A New Variant of the aadE-sat4-aphA-3 Gene Cluster Found in a Conjugative Plasmid from a MDR Campylobacter jejuni Isolate
Abstract
:1. Introduction
2. Results and Discussion
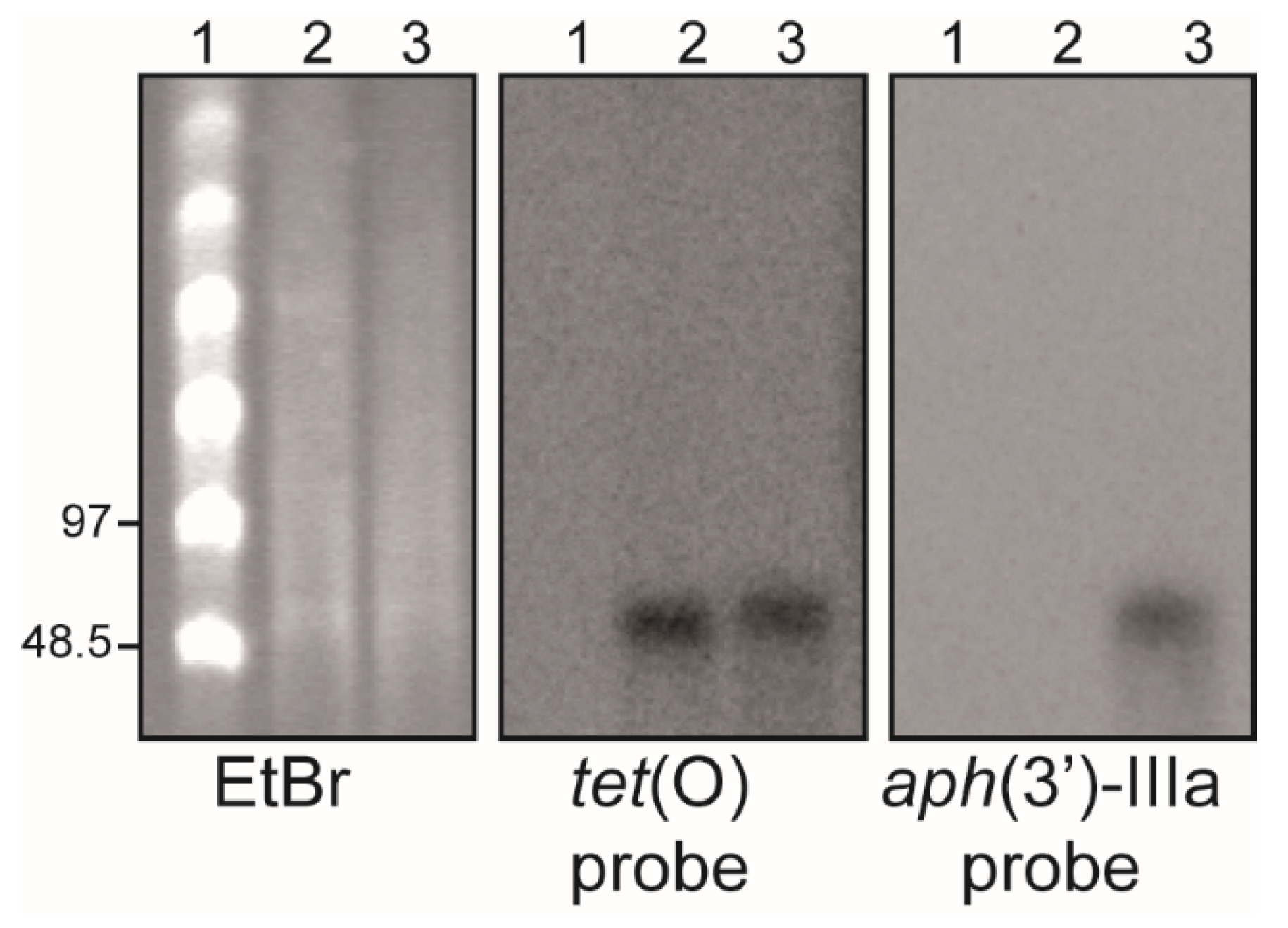
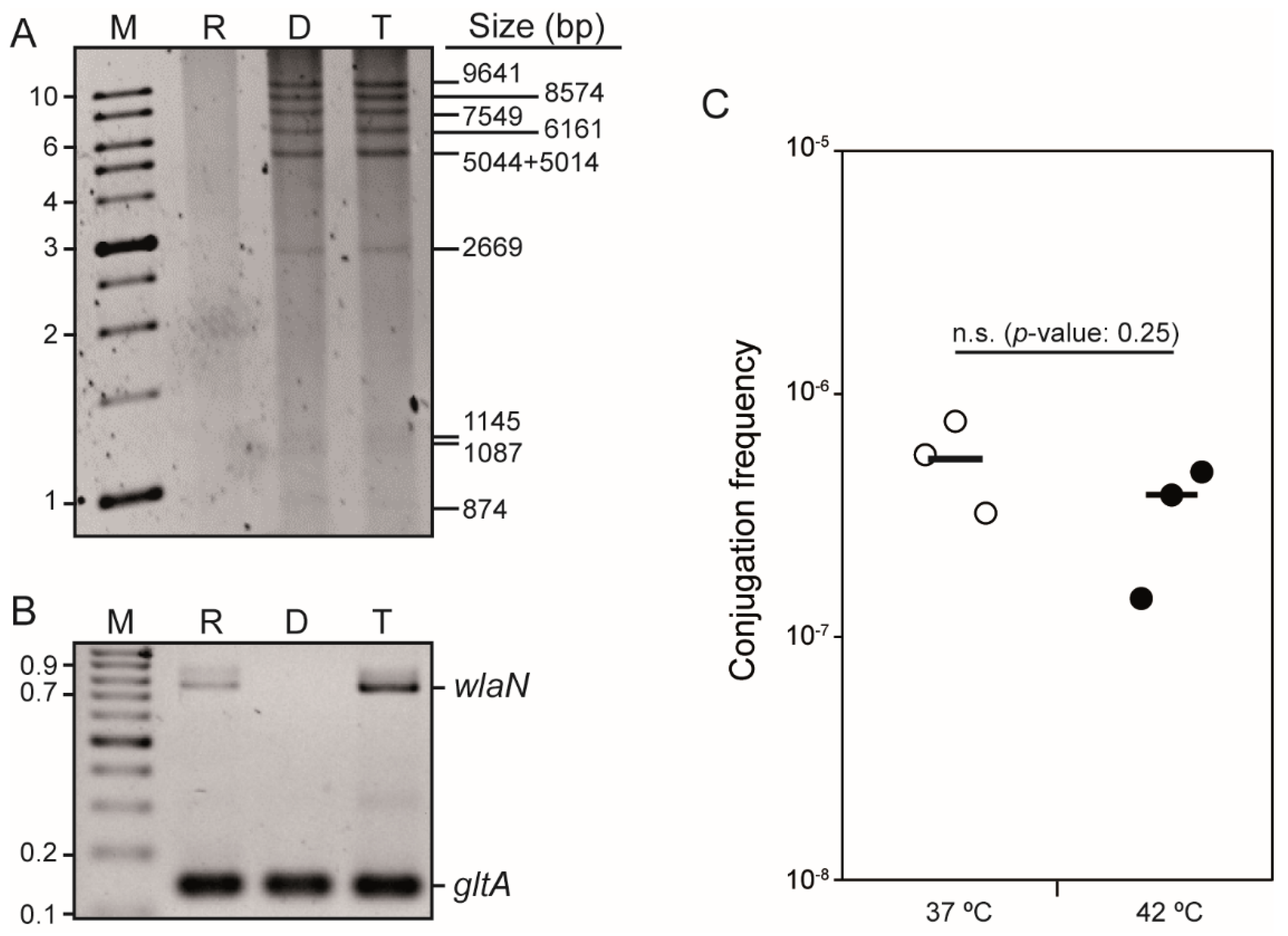
2.1. C. jejuni Strain H01 Carries a Conjugative Plasmid Providing Resistance to Tetracycline, Kanamycin, and Gentamicin
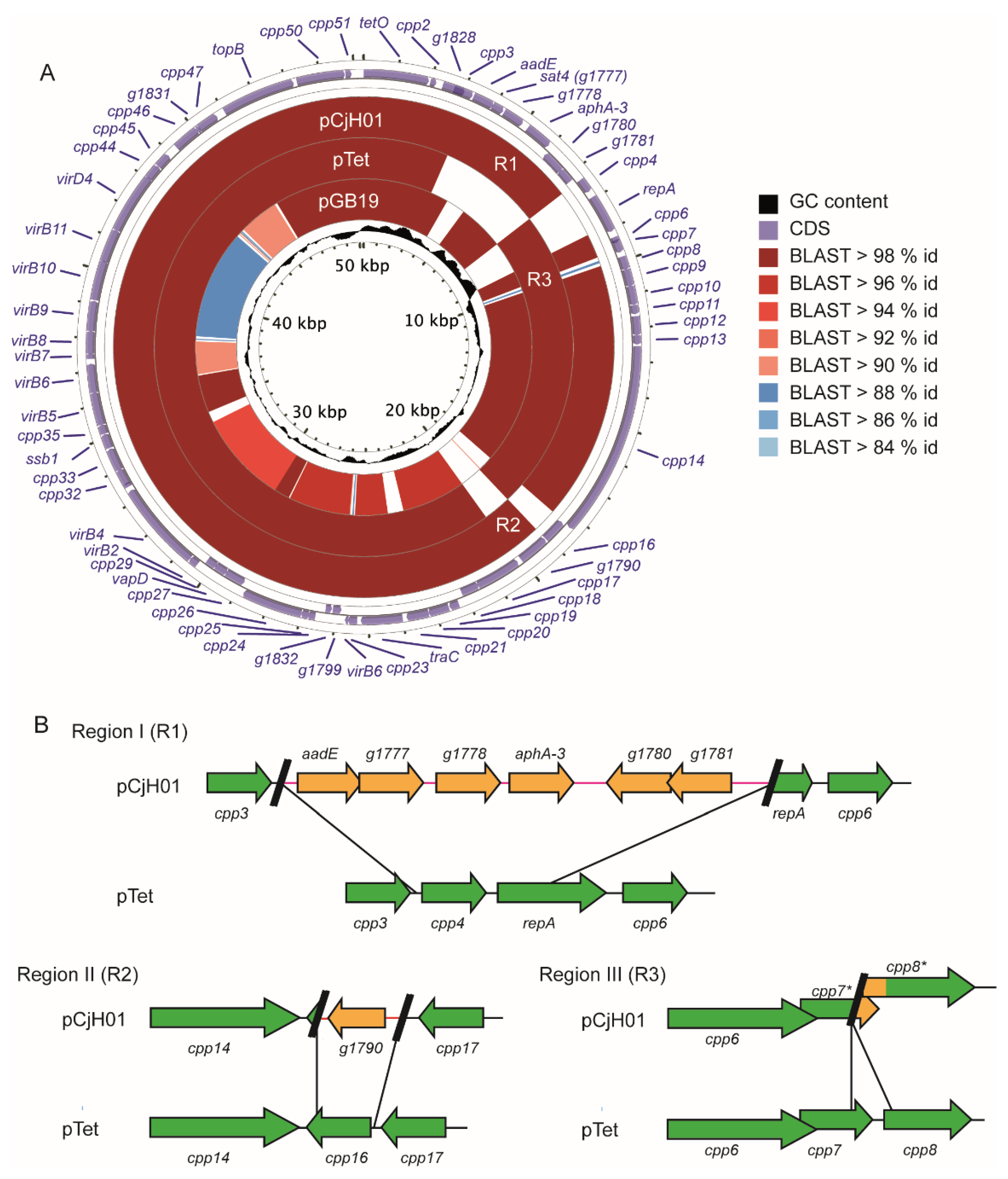
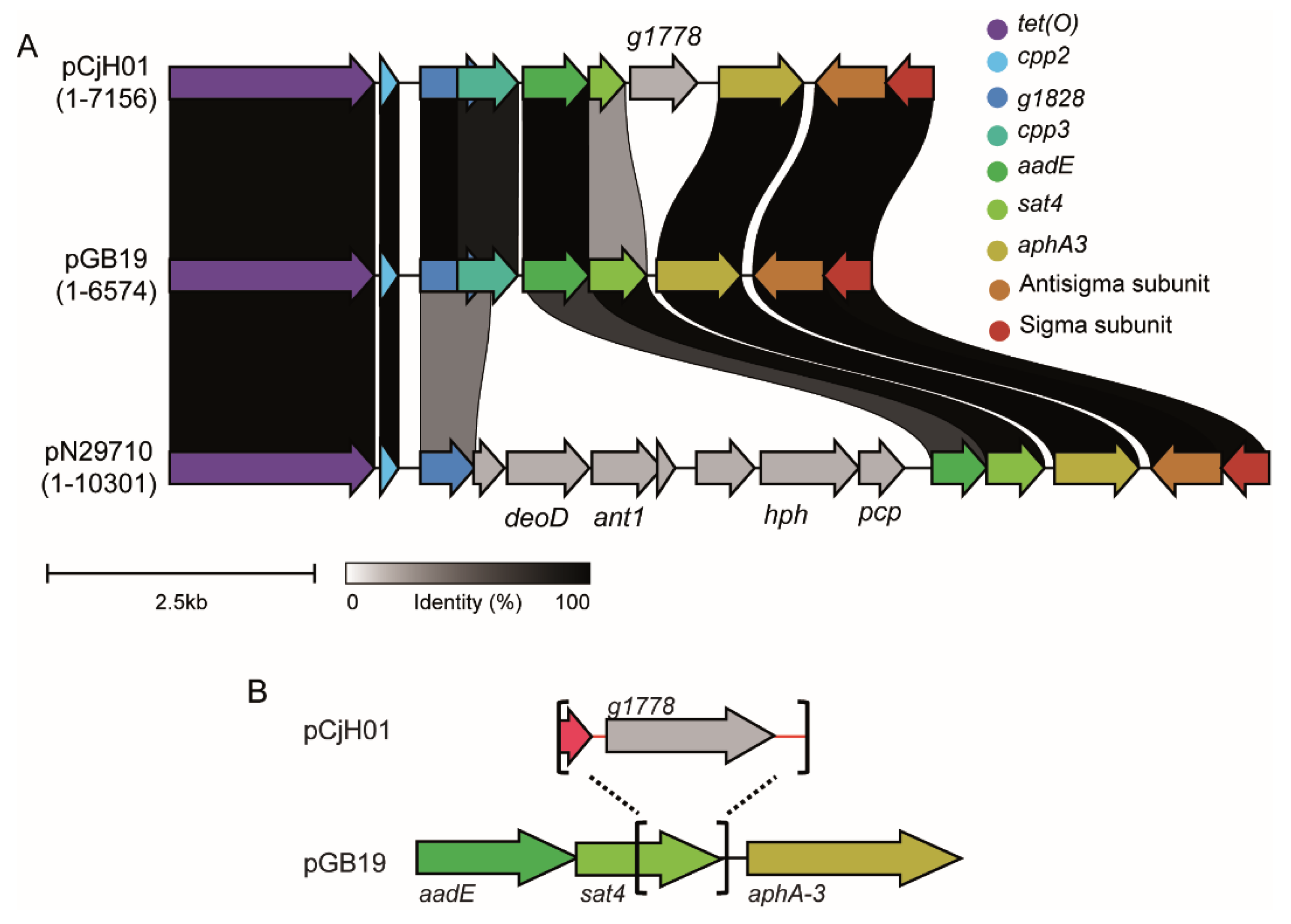
2.2. pCjH01 Is Derived from the pTet Plasmid and Carries a New Variant of the aadE-sat4-aphA-3 Gene Cluster
3. Conclusions
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Growth Conditions
4.2. Mating Experiments
4.3. DNA Techniques
4.4. S1-PFGE and DNA Hybridization
4.5. Genome Sequencing, Plasmid Assembly, and Alignment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar]
- Koga, M.; Gilbert, M.; Takahashi, M.; Li, J.; Koike, S.; Hirata, K.; Yuki, N. Comprehensive analysis of bacterial risk factors for the development of Guillain-Barré Syndrome after Campylobacter jejuni enteritis. J. Infect. Dis. 2006, 193, 547–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, K.T.; Davis, L.M.; DiRita, V.J. Campylobacter jejuni: Molecular biology and pathogenesis. Nat. Rev. Microbiol. 2007, 5, 665–679. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, e06490. [Google Scholar]
- Carattoli, A. Mini review plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Dasti, J.I.; Groß, U.; Pohl, S.; Lugert, R.; Weig, M.; Schmidt-Ott, R. Role of the plasmid-encoded tet(O) gene in tetracycline-resistant clinical isolates of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 2007, 56, 833–837. [Google Scholar] [CrossRef] [Green Version]
- Cuevas-Ferrando, E.; Guirado, P.; Miró, E.; Iglesias-Torrens, Y.; Navarro, F.; Alioto, T.S.; Gómez-Garrido, J.; Madrid, C.; Balsalobre, C. Tetracycline resistance transmission in Campylobacter is promoted at temperatures resembling the avian reservoir. Vet. Microbiol. 2020, 244, 108652. [Google Scholar] [CrossRef]
- Batchelor, R.A.; Pearson, B.M.; Friis, L.M.; Guerry, P.; Wells, J.M. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 2004, 150, 3507–3517. [Google Scholar] [CrossRef] [Green Version]
- Poly, F.; Threadgill, D.; Stintzi, A. Genomic diversity in Campylobacter jejuni: Identification of C. jejuni 81-176-specific genes. J. Clin. Microbiol. 2005, 43, 2330–2338. [Google Scholar] [CrossRef] [Green Version]
- Iglesias-Torrens, Y.; Miró, E.; Guirado, P.; Llovet, T.; Muñoz, C.; Cerdà-Cuéllar, M.; Madrid, C.; Balsalobre, C.; Navarro, F. Population structure, antimicrobial resistance, and virulence-associated genes in Campylobacter jejuni isolated from three ecological niches: Gastroenteritis patients, broilers, and wild birds. Front. Microbiol. 2018, 9, 1676. [Google Scholar] [CrossRef] [Green Version]
- Guirado, P.; Paytubi, S.; Miró, E.; Iglesias-Torrens, Y.; Navarro, F.; Cerdà-Cuéllar, M.; Stephan-Otto Attolini, C.; Balsalobre, C.; Madrid, C. Differential distribution of the wlaN and cgtB genes, associated with Guillain-Barré Syndrome, in Campylobacter jejuni isolates from humans, broiler chickens, and wild birds. Microorganisms 2020, 8, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mukherjee, S.; Hoffmann, M.; Kotewicz, M.L.; Young, S.; Abbott, J.; Luo, Y.; Davidson, M.K.; Allard, M.; McDermott, P.; et al. Whole-genome sequencing of gentamicin-resistant Campylobacter coli isolated from U.S. retail meats reveals novel plasmid-mediated aminoglycoside resistance genes. Antimicrob. Agents Chemother. 2013, 57, 5398–5405. [Google Scholar] [CrossRef] [Green Version]
- Gibreel, A.; Sköld, O.; Taylor, D.E. Characterization of plasmid-mediated aphA-3 kanamycin resistance in Campylobacter jejuni. Microb. Drug Resist. 2004, 10, 98–105. [Google Scholar] [CrossRef]
- Sougakoff, W.; Papadopoulou, B.; Nordmann, P.; Courvalin, P. Nucleotide sequence and distribution of gene tetO encoding tetracycline resistance in Campylobacter coli. FEMS Microbiol. Lett. 1987, 44, 153–159. [Google Scholar] [CrossRef]
- Shaw, K.J.; Rather, P.N.; Hare, R.S.; Miller, G.H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993, 57, 138–163. [Google Scholar] [CrossRef]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Future Microbiol. 2009, 4, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louwen, R.; Nieuwenhuis, E.E.S.; van Marrewijk, L.; Horst-Kreft, D.; de Ruiter, L.; Heikema, A.P.; van Wamel, W.J.B.; Wagenaar, J.A.; Endtz, H.P.; Samsom, J.; et al. Campylobacter jejuni translocation across intestinal epithelial cells is facilitated by ganglioside-like lipooligosaccharide structures. Infect. Immun. 2012, 80, 3307. [Google Scholar] [CrossRef] [Green Version]
- Derbise, A.; De Cespedes, G.; El Solh, N. Nucleotide sequence of the Staphylococcus aureus transposon, Tn5405, carrying aminoglycosides resistance genes. J. Basic Microbiol. 1997, 37, 379–384. [Google Scholar] [CrossRef]
- Qin, S.; Wang, Y.; Zhang, Q.; Chen, X.; Shen, Z.; Deng, F.; Wu, C.; Shen, J. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob. Agents Chemother. 2012, 56, 5332–5339. [Google Scholar] [CrossRef] [Green Version]
- Derbise, A.; Aubert, S.; El Solh, N. Mapping the regions carrying the three contiguous antibiotic resistance genes aadE, sat4, and aphA-3 in the genomes of staphylococci. Antimicrob. Agents Chemother. 1997, 41, 1024–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boerlin, P.; Burnens, A.P.; Frey, J.; Kuhnert, P.; Nicolet, J. Molecular epidemiology and genetic linkage of macrolide and aminoglycoside resistance in Staphylococcus intermedius of canine origin. Vet. Microbiol. 2001, 79, 155–169. [Google Scholar] [CrossRef]
- Werner, G.; Hildebrandt, B.; Witte, W. Aminoglycoside-streptothricin resistance gene cluster aadE-sat4-aphA-3 disseminated among multiresistant isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 2001, 45, 3267–3269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirdnoy, W.; Mason, C.J.; Guerry, P. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob. Agents Chemother. 2005, 49, 2454–2459. [Google Scholar] [CrossRef] [Green Version]
- Paget, M.S. Bacterial sigma factors and anti-sigma factors: Structure, function and distribution. Biomolecules 2015, 5, 1245. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Lai, Y.; Yang, X.; Cao, X.; Hu, Y.; Wang, X.; Wang, H. Genetic environments and related transposable elements of novel cfr(C) variants in Campylobacter coli isolates of swine origin. Vet. Microbiol. 2020, 247, 108792. [Google Scholar] [CrossRef] [PubMed]
- Burton, A.T.; DeLoughery, A.; Li, G.W.; Kearns, D.B. Transcriptional regulation and mechanism of sigN (ZpdN), a pBS32-encoded sigma factor in Bacillus subtilis. MBio 2019, 10, e01899-19. [Google Scholar] [CrossRef] [Green Version]
- Byrd, D.R.; Matson, S.W. Nicking by transesterification: The reaction catalysed by a relaxase. Mol. Microbiol. 1997, 25, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.K.; Ramachandran, G.; Ramos-Ruiz, R.; Peiró-Pastor, R.; Abia, D.; Wu, L.J.; Meijer, W.J.J. Mobility of the native Bacillus subtilis conjugative plasmid pLS20 Is regulated by intercellular signaling. PLoS Genet. 2013, 9, e1003892. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.K.; Meijer, W.J.J. Diverse regulatory circuits for transfer of conjugative elements. FEMS Microbiol. Lett. 2014, 358, 119–128. [Google Scholar] [CrossRef] [PubMed]
- López-Fuentes, E.; Torres-Tejerizo, G.; Cervantes, L.; Brom, S. Genes encoding conserved hypothetical proteins localized in the conjugative transfer region of plasmid pRet42a from Rhizobium etli CFN42 participate in modulating transfer and affect conjugation from different donors. Front. Microbiol. 2014, 5, 793. [Google Scholar] [PubMed] [Green Version]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global Epidemiology of Campylobacter Infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front. Microbiol. 2020, 11, 761. [Google Scholar] [CrossRef]
- Barton, B.M.; Harding, G.P.; Zuccarelli, A.J. A general method for detecting and sizing large plasmids. Anal. Biochem. 1995, 226, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.F.; Russell, D.W. (Eds.) Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 1–3. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Arantes, A.S.; Stothard, P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics 2012, 13, 202. [Google Scholar] [CrossRef] [Green Version]
- Gilchrist, C.L.M.; Chooi, Y.H. Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar]
- Obeng, A.S.; Rickard, H.; Sexton, M.; Pang, Y.; Peng, H.; Barton, M. Antimicrobial susceptibilities and resistance genes in Campylobacter strains isolated from poultry and pigs in Australia. J. Appl. Microbiol. 2012, 113, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Crespo, M.D.; Altermann, E.; Olson, J.; Miller, W.G.; Chandrashekhar, K.; Kathariou, S. Novel plasmid conferring kanamycin and tetracycline resistance in the turkey-derived Campylobacter jejuni strain 11601MD. Plasmid 2016, 86, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Koolman, L.; Whyte, P.; Burgess, C.; Bolton, D. Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 2015, 12, 424–432. [Google Scholar] [CrossRef] [PubMed]
| Gene (ORF) | AA(MW) | Putative Function * |
|---|---|---|
| tetO | 639 (72.5) | Tetracycline resistance ribosomal protection protein |
| cpp2 | 57 (6.7) | Conjugal transfer protein |
| g1828 | 221 (25.5) | Relaxase/mobilization nuclease-domain-containing protein |
| cpp3 | 190 (22.1) | Hypothetical protein |
| aadE | 206 (24.3) | Aminoglycoside 6 adenyltransferase. |
| sat4 | 113 (13.4) | Streptothricin acetyltransferase |
| g1778 | 209 (23.9) | Aminoglycoside phosphotransferase |
| aphA-3 | 264 (31) | Aminoglycoside O phosphotransferase APH(3′)IIIa |
| g1780 | 221 (25.1) | Anti sigma factor (Zinc-finger domain containing protein) |
| g1781 | 150 (17.1) | Sigma 70 family RNA polymerase sigma factor |
| cpp6 | 58 (7.0) | Hypothetical protein |
| cpp7 | 96 (11.9) | Hypothetical protein |
| cpp8 | 117 (13.4) | Hypothetical protein |
| cpp9 | 170 (19.6) | Hypothetical protein |
| cpp10 | 185 (22.1) | Hypothetical protein |
| cpp11 | 88 (10.6) | Hypothetical protein |
| cpp12 | 186 (21.5) | ParA hypothetical protein |
| cpp13 | 88 (10.2) | Hypothetical protein |
| cpp14 | 1932 (224.3) | DEAD/DEAH box like helicase |
| g1790 | 314 (37.3) | HTH transcriptional regulator, XRE family |
| cpp17 | 462 (54.0) | Relaxase/mobilization domain containing protein |
| cpp18 | 183 (21.1) | Hypothetical protein |
| cpp19 | 93 (11.5) | Hypothetical protein |
| cpp20 | 203 (23.9) | NTPase. Hypothetical protein |
| cpp21 | 217 (26.0) | Hypothetical protein |
| traC | 408 (47.1) | DNA primase, TraC family |
| cpp23 | 87 (9.7) | EexN family lipoprotein. IncN-type entry exclusion |
| virB6 | 32 (3.6) | Truncated VirB6 protein |
| g1799 | 85 (10.2) | Hypothetical protein |
| g1832 | 61 (7.4) | Hypothetical protein |
| cpp24 | 72 (8.1) | Type-II toxin-antitoxin system HicB family antitoxin |
| cpp25 | 67 (7.9) | Type-II toxin-antitoxin system HicA family toxin |
| cpp26 | 597 (69.0) | AAA family ATPase |
| cpp27 | 204 (26.6) | Recombinase family protein |
| vapD | 125 (15.0) | Virulence associated protein |
| cpp29 | 107 (12.6) | Hypothetical protein |
| virB2 | 87 (9.2) | TrbC/VirB2 family protein. Conjugal transfer protein TraC |
| virB4 | 992 (106.3) | VirB4 family type IV secretion/conjugal transfer ATPase |
| cpp32 | 188 (21.8) | Rha family transcriptional regulator/phage regulatory protein |
| cpp33 | 221 (25.6) | Hypothetical protein |
| ssb1 | 141 (15.8) | single-stranded DNA-binding protein |
| cpp35 | 91 (10.7) | Hypothetical protein |
| virB5 | 323 (37.2) | Type IV secretion system protein |
| virB6 | 332 (35.5) | Type IV secretion system protein. TrbL/VirB6 family protein |
| virB7 | 56 (6.3) | Hypothetical protein |
| virB8 | 220 (25) | Type IV secretion system protein |
| virB9 | 295 (34.1) | P-type conjugative transfer protein VirB9 |
| virB10 | 391 (43.1) | Type IV secretion system protein VirB10. TrbI/VirB10 family protein |
| virB11 | 330 (37.6) | P-type DNA transfer ATPase VirB11 |
| virD4 | 603 (63.7) | Type IV secretory system conjugative DNA transfer family protein |
| cpp44 | 145 (16.8) | cag pathogenicity island protein |
| cpp45 | 254 (29.4) | Hypothetical protein |
| cpp46 | 265 (30.6) | Hypothetical protein |
| g1831 | 30 (3.9) | Hypothetical protein |
| cpp47 | 206 (23.8) | Hypothetical protein |
| topB | 730 (84) | DNA topoisomerase III |
| cpp50 | 473 (57) | Hypothetical protein |
| cpp51 | 59 (7.1) | Hypothetical protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guirado, P.; Miró, E.; Iglesias-Torrens, Y.; Navarro, F.; Campoy, S.; Alioto, T.S.; Gómez-Garrido, J.; Madrid, C.; Balsalobre, C. A New Variant of the aadE-sat4-aphA-3 Gene Cluster Found in a Conjugative Plasmid from a MDR Campylobacter jejuni Isolate. Antibiotics 2022, 11, 466. https://doi.org/10.3390/antibiotics11040466
Guirado P, Miró E, Iglesias-Torrens Y, Navarro F, Campoy S, Alioto TS, Gómez-Garrido J, Madrid C, Balsalobre C. A New Variant of the aadE-sat4-aphA-3 Gene Cluster Found in a Conjugative Plasmid from a MDR Campylobacter jejuni Isolate. Antibiotics. 2022; 11(4):466. https://doi.org/10.3390/antibiotics11040466
Chicago/Turabian StyleGuirado, Pedro, Elisenda Miró, Yaidelis Iglesias-Torrens, Ferran Navarro, Susana Campoy, Tyler Scott Alioto, Jessica Gómez-Garrido, Cristina Madrid, and Carlos Balsalobre. 2022. "A New Variant of the aadE-sat4-aphA-3 Gene Cluster Found in a Conjugative Plasmid from a MDR Campylobacter jejuni Isolate" Antibiotics 11, no. 4: 466. https://doi.org/10.3390/antibiotics11040466
APA StyleGuirado, P., Miró, E., Iglesias-Torrens, Y., Navarro, F., Campoy, S., Alioto, T. S., Gómez-Garrido, J., Madrid, C., & Balsalobre, C. (2022). A New Variant of the aadE-sat4-aphA-3 Gene Cluster Found in a Conjugative Plasmid from a MDR Campylobacter jejuni Isolate. Antibiotics, 11(4), 466. https://doi.org/10.3390/antibiotics11040466







