Antimicrobial Peptides: From Design to Clinical Application
Abstract
1. Introduction
2. Antibiotic Action and Resistance
3. How Bacteria Acquire Antibiotic Resistance Genes
4. Alternative Antibiotics: Antimicrobial Peptides
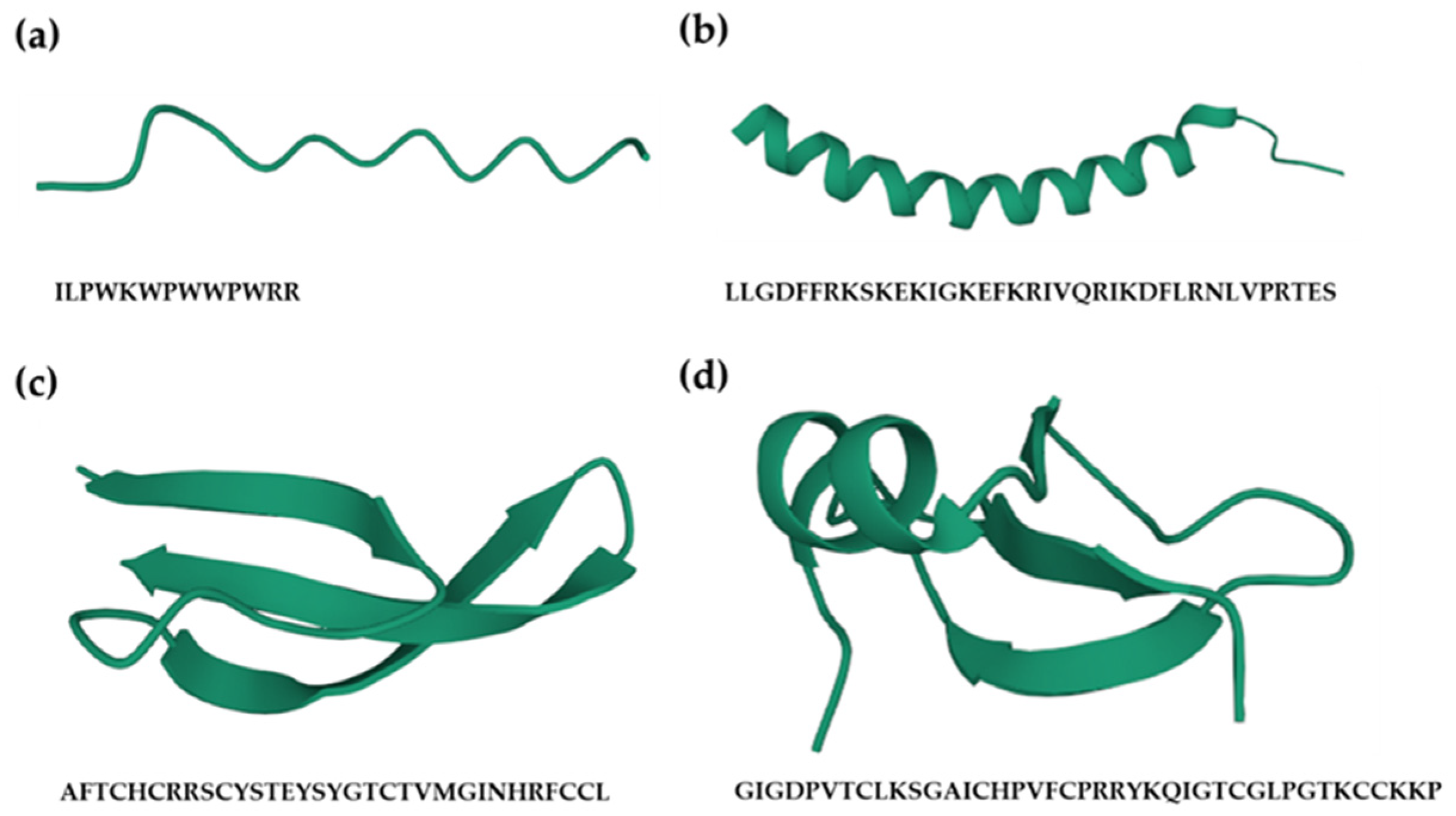
4.1. Structures of AMPs
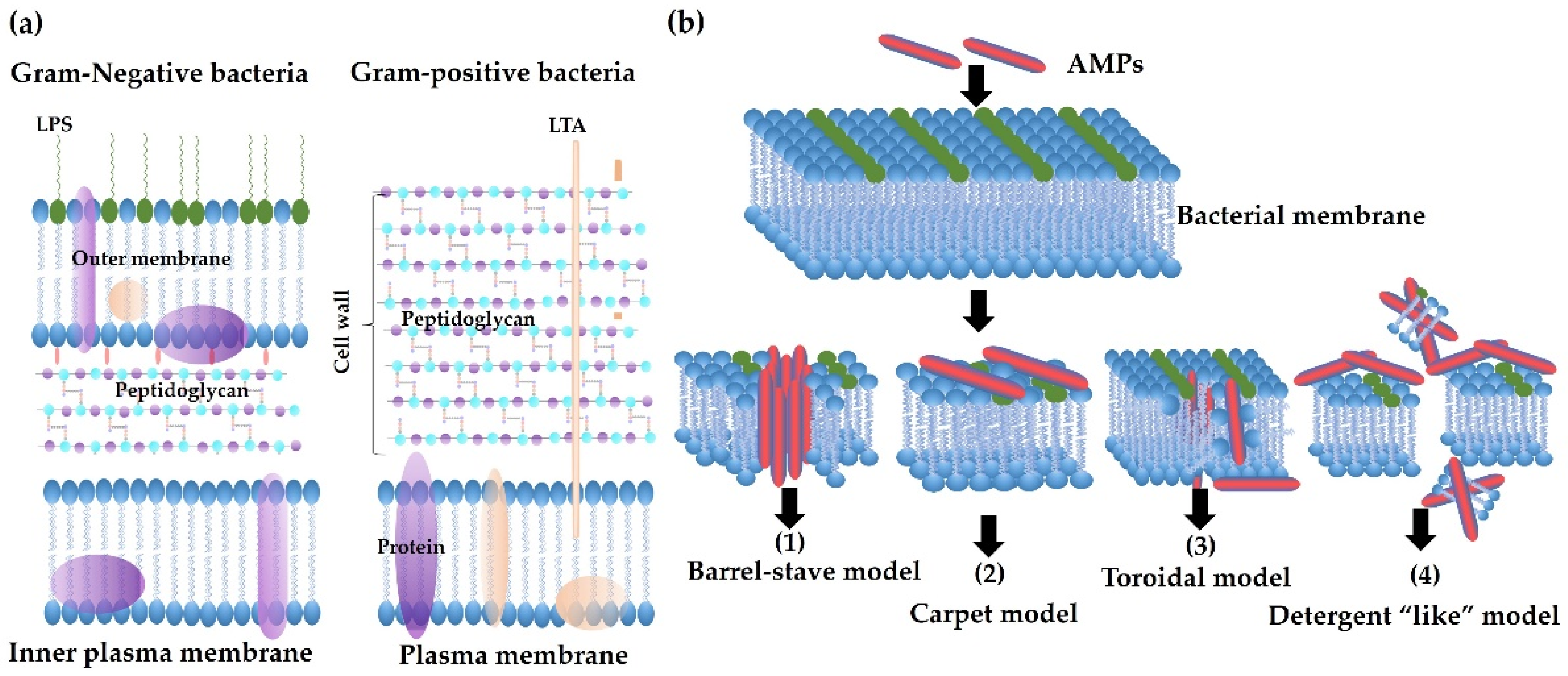
4.2. Killing Mechanisms of AMPs
4.3. Immunomodulatory Function of AMPs
4.4. Other Functions of AMPs
5. Design and Optimization of AMPs
5.1. Natural Peptides
5.2. Signaling Peptide-Derived AMPs
5.3. Structural Modification-Hybridization, Shorten, or Circulation
5.4. In Silico Design
6. Optimization of AMPs
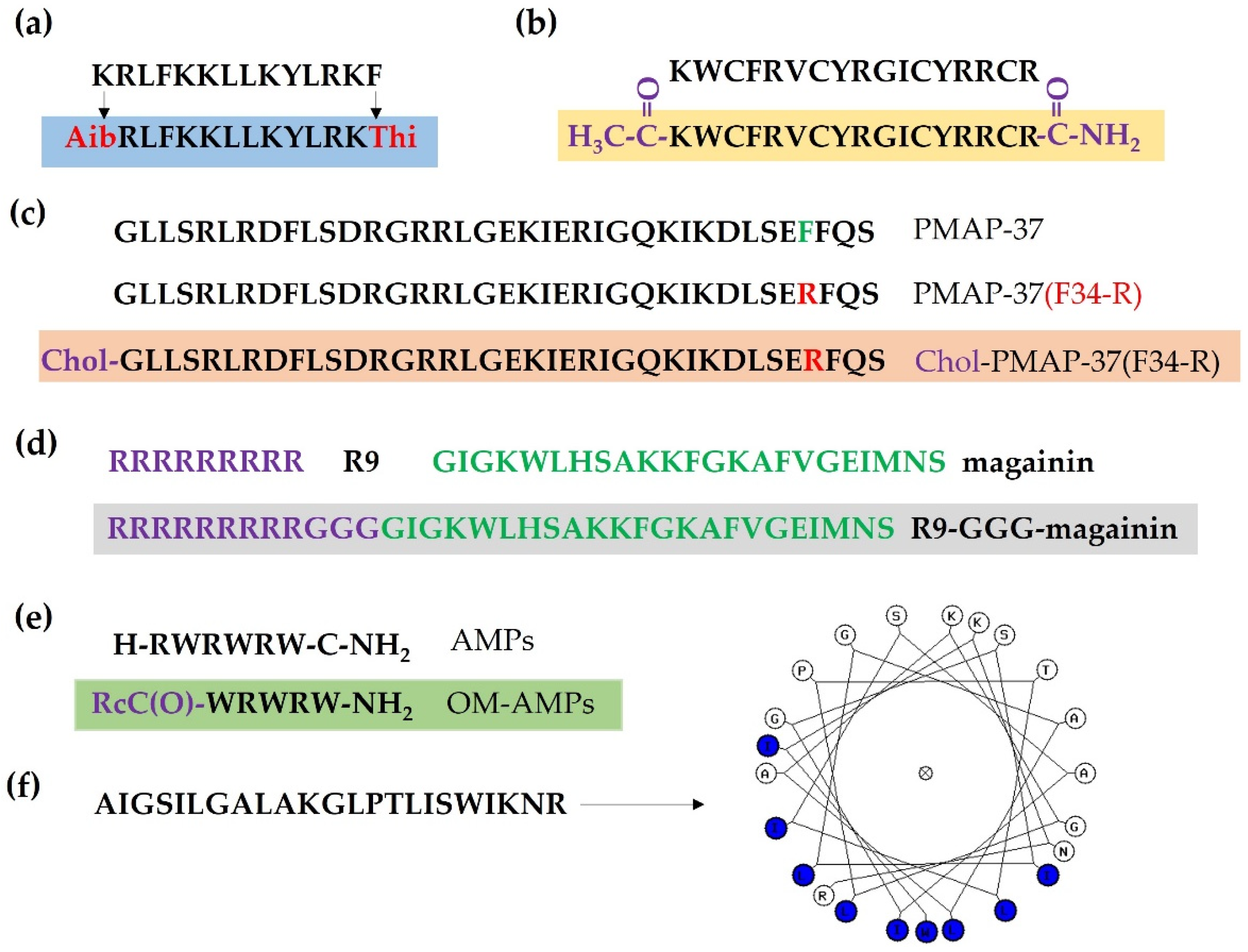
6.1. Substitution
6.2. N-Terminal Acetylation and C-Terminal Amidation
6.3. Fatty Acid Modification
6.4. Conjugation with Membrane-Binding or Penetrating Peptides
6.5. Modification of AMPs with Organometallic Agents
6.6. Structural Modification
7. Delivery
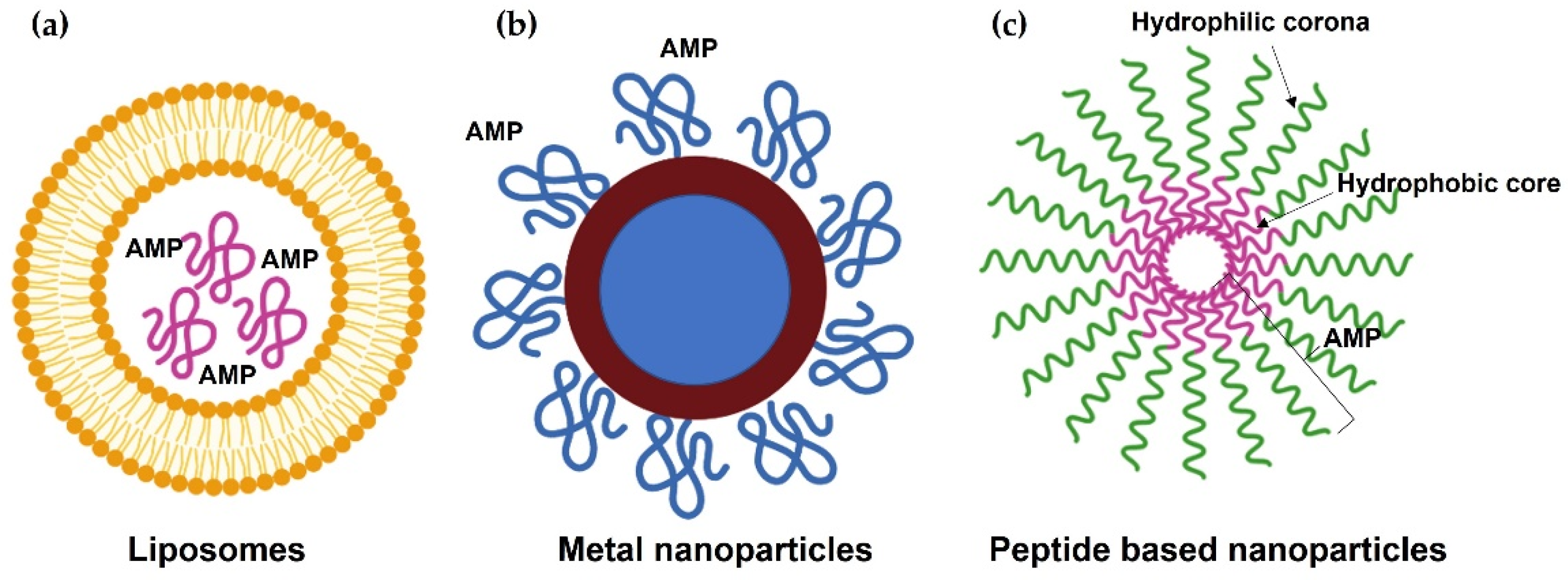
7.1. Lipid-Based Nanoparticles
7.2. Metal-Based Nanoparticles
7.3. Self-Assembling Nanoparticles
8. Clinical Application
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. [CrossRef]
- Molineri, A.I.; Camussone, C.; Zbrun, M.V.; Suárez Archilla, G.; Cristiani, M.; Neder, V.; Calvinho, L.; Signorini, M. Antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis: Systematic review and meta-analysis. Prev. Vet. Med. 2021, 188, 105261. [Google Scholar] [CrossRef] [PubMed]
- Mengistu, G.; Dejenu, G.; Tesema, C.; Arega, B.; Awoke, T.; Alemu, K.; Moges, F. Epidemiology of streptomycin resistant Salmonella from humans and animals in Ethiopia: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0244057. [Google Scholar] [CrossRef] [PubMed]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-Resistant Bacterial Infections in U.S. Hospitalized Patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Pormohammad, A.; Mehdinejadiani, K.; Gholizadeh, P.; Nasiri, M.J.; Mohtavinejad, N.; Dadashi, M.; Karimaei, S.; Safari, H.; Azimi, T. Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: A systematic review and meta-analysis. Microb. Pathog. 2020, 139, 103887. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, M.H. Resistance to tetracyclines among clinical isolates of Mycoplasma hominis and Ureaplasma species: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2021, 76, 865–875. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef]
- Meena, D.K.; Jayanthi, M. Monitoring Antibiotic Use in Public Health Care Facilities of South Indian Union Territory: A Step to Promote Rational Use of Antibiotics. Cureus 2021, 13, e18431. [Google Scholar] [CrossRef]
- Raman, G.; Avendano, E.; Berger, S.; Menon, V. Appropriate initial antibiotic therapy in hospitalized patients with gram-negative infections: Systematic review and meta-analysis. BMC Infect. Dis. 2015, 15, 395. [Google Scholar] [CrossRef]
- Segala, F.V.; Bavaro, D.F.; Di Gennaro, F.; Salvati, F.; Marotta, C.; Saracino, A.; Murri, R.; Fantoni, M. Impact of SARS-CoV-2 Epidemic on Antimicrobial Resistance: A Literature Review. Viruses 2021, 13, 2110. [Google Scholar] [CrossRef] [PubMed]
- Adebisi, Y.A.; Jimoh, N.D.; Ogunkola, I.O.; Uwizeyimana, T.; Olayemi, A.H.; Ukor, N.A.; Lucero-Prisno, D.E., 3rd. The use of antibiotics in COVID-19 management: A rapid review of national treatment guidelines in 10 African countries. Trop. Med. Health 2021, 49, 51. [Google Scholar] [CrossRef]
- Emeraud, C.; Figueiredo, S.; Bonnin, R.A.; Khecharem, M.; Ouzani, S.; Leblanc, P.E.; Jousset, A.B.; Fortineau, N.; Duranteau, J.; Dortet, L. Outbreak of CTX-M-15 Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae ST394 in a French Intensive Care Unit Dedicated to COVID-19. Pathogens 2021, 10, 1426. [Google Scholar] [CrossRef]
- Gaspari, R.; Spinazzola, G.; Teofili, L.; Avolio, A.W.; Fiori, B.; Maresca, G.M.; Spanu, T.; Nicolotti, N.; De Pascale, G.; Antonelli, M. Protective effect of SARS-CoV-2 preventive measures against ESKAPE and Escherichia coli infections. Eur. J. Clin. Investig. 2021, 51, e13687. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.M.C.; Oliveira, M.D.; Dias, R.S.; Nacif-Marçal, L.; Feio, R.N.; Ferreira, S.O.; Oliveira, L.L.; Silva, C.C.; Paula, S.O. The antimicrobial peptide HS-1 inhibits dengue virus infection. Virology 2018, 514, 79–87. [Google Scholar] [CrossRef]
- Luo, X.-L.; Li, J.-X.; Huang, H.-R.; Duan, J.-L.; Dai, R.-X.; Tao, R.-J.; Yang, L.; Hou, J.-Y.; Jia, X.-M.; Xu, J.-F. LL37 Inhibits Aspergillus fumigatus Infection via Directly Binding to the Fungus and Preventing Excessive Inflammation. Front. Immunol. 2019, 10, 283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, M.; Ericsson, A.C. Antimicrobial Peptides: Potential Application in Liver Cancer. Front. Microbiol. 2019, 10, 1257. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, M. The Role and Potential Application of Antimicrobial Peptides in Autoimmune Diseases. Front. Immunol. 2020, 11, 859. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Laneri, S.; Brancaccio, M.; Mennitti, C.; De Biasi, M.G.; Pero, M.E.; Pisanelli, G.; Scudiero, O.; Pero, R. Antimicrobial Peptides and Physical Activity: A Great Hope against COVID 19. Microorganisms 2021, 9, 1415. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.C.; Carbone, C.; Sousa, M.C.; Espina, M.; Garcia, M.L.; Sanchez-Lopez, E.; Souto, E.B. Nanomedicines for the Delivery of Antimicrobial Peptides (AMPs). Nanomaterials 2020, 10, 560. [Google Scholar] [CrossRef]
- Thapa, R.K.; Diep, D.B.; Tønnesen, H.H. Nanomedicine-based antimicrobial peptide delivery for bacterial infections: Recent advances and future prospects. J. Pharm. Investig. 2021, 51, 377–398. [Google Scholar] [CrossRef]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, S.M.; Abd El-Aziz, A.M.; Hassan, R.; Abdelmegeed, E.S. β-lactam resistance associated with β-lactamase production and porin alteration in clinical isolates of E. coli and K. pneumoniae. PLoS ONE 2021, 16, e0251594. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, M.; Park, J.; Sleno, B.; Berghuis, A.M. Structural and functional insights into esterase-mediated macrolide resistance. Nat. Commun. 2021, 12, 1732. [Google Scholar] [CrossRef] [PubMed]
- Dinos, G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017, 174, 2967–2983. [Google Scholar] [CrossRef]
- Zahedi Bialvaei, A.; Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Mardani Dashti, Y.; Soleiman-Meigooni, S. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar] [CrossRef]
- Schroeder, M.R.; Stephens, D.S. Macrolide Resistance in Streptococcus pneumoniae. Front. Cell. Infect. Microbiol. 2016, 6, 98. [Google Scholar] [CrossRef]
- Millanao, A.R.; Mora, A.Y.; Villagra, N.A.; Bucarey, S.A.; Hidalgo, A.A. Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents. Molecules 2021, 26, 7153. [Google Scholar] [CrossRef]
- Bhatnagar, K.; Wong, A. The mutational landscape of quinolone resistance in Escherichia coli. PLoS ONE 2019, 14, e0224650. [Google Scholar] [CrossRef] [PubMed]
- Podnecky, N.L.; Rhodes, K.A.; Mima, T.; Drew, H.R.; Chirakul, S.; Wuthiekanun, V.; Schupp, J.M.; Sarovich, D.S.; Currie, B.J.; Keim, P.; et al. Mechanisms of Resistance to Folate Pathway Inhibitors in Burkholderia pseudomallei: Deviation from the Norm. mBio 2017, 8, e01357-17. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting Plasmids to Limit Acquisition and Transmission of Antimicrobial Resistance. Front. Microbiol. 2020, 11, 761. [Google Scholar] [CrossRef]
- Liu, Y.; Tong, Z.; Shi, J.; Jia, Y.; Yang, K.; Wang, Z. Correlation between Exogenous Compounds and the Horizontal Transfer of Plasmid-Borne Antibiotic Resistance Genes. Microorganisms 2020, 8, 1211. [Google Scholar] [CrossRef]
- El-Badawy, M.F.; Alrobaian, M.M.; Shohayeb, M.M.; Abdelwahab, S.F. Investigation of six plasmid-mediated quinolone resistance genes among clinical isolates of pseudomonas: A genotypic study in Saudi Arabia. Infect. Drug Resist. 2019, 12, 915–923. [Google Scholar] [CrossRef]
- Aryal, S.C.; Upreti, M.K.; Sah, A.K.; Ansari, M.; Nepal, K.; Dhungel, B.; Adhikari, N.; Lekhak, B.; Rijal, K.R. Plasmid-Mediated AmpC β-Lactamase CITM and DHAM Genes Among Gram-Negative Clinical Isolates. Infect. Drug Resist. 2020, 13, 4249–4261. [Google Scholar] [CrossRef]
- Rands, C.M.; Starikova, E.V.; Brüssow, H.; Kriventseva, E.V.; Govorun, V.M.; Zdobnov, E.M. ACI-1 beta-lactamase is widespread across human gut microbiomes in Negativicutes due to transposons harboured by tailed prophages. Environ. Microbiol. 2018, 20, 2288–2300. [Google Scholar] [CrossRef]
- Xu, J.; Lin, W.; Chen, Y.; He, F. Characterization of an IMP-4-Producing Klebsiella pneumoniae ST1873 Strain Recovered from an Infant with a Bloodstream Infection in China. Infect. Drug Resist. 2020, 13, 773–779. [Google Scholar] [CrossRef]
- Ebmeyer, S.; Kristiansson, E.; Larsson, D.G.J. A framework for identifying the recent origins of mobile antibiotic resistance genes. Commun. Biol. 2021, 4, 8. [Google Scholar] [CrossRef]
- Sun, J.; Chen, C.; Cui, C.-Y.; Zhang, Y.; Liu, X.; Cui, Z.-H.; Ma, X.-Y.; Feng, Y.; Fang, L.-X.; Lian, X.-L.; et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Babakhani, S.; Oloomi, M. Transposons: The agents of antibiotic resistance in bacteria. J. Basic Microbiol. 2018, 58, 905–917. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, C.; Cui, P.; Wang, H. Detection of Tn7-Like Transposons and Antibiotic Resistance in Enterobacterales From Animals Used for Food Production With Identification of Three Novel Transposons Tn6813, Tn6814, and Tn6765. Front. Microbiol. 2020, 11, 2049. [Google Scholar] [CrossRef]
- Belaynehe, K.M.; Shin, S.W.; Yoo, H.S. Interrelationship between tetracycline resistance determinants, phylogenetic group affiliation and carriage of class 1 integrons in commensal Escherichia coli isolates from cattle farms. BMC Vet. Res. 2018, 14, 340. [Google Scholar] [CrossRef] [PubMed]
- Colavecchio, A.; Cadieux, B.; Lo, A.; Goodridge, L.D. Bacteriophages Contribute to the Spread of Antibiotic Resistance Genes among Foodborne Pathogens of the Enterobacteriaceae Family - A Review. Front. Microbiol. 2017, 8, 1108. [Google Scholar] [CrossRef] [PubMed]
- Calero-Cáceres, W.; Ye, M.; Balcázar, J.L. Bacteriophages as Environmental Reservoirs of Antibiotic Resistance. Trends Microbiol. 2019, 27, 570–577. [Google Scholar] [CrossRef]
- Anand, T.; Bera, B.C.; Vaid, R.K.; Barua, S.; Riyesh, T.; Virmani, N.; Hussain, M.; Singh, R.K.; Tripathi, B.N. Abundance of antibiotic resistance genes in environmental bacteriophages. J. Gen. Virol. 2016, 97, 3458–3466. [Google Scholar] [CrossRef]
- Gómez-Gómez, C.; Blanco-Picazo, P.; Brown-Jaque, M.; Quirós, P.; Rodríguez-Rubio, L.; Cerdà-Cuellar, M.; Muniesa, M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci. Rep. 2019, 9, 13281. [Google Scholar] [CrossRef]
- Jebri, S.; Rahmani, F.; Hmaied, F. Bacteriophages as antibiotic resistance genes carriers in agro-food systems. J. Appl. Microbiol. 2021, 130, 688–698. [Google Scholar] [CrossRef]
- Moon, K.; Jeon, J.H.; Kang, I.; Park, K.S.; Lee, K.; Cha, C.J.; Lee, S.H.; Cho, J.C. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 2020, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Imamovic, L.; Jofre, J.; Muniesa, M. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 2011, 55, 4908–4911. [Google Scholar] [CrossRef] [PubMed]
- Larrañaga, O.; Brown-Jaque, M.; Quirós, P.; Gómez-Gómez, C.; Blanch, A.R.; Rodríguez-Rubio, L.; Muniesa, M. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 2018, 115, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Chen, J.; Manohar, P.; Yu, Y.; Hua, X.; Leptihn, S. A Biological Inventory of Prophages in A. baumannii Genomes Reveal Distinct Distributions in Classes, Length, and Genomic Positions. Front. Microbiol. 2020, 11, 579802. [Google Scholar] [CrossRef] [PubMed]
- Branger, C.; Ledda, A.; Billard-Pomares, T.; Doublet, B.; Fouteau, S.; Barbe, V.; Roche, D.; Cruveiller, S.; Médigue, C.; Castellanos, M.; et al. Extended-spectrum β-lactamase-encoding genes are spreading on a wide range of Escherichia coli plasmids existing prior to the use of third-generation cephalosporins. Microb. Genom. 2018, 4, e000203. [Google Scholar] [CrossRef] [PubMed]
- Pornsukarom, S.; van Vliet, A.H.M.; Thakur, S. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genom. 2018, 19, 801. [Google Scholar] [CrossRef]
- Galata, V.; Laczny, C.C.; Backes, C.; Hemmrich-Stanisak, G.; Schmolke, S.; Franke, A.; Meese, E.; Herrmann, M.; von Müller, L.; Plum, A.; et al. Integrating Culture-based Antibiotic Resistance Profiles with Whole-genome Sequencing Data for 11,087 Clinical Isolates. Genom. Proteom. Bioinform. 2019, 17, 169–182. [Google Scholar] [CrossRef]
- Qiu, H.; Gong, J.; Butaye, P.; Lu, G.; Huang, K.; Zhu, G.; Zhang, J.; Hathcock, T.; Cheng, D.; Wang, C. CRISPR/Cas9/sgRNA-mediated targeted gene modification confirms the cause-effect relationship between gyrA mutation and quinolone resistance in Escherichia coli. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Wan, P.; Cui, S.; Ma, Z.; Chen, L.; Li, X.; Zhao, R.; Xiong, W.; Zeng, Z. Reversal of mcr-1-Mediated Colistin Resistance in Escherichia coli by CRISPR-Cas9 System. Infect. Drug Resist. 2020, 13, 1171–1178. [Google Scholar] [CrossRef]
- Burton, J.N.; Liachko, I.; Dunham, M.J.; Shendure, J. Species-Level Deconvolution of Metagenome Assemblies with Hi-C–Based Contact Probability Maps. G3 Genes|Genom.|Genet. 2014, 4, 1339–1346. [Google Scholar] [CrossRef]
- Kent, A.G.; Vill, A.C.; Shi, Q.; Satlin, M.J.; Brito, I.L. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 2020, 11, 4379. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic. Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, C.; Hansen, S.A.; Mitchell, W.J.; Zhang, M.Z.; Zhang, S. Antimicrobial efficacy and toxicity of novel CAMPs against P. aeruginosa infection in a murine skin wound infection model. BMC Microbiol. 2019, 19, 293. [Google Scholar] [CrossRef]
- Rozek, A.; Friedrich, C.L.; Hancock, R.E. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry 2000, 39, 15765–15774. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J. Biol. Chem. 2008, 283, 32637–32643. [Google Scholar] [CrossRef]
- Szyk, A.; Wu, Z.; Tucker, K.; Yang, D.; Lu, W.; Lubkowski, J. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006, 15, 2749–2760. [Google Scholar] [CrossRef]
- Hoover, D.M.; Rajashankar, K.R.; Blumenthal, R.; Puri, A.; Oppenheim, J.J.; Chertov, O.; Lubkowski, J. The structure of human beta-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 2000, 275, 32911–32918. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Vishnepolsky, B.; Grigolava, M.; Managadze, G. Physicochemical Features and Peculiarities of Interaction of AMP with the Membrane. Pharmaceuticals 2021, 14, 471. [Google Scholar] [CrossRef]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Comparative mode of action of the antimicrobial peptide melimine and its derivative Mel4 against Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 7063. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.N.; Ferre, R.; Castanho, M.A.R.B. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Seyfi, R.; Kahaki, F.A.; Ebrahimi, T.; Montazersaheb, S.; Eyvazi, S.; Babaeipour, V.; Tarhriz, V. Antimicrobial Peptides (AMPs): Roles, Functions and Mechanism of Action. Int. J. Peptide Res. Ther. 2020, 26, 1451–1463. [Google Scholar] [CrossRef]
- Pahar, B.; Madonna, S.; Das, A.; Albanesi, C.; Girolomoni, G. Immunomodulatory Role of the Antimicrobial LL-37 Peptide in Autoimmune Diseases and Viral Infections. Vaccines 2020, 8, 517. [Google Scholar] [CrossRef]
- Röhrl, J.; Yang, D.; Oppenheim, J.J.; Hehlgans, T. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 2010, 184, 6688–6694. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, C.; Zhang, X.; Zhang, M.Z.; Rottinghaus, G.E.; Zhang, S. Structure-function analysis of Avian β-defensin-6 and β-defensin-12: Role of charge and disulfide bridges. BMC Microbiol. 2016, 16, 210. [Google Scholar] [CrossRef]
- Semple, F.; MacPherson, H.; Webb, S.; Cox, S.L.; Mallin, L.J.; Tyrrell, C.; Grimes, G.R.; Semple, C.A.; Nix, M.A.; Millhauser, G.L.; et al. Human β-defensin 3 affects the activity of pro-inflammatory pathways associated with MyD88 and TRIF. Eur. J. Immunol. 2011, 41, 3291–3300. [Google Scholar] [CrossRef]
- Minns, D.; Smith, K.J.; Alessandrini, V.; Hardisty, G.; Melrose, L.; Jackson-Jones, L.; MacDonald, A.S.; Davidson, D.J.; Gwyer Findlay, E. The neutrophil antimicrobial peptide cathelicidin promotes Th17 differentiation. Nat. Commun. 2021, 12, 1285. [Google Scholar] [CrossRef]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Hwang, B.; Hwang, J.S.; Lee, J.; Lee, D.G. The antimicrobial peptide, psacotheasin induces reactive oxygen species and triggers apoptosis in Candida albicans. Biochem. Biophys. Res. Commun. 2011, 405, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Cerrato, C.P.; Künnapuu, K.; Langel, Ü. Cell-penetrating peptides with intracellular organelle targeting. Expert Opin. Drug Deliv. 2017, 14, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Buccini, D.F.; Cardoso, M.H.; Franco, O.L. Antimicrobial Peptides and Cell-Penetrating Peptides for Treating Intracellular Bacterial Infections. Front. Cell. Infect. Microbiol. 2021, 10, 612931. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Sun, J.; Nasrolahi Shirazi, A.; LaPlante, K.L.; Rowley, D.C.; Parang, K. Antibacterial activities of amphiphilic cyclic cell-penetrating peptides against multidrug-resistant pathogens. Mol. Pharm. 2014, 11, 3528–3536. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, X.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Chen, H.; Wang, X.; Wang, J. Improved antibacterial activity of a marine peptide-N2 against intracellular Salmonella typhimurium by conjugating with cell-penetrating peptides-bLFcin(6)/Tat(11). Eur. J. Med. Chem. 2018, 145, 263–272. [Google Scholar] [CrossRef]
- Yang, Y.; Xia, L.; Haapasalo, M.; Wei, W.; Zhang, D.; Ma, J.; Shen, Y. A novel hydroxyapatite-binding antimicrobial peptide against oral biofilms. Clin. Oral Investig. 2019, 23, 2705–2712. [Google Scholar] [CrossRef]
- Li, J.; Hu, S.; Jian, W.; Xie, C.; Yang, X. Plant antimicrobial peptides: Structures, functions, and applications. Bot. Stud. 2021, 62, 5. [Google Scholar] [CrossRef]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Goździcka-Józefiak, A. Plant antimicrobial peptides. Folia Microbiol. 2014, 59, 181–196. [Google Scholar] [CrossRef]
- Campos, M.L.; de Souza, C.M.; de Oliveira, K.B.S.; Dias, S.C.; Franco, O.L. The role of antimicrobial peptides in plant immunity. J. Exp. Bot. 2018, 69, 4997–5011. [Google Scholar] [CrossRef]
- Amaral, A.C.; Silva, O.N.; Mundim, N.C.C.R.; de Carvalho, M.J.A.; Migliolo, L.; Leite, J.R.S.A.; Prates, M.V.; Bocca, A.L.; Franco, O.L.; Felipe, M.S.S. Predicting antimicrobial peptides from eukaryotic genomes: In silico strategies to develop antibiotics. Peptides 2012, 37, 301–308. [Google Scholar] [CrossRef]
- Tomazou, M.; Oulas, A.; Anagnostopoulos, A.K.; Tsangaris, G.T.; Spyrou, G.M. In Silico Identification of Antimicrobial Peptides in the Proteomes of Goat and Sheep Milk and Feta Cheese. Proteomes 2019, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Yakovlev, I.A.; Lysøe, E.; Heldal, I.; Steen, H.; Hagen, S.B.; Clarke, J.L. Transcriptome profiling and in silico detection of the antimicrobial peptides of red king crab Paralithodes camtschaticus. Sci. Rep. 2020, 10, 12679. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Fensterseifer, I.C.M.; Ribeiro, S.M.; Franco, O.L. Joker: An algorithm to insert patterns into sequences for designing antimicrobial peptides. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2043–2052. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Irazazabal, L.N.; Humblot, V.; Haney, E.F.; Ribeiro, S.M.; Hancock, R.E.W.; Ladram, A.; Franco, O.L. EcDBS1R6: A novel cationic antimicrobial peptide derived from a signal peptide sequence. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129633. [Google Scholar] [CrossRef]
- Kim, H.; Jang, J.H.; Kim, S.C.; Cho, J.H. Development of a novel hybrid antimicrobial peptide for targeted killing of Pseudomonas aeruginosa. Eur. J. Med. Chem. 2020, 185, 111814. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, C.; Zhang, M.Z.; Zhang, S. Beta-defensin derived cationic antimicrobial peptides with potent killing activity against gram negative and gram positive bacteria. BMC Microbiol. 2018, 18, 54. [Google Scholar] [CrossRef]
- Yu, W.; Ning, N.; Xue, Y.; Huang, Y.; Guo, F.; Li, T.; Yang, B.; Luo, D.; Sun, Y.; Li, Z.; et al. A Chimeric Cationic Peptide Composed of Human β-Defensin 3 and Human β-Defensin 4 Exhibits Improved Antibacterial Activity and Salt Resistance. Front. Microbiol. 2021, 12, 663151. [Google Scholar] [CrossRef]
- Li, T.; Guo, F.; Wang, Q.; Fang, H.; Li, Z.; Wang, D.; Wang, H. N-terminus three residues deletion mutant of human beta-defensin 3 with remarkably enhanced salt-resistance. PLoS ONE 2015, 10, e0117913. [Google Scholar] [CrossRef][Green Version]
- Tang, Y.Q.; Yuan, J.; Osapay, G.; Osapay, K.; Tran, D.; Miller, C.J.; Ouellette, A.J.; Selsted, M.E. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science 1999, 286, 498–502. [Google Scholar] [CrossRef]
- Okella, H.; Georrge, J.J.; Ochwo, S.; Ndekezi, C.; Koffi, K.T.; Aber, J.; Ajayi, C.O.; Fofana, F.G.; Ikiriza, H.; Mtewa, A.G.; et al. New Putative Antimicrobial Candidates: In silico Design of Fish-Derived Antibacterial Peptide-Motifs. Front. Bioeng. Biotechnol. 2020, 8, 604041. [Google Scholar] [CrossRef]
- Yin, Q.; Wu, S.; Wu, L.; Wang, Z.; Mu, Y.; Zhang, R.; Dong, C.; Zhou, B.; Zhao, B.; Zheng, J.; et al. A novel in silico antimicrobial peptide DP7 combats MDR Pseudomonas aeruginosa and related biofilm infections. J. Antimicrob. Chemother. 2020, 75, 3248–3259. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Idicula-Thomas, S. Collection of antimicrobial peptides database and its derivatives: Applications and beyond. Protein Sci. A Publ. Protein Soc. 2020, 29, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Jhong, J.H.; Yao, L.; Pang, Y.; Li, Z.; Chung, C.R.; Wang, R.; Li, S.; Li, W.; Luo, M.; Ma, R.; et al. dbAMP 2.0: Updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data. Nucleic Acids Res. 2022, 50, D460–D470. [Google Scholar] [CrossRef] [PubMed]
- Lertampaiporn, S.; Vorapreeda, T.; Hongsthong, A.; Thammarongtham, C. Ensemble-AMPPred: Robust AMP Prediction and Recognition Using the Ensemble Learning Method with a New Hybrid Feature for Differentiating AMPs. Genes 2021, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Zhu, N.; Zhu, Y.; Liu, T.; Gou, S.; Xie, J.; Yao, J.; Ni, J. Antimicrobial peptides conjugated with fatty acids on the side chain of D-amino acid promises antimicrobial potency against multidrug-resistant bacteria. Eur. J. Pharm. Sci. 2020, 141, 105123. [Google Scholar] [CrossRef]
- Lu, J.; Xu, H.; Xia, J.; Ma, J.; Xu, J.; Li, Y.; Feng, J. D- and Unnatural Amino Acid Substituted Antimicrobial Peptides With Improved Proteolytic Resistance and Their Proteolytic Degradation Characteristics. Front. Microbiol. 2020, 11, 563030. [Google Scholar] [CrossRef]
- Panahi Chegini, P.; Nikokar, I.; Tabarzad, M.; Faezi, S.; Mahboubi, A. Effect of Amino Acid Substitutions on Biological Activity of Antimicrobial Peptide: Design, Recombinant Production, and Biological Activity. Iran. J. Pharm. Res. 2019, 18, 157–168. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- D’Souza, A.R.; Necelis, M.R.; Kulesha, A.; Caputo, G.A.; Makhlynets, O.V. Beneficial Impacts of Incorporating the Non-Natural Amino Acid Azulenyl-Alanine into the Trp-Rich Antimicrobial Peptide buCATHL4B. Biomolecules 2021, 11, 421. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, C.; Zhang, M.Z.; Zhang, S. Novel synthetic analogues of avian β-defensin-12: The role of charge, hydrophobicity, and disulfide bridges in biological functions. BMC Microbiol. 2017, 17, 43. [Google Scholar] [CrossRef]
- Kuzmin, D.V.; Emelianova, A.A.; Kalashnikova, M.B.; Panteleev, P.V.; Ovchinnikova, T.V. Effect of N- and C-Terminal Modifications on Cytotoxic Properties of Antimicrobial Peptide Tachyplesin I. Bull. Exp. Biol. Med. 2017, 162, 754–757. [Google Scholar] [CrossRef] [PubMed]
- Soleymani-Goloujeh, M.; Nokhodchi, A.; Niazi, M.; Najafi-Hajivar, S.; Shahbazi-Mojarrad, J.; Zarghami, N.; Zakeri-Milani, P.; Mohammadi, A.; Karimi, M.; Valizadeh, H. Effects of N-terminal and C-terminal modification on cytotoxicity and cellular uptake of amphiphilic cell penetrating peptides. Artif. Cells Nanomed. Biotechnol. 2018, 46, 91–103. [Google Scholar] [CrossRef]
- Liu, Y.; Li, S.; Shen, T.; Chen, L.; Zhou, J.; Shi, S.; Wang, Y.; Zhao, Z.; Liao, C.; Wang, C. N-terminal Myristoylation Enhanced the Antimicrobial Activity of Antimicrobial Peptide PMAP-36PW. Front. Cell. Infect. Microbiol. 2020, 10, 450. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shen, T.; Liu, Y.; Zhou, J.; Shi, S.; Wang, Y.; Zhao, Z.; Yan, Z.; Liao, C.; Wang, C. Enhancing the antibacterial activity of antimicrobial peptide PMAP-37(F34-R) by cholesterol modification. BMC Vet. Res. 2020, 16, 419. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, X.; Da, T.; Mao, R.; Hao, Y.; Yang, N.; Wang, X.; Li, Z.; Wang, X.; Wang, J. Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli. Commun. Biol. 2020, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lim, S.I.; Shin, S.H.; Lim, Y.; Koh, J.W.; Yang, S. Conjugation of Cell-Penetrating Peptides to Antimicrobial Peptides Enhances Antibacterial Activity. ACS Omega 2019, 4, 15694–15701. [Google Scholar] [CrossRef] [PubMed]
- Albada, B.; Metzler-Nolte, N. Highly Potent Antibacterial Organometallic Peptide Conjugates. Acc. Chem. Res. 2017, 50, 2510–2518. [Google Scholar] [CrossRef]
- Moiola, M.; Memeo, M.G.; Quadrelli, P. Stapled Peptides-A Useful Improvement for Peptide-Based Drugs. Molecules 2019, 24, 3654. [Google Scholar] [CrossRef]
- Zhang, S.-K.; Song, J.-W.; Gong, F.; Li, S.-B.; Chang, H.-Y.; Xie, H.-M.; Gao, H.-W.; Tan, Y.-X.; Ji, S.-P. Design of an α-helical antimicrobial peptide with improved cell-selective and potent anti-biofilm activity. Sci. Rep. 2016, 6, 27394. [Google Scholar] [CrossRef]
- Vicente, F.E.M.; González-Garcia, M.; Diaz Pico, E.; Moreno-Castillo, E.; Garay, H.E.; Rosi, P.E.; Jimenez, A.M.; Campos-Delgado, J.A.; Rivera, D.G.; Chinea, G.; et al. Design of a Helical-Stabilized, Cyclic, and Nontoxic Analogue of the Peptide Cm-p5 with Improved Antifungal Activity. ACS Omega 2019, 4, 19081–19095. [Google Scholar] [CrossRef]
- Liu, B.; Huang, H.; Yang, Z.; Liu, B.; Gou, S.; Zhong, C.; Han, X.; Zhang, Y.; Ni, J.; Wang, R. Design of novel antimicrobial peptide dimer analogues with enhanced antimicrobial activity in vitro and in vivo by intermolecular triazole bridge strategy. Peptides 2017, 88, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Greber, K.E.; Zielińska, J.; Nierzwicki, Ł.; Ciura, K.; Kawczak, P.; Nowakowska, J.; Bączek, T.; Sawicki, W. Are the short cationic lipopeptides bacterial membrane disruptors? Structure-Activity Relationship and molecular dynamic evaluation. Biochim. Biophys. Acta Biomembr. 2019, 1861, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Greber, K.E.; Ciura, K.; Belka, M.; Kawczak, P.; Nowakowska, J.; Bączek, T.; Sawicki, W. Characterization of antimicrobial and hemolytic properties of short synthetic cationic lipopeptides based on QSAR/QSTR approach. Amino Acids 2018, 50, 479–485. [Google Scholar] [CrossRef]
- Ciura, K.; Ptaszyńska, N.; Kapica, H.; Pastewska, M.; Łęgowska, A.; Rolka, K.; Kamysz, W.; Sawicki, W.; Greber, K.E. Can Immobilized Artificial Membrane Chromatography Support the Characterization of Antimicrobial Peptide Origin Derivatives? Antibiotics 2021, 10, 1237. [Google Scholar] [CrossRef] [PubMed]
- Petkov, P.; Lilkova, E.; Ilieva, N.; Litov, L. Self-Association of Antimicrobial Peptides: A Molecular Dynamics Simulation Study on Bombinin. Int. J. Mol. Sci. 2019, 20, 5450. [Google Scholar] [CrossRef] [PubMed]
- Ciornei, C.D.; Sigurdardóttir, T.; Schmidtchen, A.; Bodelsson, M. Antimicrobial and chemoattractant activity, lipopolysaccharide neutralization, cytotoxicity, and inhibition by serum of analogs of human cathelicidin LL-37. Antimicrob. Agents Chemother. 2005, 49, 2845–2850. [Google Scholar] [CrossRef]
- Carratalá, J.V.; Serna, N.; Villaverde, A.; Vázquez, E.; Ferrer-Miralles, N. Nanostructured antimicrobial peptides: The last push towards clinics. Biotechnol. Adv. 2020, 44, 107603. [Google Scholar] [CrossRef]
- Klubthawee, N.; Bovone, G.; Marco-Dufort, B.; Guzzi, E.A.; Aunpad, R.; Tibbitt, M.W. Biopolymer Nano-network for Antimicrobial Peptide Protection and Local Delivery. Adv. Healthc. Mater. 2021, e2101426. [Google Scholar] [CrossRef]
- Pranantyo, D.; Raju, C.; Si, Z.; Xu, X.; Pethe, K.; Kang, E.T.; Chan-Park, M.B. Nontoxic Antimicrobial Cationic Peptide Nanoconstructs with Bacteria-Displaceable Polymeric Counteranions. Nano Lett. 2021, 21, 899–906. [Google Scholar] [CrossRef]
- Olusanya, T.O.B.; Haj Ahmad, R.R.; Ibegbu, D.M.; Smith, J.R.; Elkordy, A.A. Liposomal Drug Delivery Systems and Anticancer Drugs. Molecules 2018, 23, 907. [Google Scholar] [CrossRef]
- Yang, K.; Gitter, B.; Rüger, R.; Wieland, G.D.; Chen, M.; Liu, X.; Albrecht, V.; Fahr, A. Antimicrobial peptide-modified liposomes for bacteria targeted delivery of temoporfin in photodynamic antimicrobial chemotherapy. Photochem. Photobiol. Sci. 2011, 10, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Makowski, M.; Silva Í, C.; Pais do Amaral, C.; Gonçalves, S.; Santos, N.C. Advances in Lipid and Metal Nanoparticles for Antimicrobial Peptide Delivery. Pharmaceutics 2019, 11, 588. [Google Scholar] [CrossRef] [PubMed]
- Bruna, T.; Maldonado-Bravo, F.; Jara, P.; Caro, N. Silver Nanoparticles and Their Antibacterial Applications. Int. J. Mol. Sci. 2021, 22, 7202. [Google Scholar] [CrossRef] [PubMed]
- de Lacerda Coriolano, D.; de Souza, J.B.; Bueno, E.V.; Medeiros, S.; Cavalcanti, I.D.L.; Cavalcanti, I.M.F. Antibacterial and antibiofilm potential of silver nanoparticles against antibiotic-sensitive and multidrug-resistant Pseudomonas aeruginosa strains. Braz. J. Microbiol. 2021, 52, 267–278. [Google Scholar] [CrossRef]
- Jin, Y.; Yang, Y.; Duan, W.; Qu, X.; Wu, J. Synergistic and On-Demand Release of Ag-AMPs Loaded on Porous Silicon Nanocarriers for Antibacteria and Wound Healing. ACS Appl. Mater. Interfaces 2021, 13, 16127–16141. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Shukla, S.K.; Pandey, M.; Pandey, A.; Pathak, A.; Dikshit, A. Synthesis and antimicrobial effects of colloidal gold nanoparticles against prevalent waterborne bacterial pathogens. Cogent. Chem. 2016, 2, 1192522. [Google Scholar] [CrossRef]
- Rai, A.; Pinto, S.; Velho, T.R.; Ferreira, A.F.; Moita, C.; Trivedi, U.; Evangelista, M.; Comune, M.; Rumbaugh, K.P.; Simões, P.N.; et al. One-step synthesis of high-density peptide-conjugated gold nanoparticles with antimicrobial efficacy in a systemic infection model. Biomaterials 2016, 85, 99–110. [Google Scholar] [CrossRef]
- Casciaro, B.; Moros, M.; Rivera-Fernández, S.; Bellelli, A.; de la Fuente, J.M.; Mangoni, M.L. Gold-nanoparticles coated with the antimicrobial peptide esculentin-1a(1-21)NH(2) as a reliable strategy for antipseudomonal drugs. Acta Biomater. 2017, 47, 170–181. [Google Scholar] [CrossRef]
- Gottschalk, F.; Sonderer, T.; Scholz, R.W.; Nowack, B. Modeled environmental concentrations of engineered nanomaterials (TiO(2), ZnO, Ag, CNT, Fullerenes) for different regions. Environ. Sci. Technol. 2009, 43, 9216–9222. [Google Scholar] [CrossRef]
- Yu, C.Y.; Huang, W.; Li, Z.P.; Lei, X.Y.; He, D.X.; Sun, L. Progress in Self-assembling Peptide-based Nanomaterials for Biomedical Applications. Curr. Top. Med. Chem. 2016, 16, 281–290. [Google Scholar] [CrossRef]
- Lei, R.; Hou, J.; Chen, Q.; Yuan, W.; Cheng, B.; Sun, Y.; Jin, Y.; Ge, L.; Ben-Sasson, S.A.; Chen, J.; et al. Self-Assembling Myristoylated Human α-Defensin 5 as a Next-Generation Nanobiotics Potentiates Therapeutic Efficacy in Bacterial Infection. ACS Nano 2018, 12, 5284–5296. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Jian, Q.; Li, G.; Shao, C.; Zhu, Y.; Yuan, X.; Chen, H.; Shan, A. Self-Assembling Peptide Dendron Nanoparticles with High Stability and a Multimodal Antimicrobial Mechanism of Action. ACS Nano 2021, 15, 15824–15840. [Google Scholar] [CrossRef] [PubMed]
- Pentlavalli, S.; Coulter, S.; Laverty, G. Peptide Nanomaterials for Drug Delivery Applications. Curr. Protein Pept. Sci. 2020, 21, 401–412. [Google Scholar] [CrossRef]
- Dijksteel, G.S.; Ulrich, M.M.W.; Middelkoop, E.; Boekema, B.K.H.L. Review: Lessons Learned From Clinical Trials Using Antimicrobial Peptides (AMPs). Front. Microbiol. 2021, 12, 287. [Google Scholar] [CrossRef] [PubMed]
- Nutman, A.; Lellouche, J.; Temkin, E.; Daikos, G.; Skiada, A.; Durante-Mangoni, E.; Dishon-Benattar, Y.; Bitterman, R.; Yahav, D.; Daitch, V.; et al. Colistin plus meropenem for carbapenem-resistant Gram-negative infections: In vitro synergism is not associated with better clinical outcomes. Clin. Microbiol. Infect. 2020, 26, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Daikos, G.L.; Durante-Mangoni, E.; Yahav, D.; Carmeli, Y.; Benattar, Y.D.; Skiada, A.; Andini, R.; Eliakim-Raz, N.; Nutman, A.; et al. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: An open-label, randomised controlled trial. Lancet Infect. Dis. 2018, 18, 391–400. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Vargas Buonfiglio, L.G.; Vanegas Calderon, O.G.; Cano, M.; Simmering, J.E.; Polgreen, P.M.; Zabner, J.; Gerke, A.K.; Comellas, A.P. Seasonal Antimicrobial Activity of the Airway: Post-Hoc Analysis of a Randomized Placebo-Controlled Double-Blind Trial. Nutrients 2020, 12, 2602. [Google Scholar] [CrossRef]
- Han, J.E.; Alvarez, J.A.; Jones, J.L.; Tangpricha, V.; Brown, M.A.; Hao, L.; Brown, L.A.S.; Martin, G.S.; Ziegler, T.R. Impact of high-dose vitamin D(3) on plasma free 25-hydroxyvitamin D concentrations and antimicrobial peptides in critically ill mechanically ventilated adults. Nutrition 2017, 38, 102–108. [Google Scholar] [CrossRef]
- Håkansson, J.; Ringstad, L.; Umerska, A.; Johansson, J.; Andersson, T.; Boge, L.; Rozenbaum, R.T.; Sharma, P.K.; Tollbäck, P.; Björn, C.; et al. Characterization of the in vitro, ex vivo, and in vivo Efficacy of the Antimicrobial Peptide DPK-060 Used for Topical Treatment. Front. Cell. Infect. Microbiol. 2019, 9, 174. [Google Scholar] [CrossRef]
- Niemeyer-van der Kolk, T.; van der Wall, H.; Hogendoorn, G.K.; Rijneveld, R.; Luijten, S.; van Alewijk, D.; van den Munckhof, E.H.A.; de Kam, M.L.; Feiss, G.L.; Prens, E.P.; et al. Pharmacodynamic Effects of Topical Omiganan in Patients With Mild to Moderate Atopic Dermatitis in a Randomized, Placebo-Controlled, Phase II Trial. Clin. Transl. Sci. 2020, 13, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Peek, N.F.A.W.; Nell, M.J.; Brand, R.; Jansen-Werkhoven, T.; van Hoogdalem, E.J.; Verrijk, R.; Vonk, M.J.; Wafelman, A.R.; Valentijn, A.R.P.M.; Frijns, J.H.M.; et al. Ototopical drops containing a novel antibacterial synthetic peptide: Safety and efficacy in adults with chronic suppurative otitis media. PloS ONE 2020, 15, e0231573. [Google Scholar] [CrossRef] [PubMed]
- Farsi, F.; Ebrahimi-Daryani, N.; Golab, F.; Akbari, A.; Janani, L.; Karimi, M.Y.; Irandoost, P.; Alamdari, N.M.; Agah, S.; Vafa, M. A randomized controlled trial on the coloprotective effect of coenzyme Q10 on immune-inflammatory cytokines, oxidative status, antimicrobial peptides, and microRNA-146a expression in patients with mild-to-moderate ulcerative colitis. Eur. J. Nutr. 2021, 60, 3397–3410. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ai, L.; Zhang, Y.; Cheng, J.; Yu, H.; Li, C.; Zhang, D.; Pan, Y.; Lin, L. The Effects of Antimicrobial Peptide Nal-P-113 on Inhibiting Periodontal Pathogens and Improving Periodontal Status. BioMed Res. Int. 2018, 2018, 1805793. [Google Scholar] [CrossRef]
- Zupin, L.; Polesello, V.; Segat, L.; Kamada, A.J.; Kuhn, L.; Crovella, S. DEFB1 polymorphisms and HIV-1 mother-to-child transmission in Zambian population. J. Matern.-Fetal Neonatal Med. 2019, 32, 2805–2811. [Google Scholar] [CrossRef]
- Park, H.J.; Cho, J.H.; Kim, H.J.; Han, S.H.; Jeong, S.H.; Byun, M.K. Colistin monotherapy versus colistin/rifampicin combination therapy in pneumonia caused by colistin-resistant Acinetobacter baumannii: A randomised controlled trial. J. Glob. Antimicrob. Resist. 2019, 17, 66–71. [Google Scholar] [CrossRef]
- El Shazely, B.; Yu, G.; Johnston, P.R.; Rolff, J. Resistance Evolution Against Antimicrobial Peptides in Staphylococcus aureus Alters Pharmacodynamics Beyond the MIC. Front. Microbiol. 2020, 11, 103. [Google Scholar] [CrossRef]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K.; et al. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019, 10, 4538. [Google Scholar] [CrossRef]
- Zhu, Y.; Hao, W.; Wang, X.; Ouyang, J.; Deng, X.; Yu, H.; Wang, Y. Antimicrobial peptides, conventional antibiotics, and their synergistic utility for the treatment of drug-resistant infections. Med. Res. Rev. 2022. [Google Scholar] [CrossRef]
- Pizzolato-Cezar, L.R.; Okuda-Shinagawa, N.M.; Machini, M.T. Combinatory Therapy Antimicrobial Peptide-Antibiotic to Minimize the Ongoing Rise of Resistance. Front. Microbiol. 2019, 10, 1703. [Google Scholar] [CrossRef]
| Antibiotics/Classes | Mode of Action | Bacteria | Mechanism of Resistance | References |
|---|---|---|---|---|
| Penicillin and carbapenem (beta-lactam) | Inhibiting bacterial cell wall synthesis | Escherichia coli and Klebsiella pneumoniae | Producing beta-lactamase and carbapenemase and porin alteration | [26] |
| Macrolides | Inhibiting protein synthesis by binding to the 50S ribosomal subunit | K. pneumoniae | Producing erythromycin esterases (Eres) such as EreA and EreC | [27,34] |
| Ticarcillin (beta-lactam) and ciprofloxacin (quinolone) | Inhibiting bacterial cell wall and protein synthesis | Pseudomonas aeruginosa | Resistance-nodulation-division (RND) efflux pumps | [29] |
| Macrolides | Inhibiting protein synthesis | Streptococcus pneumoniae | Ribosomal demethylation, expelling by efflux pump, and target site mutation | [30] |
| Quinolones | Inhibiting nucleic acid synthesis | K. pneumoniae and Clostridium perfringens | Mutations in the genes that encode gyrase and topoisomerase IV | [31,32] |
| Trimethoprim-sulfamethoxazole | Inhibiting folate synthesis | Burkholderia pseudomallei | Structural modification of dihydrofolate reductase (DHFR) or dihydropteroic acid synthase (DHPS) | [33] |
| Bacterial Strains | Mobile Genetic Elements | Resistance to Antibiotics | References |
|---|---|---|---|
| E. coli, K. pneumoniae, and A. baumannii | Plasmid-encoded tigecycline resistance tet(X3) and tet(X4) genes. | Tigecycline | [42,43] |
| Pseudomonas spp. | Plasmid-mediated quinolone resistance (qnr) genes such as qnrD, qnrS, and aac(6’)-Ib-cr. | Quinolone | [37] |
| Gram-negative bacteria such as E. coli and P. aeruginosa | Plasmid-mediated AmpC β-lactamases genes blaCITM and blaDHAM genes | Beta-lactam antibiotics such as ceftazidime, cefepime, and cefoxitin | [38] |
| Enterobacterales | Tn7-like transposons such as Tn6813, Tn6814, and Tn6765. | Sulfamethoxazole and streptomycin | [45] |
| Acidaminococcus intestine | Beta-lactamase encoded gene aci-1 is found in transposons of in human microbiota, which causes resistance to β-lactam antibiotics. | Beta-lactam antibiotics such as penicillin | [39] |
| E. coli | Class 1 integrons associated with tetracycline-resistant genes tet(A) and tet(B). | Tetracycline | [40] |
| Bacteria such as E. coli and Enterobacteriaceae | Bacteriophage-carried resistance genes such as blaTEM, qnrA, mecA, and sul1. | Penicillin, quinolone, methicillin, sulfonamide | [50,53] |
| E. coli | ARGs were found in agricultural soil and fresh vegetables such as lettuce and cucumber, including blaTEM and qnrA. | Penicillin and quinolone | [54] |
| A. baumannii | Phage-carried antimicrobial resistance genes carbapenemase gene OXA-23 and New Delhi metallobeta-lactamase 1 (NDM-1). | Beta-lactam antibiotics such as carbapenem | [55] |
| E. coli | ESBL-encoding genes (e.g., blaCTX-M-15) in E. coli include at least three types of mobile elements including plasmids, bacteriophages, and transposon. | Beta-lactam antibiotics such as carbapenem | [56] |
| Trial Number | Phase | Results | Reference |
|---|---|---|---|
| NCT01959113 | 1 | AMPs secreted by commensal coagulase-negative Staphylococcus in healthy skin displayed selectively antimicrobial activity against S. aureus. | [148] |
| NCT01967628 | 1 | Supplementation of vitamin D3 during increased AMP (e.g., LL-37) concentration in airway surface liquid in the Winter and Spring seasons. | [149] |
| NCT01372995 | 2 | Treatment with a high-dose vitamin D3 can increase the expression of human cationic antimicrobial protein (hCAP18) mRNA in plasma. | [150] |
| NCT01447017 NCT01522391 | 2 | DPK 060, an antimicrobial peptide derived from the endogenous protein kininogen, was an effective and safe drug candidate for the topical treatment of microbial infections. | [151] |
| NCT02456480 | 2 | Treatment with topical omiganan, an indolicidin analog, significantly improved the local objective scoring atopic dermatitis index in patients. | [152] |
| ISRCTN12149720 | 2 | Treatment of anti-biofilm peptide P60.4Ac-containing ototopical drops was safe and well-tolerated, with 47% of successful cases for patients suffering from chronic suppurative otitis media. | [153] |
| IRCT20090822002365N17 | 3 | Supplementation of CoQ10 dramatically increased serum levels of cathelicidin LL-37. | [154] |
| ChiCTR-OIC-16010250 | 3 | Nal-P-113, an AMP P-113 with histidine residues replaced by β-naphthylalanine, can restrain the growth of Streptococcus gordonii, Fusobacterium nucleatum, and Porphyromonas gingivalis and biofilm formation at a concentration of 20 μg/mL. | [155] |
| NCT00310726 | None | Polymorphisms in the human β-defensin 1 gene were negatively and significantly associated with HIV-1 infection in the Zambian population. | [156] |
| NCT03622918 | None | The colistin/rifampicin combination treatment induced a higher microbiological response rate in patients with pneumonia induced by colistin-resistant Acinetobacter baumannii. | [157] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Yang, M. Antimicrobial Peptides: From Design to Clinical Application. Antibiotics 2022, 11, 349. https://doi.org/10.3390/antibiotics11030349
Zhang C, Yang M. Antimicrobial Peptides: From Design to Clinical Application. Antibiotics. 2022; 11(3):349. https://doi.org/10.3390/antibiotics11030349
Chicago/Turabian StyleZhang, Chunye, and Ming Yang. 2022. "Antimicrobial Peptides: From Design to Clinical Application" Antibiotics 11, no. 3: 349. https://doi.org/10.3390/antibiotics11030349
APA StyleZhang, C., & Yang, M. (2022). Antimicrobial Peptides: From Design to Clinical Application. Antibiotics, 11(3), 349. https://doi.org/10.3390/antibiotics11030349






