; Ai, X.; Hussain, H.I.; Huang, L.; Dai, M.; Wang, Y.; Liu, Z.; Yuan, Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014, 5, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhaji, N.B.; Isola, T.O. Antimicrobial usage by pastoralists in food animals in North-central Nigeria: The associated socio-cultural drivers for antimicrobials misuse and public health implications. One Health 2018, 6, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Van, T.T.H.; Yidana, Z.; Smooker, P.M.; Coloe, P.J. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J. Glob. Antimicrob. Resist. 2020, 20, 170–177. [Google Scholar] [CrossRef]
- Commission Regulation (EC) No. 889/2008 Laying Down Detailed Rules for the Implementation of Council Regulation (EC) No. 834/2007 on Organic Production and Labelling of Organic Products with Regard to Organic Production, Labelling and Control. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32008R0889 (accessed on 7 March 2022).
- European Commission. Eggs—Dashboard. 2020. Available online: https://ec.europa.eu/info/sites/default/files/food-farming-fisheries/farming/documents/eggs-dashboard_en.pdf (accessed on 7 March 2022).
- Nolan, L.K.; Vaillancourt, J.P.; Barbieri, N.L.; Logue, C.M. Colibacillosis. In Diseases of Poultry, 14th ed.; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., Eds.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2020; pp. 770–830. [Google Scholar]
- Arwoh, M.K.; Kwanga, J.K.P.; Hendriksen, R.S.; Okolocha, E.C.; Thakur, S. Genetic relatedness of multidrug resistant Escherichia coli isolated from humans, chickens and poultry environments. Antimicrob. Resist. Infect. Control 2021, 10, 1–13. [Google Scholar] [CrossRef]
- Voets, G.M.; Fluit, A.C.; Scharringa, J.; Schapendonk, C.; van den Munckhof, T.; Leverstein-van Hall, M.A.; Stuart, J.C. Identical plasmid AmpC beta-lactamase genes and plasmid types in E. coli isolates from patients and poultry meat in the Netherlands. Int. J. Food Microbiol. 2013, 167, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.R.; Collignon, P.; Aarestrup, F.M.; McEwen, S.A.; Hendriksen, R.S.; Hald, T.; Wegener, H.C. Association between antimicrobial resistance in Escherichia coli isolates from food animals and blood stream isolates from humans in Europe: An ecological study. Foodborne Pathog. Dis. 2011, 8, 1295–1301. [Google Scholar] [CrossRef]
- Kluytmans, J.A.; Overdevest, I.T.; Willemsen, I.; Kluytmans-van den Bergh, M.F.; van der Zwaluw, K.; Heck, M.; Rijnsburger, M.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M.; Johnston, B.D.; et al. Extended-spectrum beta-lactamase-producing Escherichia coli from retail chicken meat and humans: Comparison of strains, plasmids, resistance genes, and virulence factors. Clin. Infect. Dis. 2013, 56, 478–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nayme, K.; Barguigua, A.; Diawara, I.; El Otmani, F.; Elmdaghri, N.; Zerouali, K.; Timinouni, M. Occurence of genes encoding anminoglycoside-modifying enzymes in Escherichia coli isolates from chicken meat. Brit. Poult. Sci. 2019, 60, 798–801. [Google Scholar] [CrossRef]
- Scott, H.; Campbell, L.; Harvey, R.; Bischoff, K.; Alali, W.; Barling, K.; Anderson, L. Patterns of Antimicrobial Resistance among Commensal Escherichia coli Isolated from Integrated Multi-site Housing and Worker Cohorts of Humans and Swine. Foodborne Pathog. Dis. 2005, 2, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Blickenstaff, K.; Bodeis-Jones, S.; Gaines, S.A.; Tong, E.; Mcdermott, P.F. Comparison of the Prevalences and Antimicrobial Resistances of Escherichia coli Isolates from Different Retail Meats in the United States, 2002 to 2008. Appl. Environ. Microbiol. 2012, 78, 1701–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, J.M.; Vazquez, B.I.; Fente, C.A.; Barros-Velazquez, J.; Cepeda, A.; Franco, C.M. Evolution of resistance in poultry intestinal Escherichia coli during three commonly used antimicrobial therapeutic treatments in poultry. Poult. Sci. 2008, 87, 1643–1648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Shen, Z.; Zhang, C.; Song, L.; Wang, B.; Shang, J.; Yue, X.; Qu, Z.; Li, X.; Zheng, Y.; et al. Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Vet. Microbiol. 2017, 203, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Trung, N.V.; Jamrozy, D.; Matamoros, S.; Carrique-Mas, J.J.; Mai, H.H.; Hieu, T.Q.; Mai, N.T.N.; Wagenaar, J.A.; Thwaites, G.; Parkhill, J.; et al. Limited contribution of non-intensive chicken farming to ESBL-producing Escherichia coli colonization in humans in Vietnam: An epidemiological and genomic analysis. J. Antimicrob. Chemother. 2019, 74, 561–570. [Google Scholar] [CrossRef] [Green Version]
- Randall, L.P.; Horton, R.H.; Chanter, J.I.; Lemma, F.; Evans, S.J. A decline in the occurrence of extended-spectrum ß-lactamase-producing Escherichia coli in retail chicken meat in the UK between 2013 and 2018. J. Appl. Microbiol. 2020, 130, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Gomis, J.; Marin, P.; Otal, J.; Galecio, J.S.; Martinez-Conesa, C.; Cubero, M.J. Resistance patterns to C and D antibiotic categories for veterinary use of Campylobacter spp., Escherichia coli and Enterococcus spp. commensal isolates from laying hen farms in Spain during 2018. Prev. Vet. Med. 2021, 186, 105222. [Google Scholar] [CrossRef] [PubMed]
- Commission Implementing Decision of 12 November 2013 on the Monitoring and Reporting of Antimicrobial Resistance in Zoonotic and Commensal Bacteria. Off. J. Eur. Union. 2013. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2013:303:0026:0039:EN:PDF (accessed on 7 March 2022).
- Schwaiger, K.; Schmied, E.-M.V.; Bauer, J. Comparative analysis of antibiotic resistance characteristics of Gram-negative bacteria isolated from laying hens and eggs in conventional and organic keeping systems in Bavaria, Germany. Zoonoses Public Health 2008, 55, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Koga, V.L.; Scandorieiro, S.; Vespero, E.C.; Oba, A.; de Brito, B.G.; de Brito, K.C.T.; Nakazato, G.; Kobayashi, R.K.T. Comparison of antibiotic resistance and virulence factors among Escherichia coli isolated from conventional and free-range poultry. BioMed Res. Int. 2015, 618752. [Google Scholar] [CrossRef] [Green Version]
- Pesciaroli, M.; Magistrali, C.F.; Filippini, G.; Epifanio, E.M.; Lovito, C.; Marchi, L.; Maresca, C.; Massacci, F.M.; Orsini, S.; Scoccia, E.; et al. Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than for conventional broilers. Int. J. Food Microbiol. 2020, 314, 108293. [Google Scholar] [CrossRef] [PubMed]
- Catry, B.; Laevens, H.; Devriese, L.A.; Opsomer, G.; de Kruif, A. Antimicrobial resistance in lifestock. J. Vet. Pharmacol. Therap. 2003, 26, 81–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Commission. Agriculture and Rural Development. 2021. Available online: https://ec.europa.eu/info/sites/default/files/food-farming-fisheries/farming/documents/eggs-dashboard_en (accessed on 7 March 2022).
- Dawadi, P.; Bista, S.; Bista, S. Prevalence of colistin-resistant Escherichia coli from poultry in South Asian developing countries. Vet. Med. Int. 2021, 6398838. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Schouler, C.; Souillard, R.; Yousfi, L.; Le Devendec, L.; Lucas, C.; Bougeard, S.; Keita, A.; Fach, P.; Galliot, P.; et al. Diversity of Escherichia coli strains isolated from day-old broiler chicks, their environment and colibacillosis lesions in 80 flocks in France. Vet. Microbiol. 2021, 252, 108923. [Google Scholar] [CrossRef] [PubMed]
- Paudel, S.; Stessl, B.; Hess, C.; Zloch, A.; Hess, M. High genetic diversity among extraintestinal Escherichia coli isolates in pullets and layers revealed by a longitudinal study. BMC Vet. Res. 2016, 12, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Stock, I.; Wiedemann, B. Natural Antibiotic susceptibility of Escherichia coli, Shigella, E. vulneris, and E. hermannii strains. Diagn. Microb. Infect. Dis. 1999, 33, 187–199. [Google Scholar] [CrossRef]
- Oosterik, L.H.; Peeters, L.; Mutuku, I.; Goddeeris, B.M.; Butaye, P. Susceptibility of avian pathogenic Escherichia coli from laying hens in Belgium to antibiotics and disinfectants and integrin prevalence. Avian Dis. 2014, 58, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Husna, A.; Elshabrawy, H.A.; Alam, J.; Runa, N.J.; Badruzzaman, A.T.M.; Banu, N.A.; Al Mamun, M.; Paul, B.; Das, S.; et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci. Rep. 2020, 20, 21999. [Google Scholar] [CrossRef]
- Baron, S.; Jouy, E.; Larvor, E.; Eono, F.; Bougeard, S.; Kempf, I. Impact of third-generation-cephalosporin administration in hatcheries on fecal Escherichia coli antimicrobial resistance in broilers and layers. Antimicrob. Agents Chemother. 2014, 58, 5428–5434. [Google Scholar] [CrossRef] [Green Version]
- Seo, K.W.; Shim, J.B.; Kim, Y.B.; Son, S.H.; Noh, E.B.; Yoon, S.; Lim, S.K.; Lee, Y.J. Impacts and characteristics of antimicrobial resistance of Escherichia coli isolates by administration of third-generation cephalosporins in layer hatcheries. Vet. Microbiol. 2020, 243, 108643. [Google Scholar] [CrossRef]
- Bennett, P.M. Plasmid encoded antibiotic resistance: Acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 2008, 153, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Zou, M.; Ma, P.P.; Liu, W.S.; Liang, X.; Li, X.Y.; Li, Y.Z.; Liu, B.T. Prevalence and antibiotic resistance characteristics of extraintestinal pathogenic Escherichia coli among healthy chickens from farms and live poultry markets in China. Animals 2021, 11, 1112. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Apostolakos, I.; Piccirillo, A. A review on the current situation and challenges of colistin resistance in poultry production. Avian Pathol. 2018, 47, 546–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kempf, I.; Fleury, M.A.; Dirder, D.; Bruneau, M.; Sanders, P.; Chauvin, C.; Madec, J.Y.; Jouy, E. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int. J. Antimicrob. Agents 2013, 42, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Jochum, J.M.; Redweik, G.A.J.; Ott, L.C.; Mellata, M. Bacteria broadly-resistant to last resort antibiotics detected in commercial chicken farms. Microorganisms 2021, 9, 141. [Google Scholar] [CrossRef]
- World Health Organization. WHO Releases the 2010 AWaRE Classification Antibiotics. 2019. Available online: https://www.who.int/news/item/01-10-2019-who-releases-the-2019-aware-classification-antibiotics (accessed on 7 March 2022).
- Kmet, V.; Kmetova, M. High level of quinolone resistance in Escherichia coli from healty chicken broilers. Folia Microbiol. 2009, 55, 79–82. [Google Scholar] [CrossRef]
- Bortolaia, V.; Bisgaard, M.; Bojesen, A.M. Distribution and possible transmission of ampicillin- and nalidixic acid-resistant Escherichia coli within the broiler industry. Vet. Microbiol. 2010, 142, 379–386. [Google Scholar] [CrossRef]
- Callens, B.; Cargnel, M.; Sarrazin, S.; Dewulf, J.; Hoet, B.; Vermeersch, K.; Wattiau, P.; Welby, S. Associations between a decreased veterinary antimicrobial use and resistance in commensal Escherichia coli from Belgian livestock species (2011–2015). Prev. Vet. Med. 2018, 157, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Temmerman, R.; Garmyn, A.; Antonissen, G.; Vananterwerpen, G.; Vanrobaeys, M.; Haesebrouck, F.; Devreese, M. Evaluation of fluoropuinolone resistance in clinical avain pathogenic Escherichia coli isolates from Flanders (Belgium). Antibiotics 2020, 9, 800. [Google Scholar] [CrossRef]
- Kasuba, J.; Appala, K.; Agga, G.E.; Loughrin, J.; Conte, E.D. Anaerobic digestion of livestock and poultry manures spiked with tetracycline antibiotics. J. Environ. Sci. Health B 2020, 55, 135–147. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, C.G.; Zhong, X.H. Survey on sulfonamide antibiotic-resistant genotype and phenotype of avian Escherichia coli in North China. Poult. Sci. 2012, 91, 884–887. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Sandvang, D.; Andersen, S.R.; Seyfarht, A.M.; Porsbo, L.J.; Frimodt-Moller, N.; Heuer, O.E. Detection of sul1, sul2 and sul3 in sulphonamide-resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 2006, 106, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.A.; Garcia-Soto, S.; Hernandez, M.; Barcena, C.; Rodriguez-Lazaro, D.; Ugarte-Ruiz, M.; Dominguez, L. Day-old chicks are a source of antimicrobial resistant bacteria for laying hen farms. Vet. Microbiol. 2019, 230, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Massaccesi, L.; Albini, E.; Massacci, F.R.; Orsini, S.; Tofai, S.; Blasi, F.; Marchi, L.; Petzzotti, G.; Magistrali, C.F. Longitudinal study on antibiotic susceptibility in commensal E. coli from geese raised in free-range production systems. Poult. Sci. 2021, 100, 101230. [Google Scholar] [CrossRef] [PubMed]
- Daehre, K.; Projahn, M.; Semmler, T.; Roesler, U.; Friese, A. Extended-spectrum beta-lactamase-/AmpC beta-lactamase-producing Enterobacteriaceae in broiler farms: Transmission dynamics at farm level. Microb. Drug Resist. 2018, 24, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, J.; Pusz, P.; Bok, E.; Stosik, M.; Baldy-Chudzik, K. The phenotypic and genotypic characteristics of antibiotic resistance in Escherichia coli populations isolated from farm animals with different exposure to antimicrobial agents. Polish J. Microbiol. 2013, 62, 173–179. [Google Scholar] [CrossRef]
- Konicek, C.; Vodrazka, P.; Bartak, P.; Knotek, Z.; Hess, C.; Racka, K.; Hess, M.; Troxler, S. Detection of zoonotic pathogens in wild birds in the cross-border region Austria-Czech Republic. J. Wild. Dis. 2016, 52, 850–886l. [Google Scholar] [CrossRef] [Green Version]
- Barrera, S.; Cardenas, P.; Graham, J.P.; Trueba, G. Changes in dominant Escherichia coli and antimicrobial resistance after 24 h in fecal matter. Microbiologyopen 2019, 8, e00643. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Wang, J.; Zhu, L.; Yang, L.; Yang, R. Distribution characteristics of antibiotic resistant bacteria and genes in fresh and composted manures of livestock farms. Sci. Total Environ. 2019, 695, 1–9. [Google Scholar] [CrossRef]
- Juricova, H.; Matiascovicova, J.; Kubasova, T.; Cejkova, D.; Rychlik, I. The distribution of antibiotic resistance genes in chicken gut microbiota community. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Ballou, A.L.; Ali, R.A.; Mendoza, M.A.; Ellis, J.C.; Hassan, H.M.; Croom, W.J.; Koci, M.D. Development of the chick microbiome: How early exposure influences future microbial diversity. Front. Vet. Sci. 2016, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Moore, R.J.; Stanley, D.; Chousalkar, K.K. The gut microbiota of laying hens and its manipulation with prebiotic and probiotics to enhance gut health and food safety. Appl. Environ. Microbiol. 2020, 86, e00600-20. [Google Scholar] [CrossRef] [PubMed]
- Zloch, A.; Kuchling, S.; Hess, M.; Hess, C. Influence of alternative husbandry systems on postmortem findings and prevalence of important bacteria and parasites in layers monitored from end of rearing until slaughter. Vet. Rec. 2018, 182, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wanger, A.; Chavez, V.; Huang, R.S.P.; Wahed, A.; Actor, J.K.; Dasgupta, A. Chapter 4—Media for the clinical microbiology laboratory. Microbiol. Mol. Diagn. Pathol. 2017, 51–60. [Google Scholar] [CrossRef]
- CLSI. Methods for Antimicrobial Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria Isolated from Animals, 1st ed.; Supplement VET06; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- CLSI. Perfromance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; Standard VET01; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CASFM—Comité de l’Antibiogramme de la Société Française de Microbiologie. Recommendations Vétérinaires. 2018. Available online: https://www.sfm-microbiologie.org/ (accessed on 7 March 2022).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistnat, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
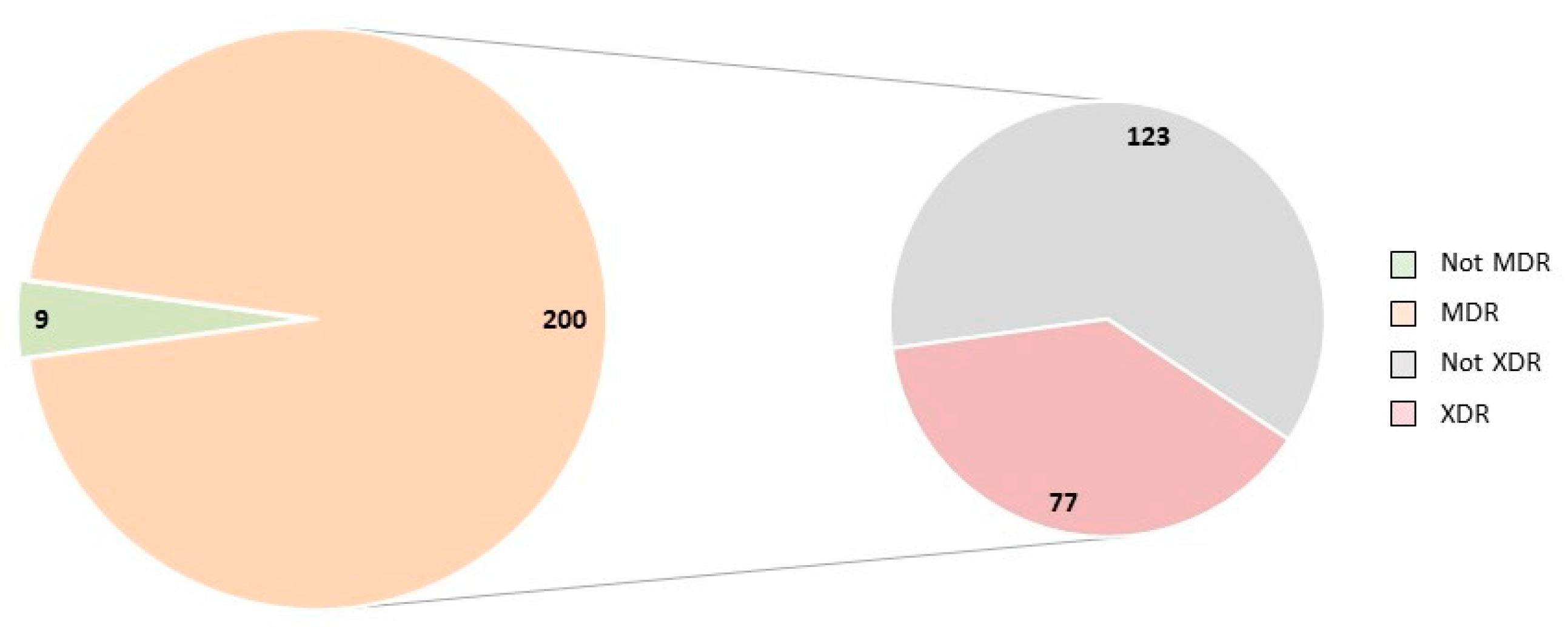

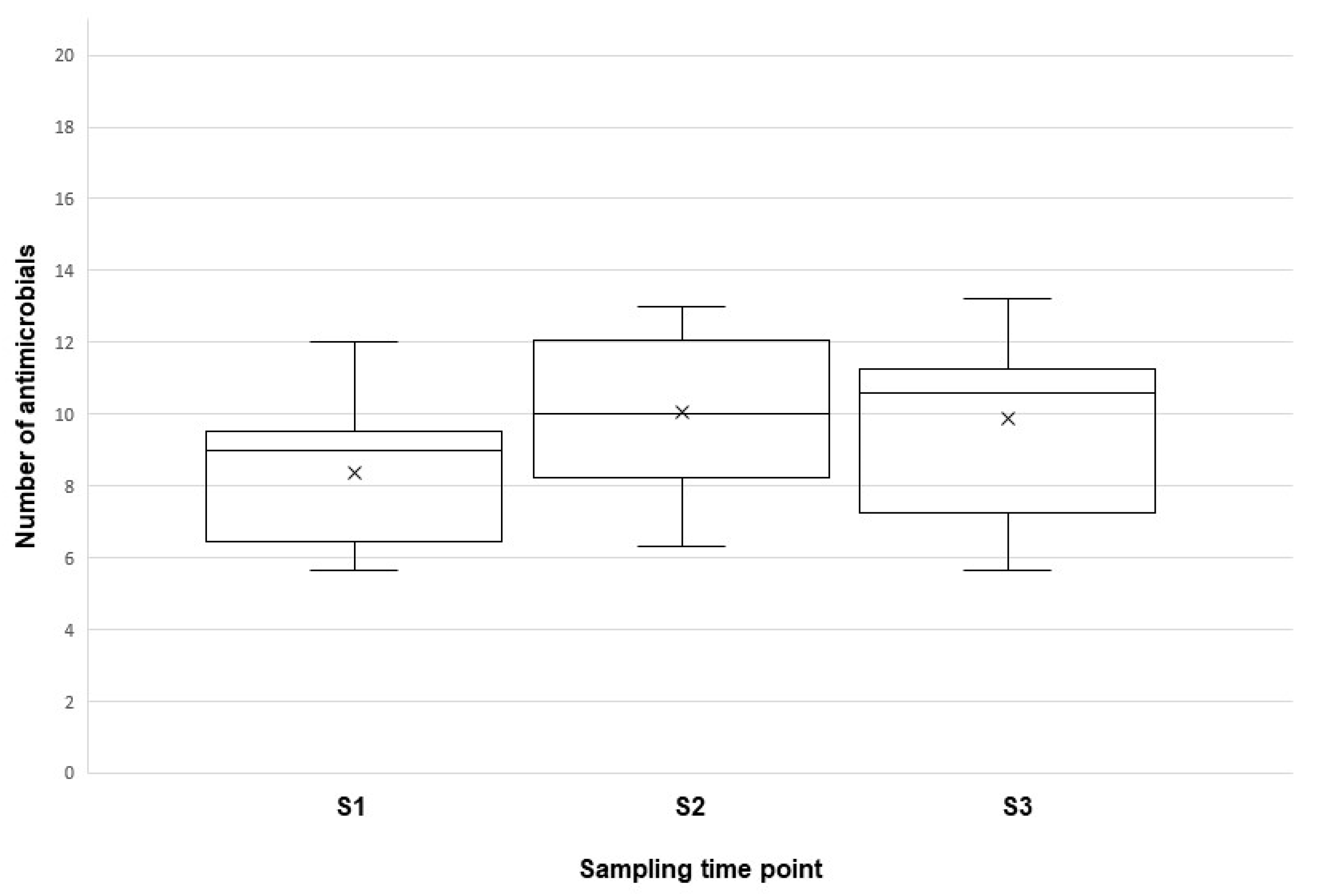
| Flock No. | Pathological Lesions in Ovary/Oviduct | ||
|---|---|---|---|
| S1 | S2 | S3 | |
| 1 | 0/5 b | 2/5 | 0/5 |
| 2 | 0/5 | 0/5 | 0/5 |
| 3 | 0/5 | 0/5 | 2/5 |
| 4 | 0/5 | 1/5 | 4/5 |
| 5 | 0/5 | 0/5 | 0/5 |
| 6 | 0/5 | 0/5 | 0/5 |
| 7 | 0/5 | 0/5 | 1/5 |
| 8 | 0/5 | 0/5 | 0/5 |
| 9 | 0/5 | 0/5 | 1/5 |
| 10 | 0/5 | 0/5 | 2/5 |
| 11 | 0/5 | 0/5 | 0/5 |
| 12 | 0/5 | 0/5 | 4/5 |
| 13 a | 0/5 | 0/5 | 0/5 |
| 14 a | 0/5 | 2/5 | 1/5 |
| 15 | 0/5 | 0/5 | 2/5 |
| 16 | 0/5 | 0/5 | 2/5 |
| 17 | 0/5 | 0/5 | 4/5 |
| 18 | 0/5 | 0/5 | 0/5 |
| Flock | Time Point | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | |||||||||||||
| Heart | Liver | Lung | Ovary | Oviduct | Heart | Liver | Lung | Ovary | Oviduct | Heart | Liver | Lung | Ovary | Oviduct | |
| 1 | 5 (5 × O1) | 1 | 1 | 1 (O2) | 1 | 1 | 4 | ||||||||
| 2 | 4 | 4 (1 × O1) | 1 (O1) | 4 (3 × O1) | |||||||||||
| 3 | 1 | 4 | |||||||||||||
| 4 | 1 | 1 | 1 | 2 | 3 | 9 | 6 | ||||||||
| 5 | 2 | 2 | 6 | 4 (1 × O1) | 3 | ||||||||||
| 6 | 2 | 3 | 3 (1 × O1) | ||||||||||||
| 7 | 2 | 2 | 3 (1 × O1) | ||||||||||||
| 8 | 3 | 3 | 3 | 6 | 3 | ||||||||||
| 9 | 1 | 3 (1 × O1) | 1 (O1) | 1 | 1 | 5 (2 × O1) | 2 | ||||||||
| 10 | 5 (4 × O1) | 2 (1 × O1) | 2 (2 × O1) | ||||||||||||
| 11 | 5 (4 × O1) | 2 | 1 | ||||||||||||
| 12 | 4 (1 × O1) | 1 | 4 | 1 | 1 | 2 | 1 | 1 (O2) | |||||||
| 13 | 2 (1 × O2) | 2 | 1 | 3 | 1 | ||||||||||
| 14 | 3 (2 × O2) | 6 | 3 | 3 | |||||||||||
| 15 | 2 | 3 | 5 | ||||||||||||
| 16 | 3 | 3 | |||||||||||||
| 17 | 15 | ||||||||||||||
| 18 | 3 | ||||||||||||||
| Total | 3 | 2 | 8 | 23 | 11 | 3 | 4 | 4 | 40 | 19 | 0 | 12 | 7 | 49 | 24 |
| Class | Antimicrobial Substance | Concentrations (µg/mL) | MIC (µg/mL) | % of Resistant Isolates (n = 209) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillin, penicillin combination | Amoxicillin | 4 | 8 | 16 | 32 | ≥32 | 67.94% | ||||||
| Amoxicillin/clavulanate | 4/2 | 8/4 | 16/8 | 32/16 | ≥32/16 | 2.39% | |||||||
| Ampicillin | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | >16 | 17.70% | ||||
| Oxacillin | 0.25 | >0.2 | 100.00% | ||||||||||
| Cephalosporin | Cefazolin (1st generation) | 2 | 4 | >4 | 20.10% | ||||||||
| Cefoxitin (2nd generation) | 4 | >4 | 55.98% | ||||||||||
| Cefotaxim (3rd generation) | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ≥4 | 9.57% | |||
| Ceftazidim (3rd generation) | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ≥16 | 82.30% | |||
| Chloramphenicol | Chloramphenicol | 4 | 8 | 16 | 32 | ≥32 | 33.97% | ||||||
| Polypeptide | Colistin | 0.0313 | 0.063 | 0.13 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ≥4 | 73.68% |
| Quinolone | Enrofloxacin | 0.125 | 0.25 | 0.5 | 1 | 2 | ≥2 | 11.96% | |||||
| Nalidixic acid | 4 | 8 | 16 | 32 | 64 | >64 | 91.87% | ||||||
| Aminoglycoside | Gentamicin | 1 | 2 | 4 | 8 | 16 | ≥8 | 6.22% | |||||
| Neomycin | 4 | 8 | 16 | 32 | ≥16 | 11.48% | |||||||
| Streptomycin | 8 | 16 | 32 | 64 | ≥32 | 42.58% | |||||||
| Carbapenem | Imipenem | 1 | 2 | 4 | ≥4 | 9.09% | |||||||
| Tetracycline | Tetracycline | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ≥16 | 53.59% | |||
| Diaminopyrimidine, sulfamethoxazole, and combinations | Trimethoprim | 8 | 16 | ≥16 | 37.80% | ||||||||
| Sulfamethoxazole | 256 | 512 | ≥512 | 95.22% | |||||||||
| Trimethoprim/sulfamethoxazole | 0.5/9.5 | 1/19 | 2/38 | 4/76 | ≥4/76 | 14.83% | |||||||
| Macrolide | Tylosin | 1 | 2 | 4 | 8 | 16 | ≥16 | 100.00% | |||||
| Time Point | Organ | Antimicrobials | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMP | AMX | CAZ | CEZ | CMP | COL | COX | CTX | ENR | GEN | IMP | NAL | NEO | SMO | STR | T/S | TET | TRP | ||
| 1 | oviduct | S | R | R | R | I | S | S | R | S | I | S | R | R | S | R | I | S | R | S |
| 1 | oviduct | S | S | S | R | I | S | R | S | S | I | S | S | R | S | S | R | S | S | S |
| 1 | oviduct | S | S | S | R | S | S | R | S | S | I | S | S | R | S | R | I | S | S | S |
| 1 | oviduct | S | S | S | S | I | S | S | R | S | R | R | R | R | R | R | R | S | I | S |
| 1 | oviduct | S | S | S | S | I | S | S | S | S | S | S | S | S | S | R | S | S | S | S |
| 2 | heart | S | S | R | R | S | S | R | R | S | I | S | S | R | S | R | S | R | R | R |
| 2 | lung | S | S | R | R | S | I | R | R | S | I | S | S | R | I | R | I | R | R | R |
| 2 | ovary | S | I | R | R | S | R | R | R | S | I | S | S | R | I | R | R | R | R | R |
| 2 | ovary | S | S | I | R | S | R | R | S | S | I | S | S | R | I | R | S | R | R | R |
| 2 | ovary | S | S | R | R | S | I | R | R | S | I | S | S | R | S | R | I | S | R | R |
| 2 | oviduct | S | S | S | R | S | S | R | R | R | I | I | R | R | S | R | R | R | R | R |
| 3 | liver | S | S | R | R | S | R | R | S | S | R | S | S | R | R | R | R | S | R | R |
| 3 | ovary | R | R | R | R | R | R | R | S | R | R | R | S | R | R | R | R | R | R | R |
| 3 | ovary | S | S | R | R | S | R | R | R | S | I | I | S | R | I | R | I | S | R | R |
| 3 | ovary | S | S | R | R | S | R | R | S | I | I | S | S | R | I | R | R | S | R | R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hess, C.; Troxler, S.; Jandreski-Cvetkovic, D.; Zloch, A.; Hess, M. Escherichia coli Isolated from Organic Laying Hens Reveal a High Level of Antimicrobial Resistance despite No Antimicrobial Treatments. Antibiotics 2022, 11, 467. https://doi.org/10.3390/antibiotics11040467
Hess C, Troxler S, Jandreski-Cvetkovic D, Zloch A, Hess M. Escherichia coli Isolated from Organic Laying Hens Reveal a High Level of Antimicrobial Resistance despite No Antimicrobial Treatments. Antibiotics. 2022; 11(4):467. https://doi.org/10.3390/antibiotics11040467
Chicago/Turabian StyleHess, Claudia, Salome Troxler, Delfina Jandreski-Cvetkovic, Angelika Zloch, and Michael Hess. 2022. "Escherichia coli Isolated from Organic Laying Hens Reveal a High Level of Antimicrobial Resistance despite No Antimicrobial Treatments" Antibiotics 11, no. 4: 467. https://doi.org/10.3390/antibiotics11040467
APA StyleHess, C., Troxler, S., Jandreski-Cvetkovic, D., Zloch, A., & Hess, M. (2022). Escherichia coli Isolated from Organic Laying Hens Reveal a High Level of Antimicrobial Resistance despite No Antimicrobial Treatments. Antibiotics, 11(4), 467. https://doi.org/10.3390/antibiotics11040467







