Determination of the Relationships between the Chemical Structure and Antimicrobial Activity of a GAPDH-Related Fish Antimicrobial Peptide and Analogs Thereof
Abstract
:1. Introduction
2. Results
2.1. Physicochemical Properties
2.2. Antimicrobial Activity
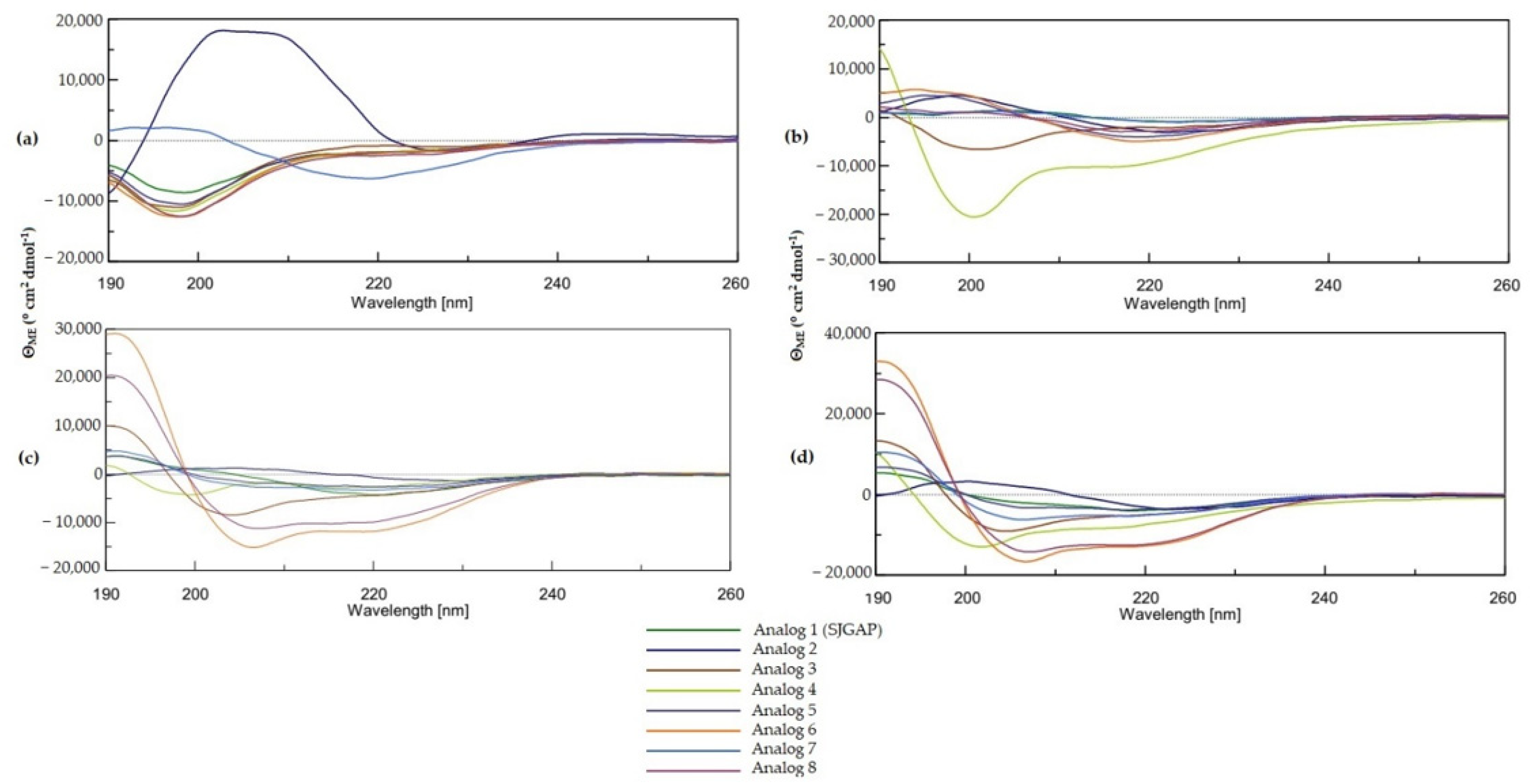
2.3. Secondary Structure
2.3.1. Circular Dichroism (CD) Measurements
2.3.2. Secondary Structure Predictions
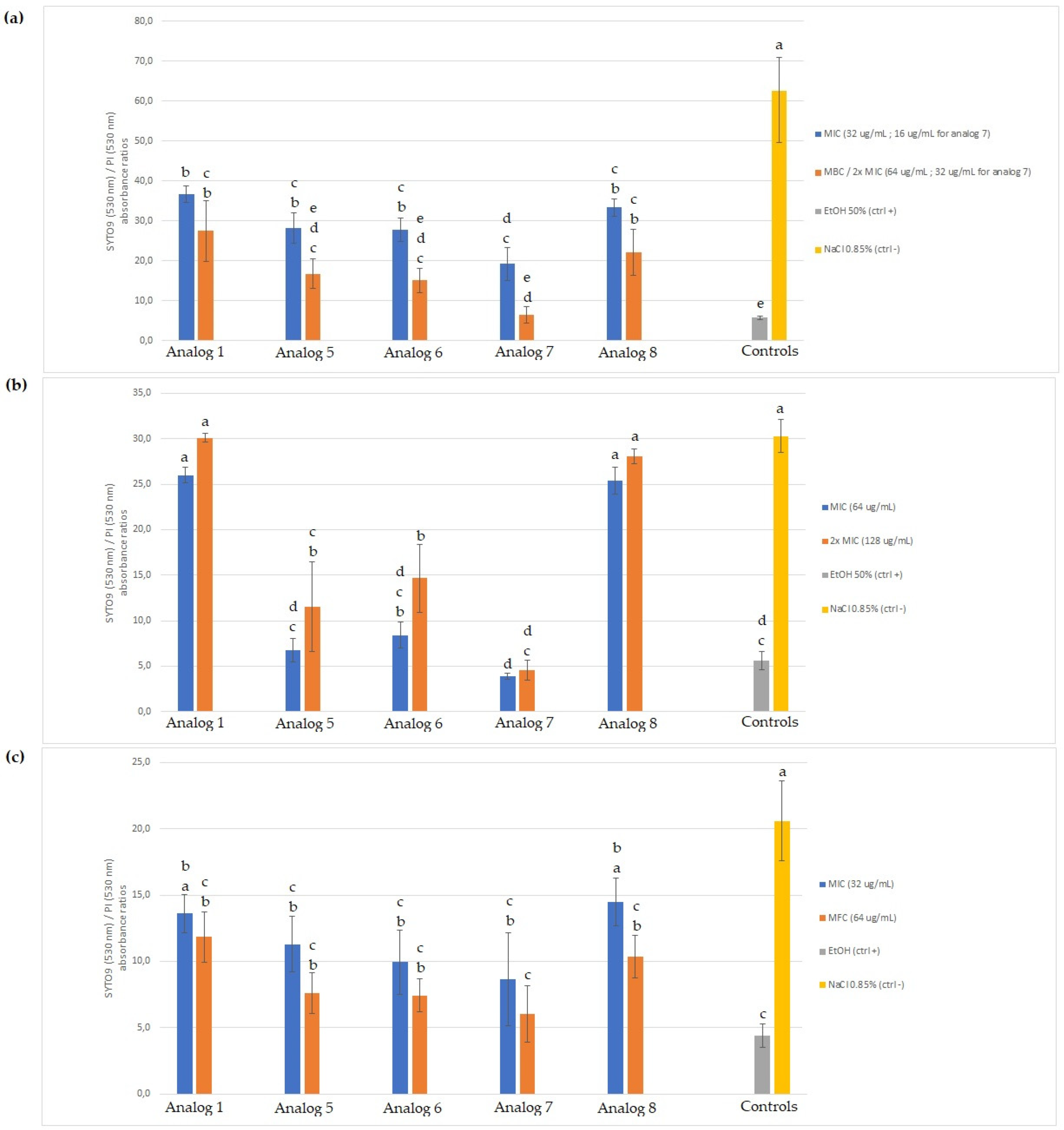
2.4. Membrane Permeabilization
2.4.1. SYTO 9/Propidium Iodide (PI) Staining
2.4.2. 260 nm-Absorbing Intracellular Material Leakage
3. Discussion
3.1. Antimicrobial Activity and Peptides’ Sequences
3.2. Secondary Structure
3.3. Membrane Permeabilization
4. Materials and Methods
4.1. Peptide Design and Synthesis
4.2. Determination of Peptides Physicochemical Properties
4.3. Antimicrobial Activity Assays
4.3.1. Microbial Strains and Culture Conditions
4.3.2. Minimal Inhibitory and Bactericidal Concentrations for Bacterial Strains
4.3.3. Minimal Inhibitory and Fungicidal Concentrations for Fungal Strains
4.4. CD Measurements
4.5. Secondary Structure Predictions
4.6. Membrane Permeabilization Assays
4.6.1. SYTO 9 and PI Assays
4.6.2. The 260 nm Absorbing Material (DNA/RNA) Leak Assays
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 28 January 2022).
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides with Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrêa, J.A.; Evangelista, A.G.; de Melo Nazareth, T.; Luciano, F.B. Fundamentals on the molecular mechanism of action of antimicrobial peptides. Materialia 2019, 8, 100494. [Google Scholar] [CrossRef]
- Koehbach, J.; Craik, D.J. The Vast Structural Diversity of Antimicrobial Peptides. Trends Pharmacol. Sci. 2019, 40, 517–528. [Google Scholar] [CrossRef]
- Ageitos, J.M.; Sanchez-Perez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Li, Y.; Xiang, Q.; Zhang, Q.; Huang, Y.; Su, Z. Overview on the recent study of antimicrobial peptides: Origins, functions, relative mechanisms and application. Peptides 2012, 37, 207–215. [Google Scholar] [CrossRef]
- da Costa, J.P.; Cova, M.; Ferreira, R.; Vitorino, R. Antimicrobial peptides: An alternative for innovative medicines? Appl. Microbiol. Biotechnol. 2015, 99, 2023–2040. [Google Scholar] [CrossRef]
- Cantisani, M.; Leone, M.; Mignogna, E.; Kampanaraki, K.; Falanga, A.; Morelli, G.; Galdiero, M.; Galdiero, S. Structure-activity relations of myxinidin, an antibacterial peptide derived from the epidermal mucus of hagfish. Antimicrob. Agents Chemother. 2013, 57, 5665–5673. [Google Scholar] [CrossRef] [Green Version]
- Ben said, L.; Fliss, I.; Offret, C.; Beaulieu, L. Antimicrobial Peptides: The New Generation of Food Additives. In Encyclopedia of Food Chemistry; Melton, L., Shahidi, F., Varelis, P., Eds.; Academic Press: Oxford, UK, 2019; pp. 576–582. [Google Scholar] [CrossRef]
- Perez Espitia, P.J.; de Fátima Ferreira Soares, N.; dos Reis Coimbra, J.S.; de Andrade, N.J.; Souza Cruz, R.; Alves Medeiros, E.A. Bioactive Peptides: Synthesis, Properties, and Applications in the Packaging and Preservation of Food. Compr. Rev. Food Sci. Food Saf. 2012, 11, 187–204. [Google Scholar] [CrossRef]
- Palmieri, G.; Balestrieri, M.; Proroga, Y.T.; Falcigno, L.; Facchiano, A.; Riccio, A.; Capuano, F.; Marrone, R.; Neglia, G.; Anastasio, A. New antimicrobial peptides against foodborne pathogens: From in silico design to experimental evidence. Food Chem. 2016, 211, 546–554. [Google Scholar] [CrossRef]
- de Castro, R.J.S.; Sato, H.H. Biologically active peptides: Processes for their generation, purification and identification and applications as natural additives in the food and pharmaceutical industries. Food Res. Int. 2015, 74, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.P.; Sousa, R.C.S.; Otoni, C.G.; Moraes, A.R.F.; Souza, V.G.L.; Medeiros, E.A.A.; Espitia, P.J.P.; Pires, A.C.S.; Coimbra, J.S.R.; Soares, N.F.F. Nisin and other antimicrobial peptides: Production, mechanisms of action, and application in active food packaging. Innov. Food Sci. Emerg. Technol. 2018, 48, 179–194. [Google Scholar] [CrossRef]
- Food Safety. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 28 January 2022).
- Rai, M.; Pandit, R.; Gaikwad, S.; Kovics, G. Antimicrobial peptides as natural bio-preservative to enhance the shelf-life of food. J. Food Sci. Technol. 2016, 53, 3381–3394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef]
- Shwaiki, L.N.; Arendt, E.K.; Lynch, K.M.; Thery, T.L.C. Inhibitory effect of four novel synthetic peptides on food spoilage yeasts. Int. J. Food Microbiol. 2019, 300, 43–52. [Google Scholar] [CrossRef]
- Thery, T.; Shwaiki, L.N.; O’Callaghan, Y.C.; O’Brien, N.M.; Arendt, E.K. Antifungal activity of a de novo synthetic peptide and derivatives against fungal food contaminants. J. Pept. Sci. 2019, 25, e3137. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of Bacteriocins and Protective Cultures in Dairy Food Preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol. Rev. 2020, 45, fuaa039. [Google Scholar] [CrossRef]
- Shabir, U.; Ali, S.; Magray, A.R.; Ganai, B.A.; Firdous, P.; Hassan, T.; Nazir, R. Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microb. Pathog. 2018, 114, 50–56. [Google Scholar] [CrossRef]
- Najafian, L.; Babji, A.S. A review of fish-derived antioxidant and antimicrobial peptides: Their production, assessment, and applications. Peptides 2012, 33, 178–185. [Google Scholar] [CrossRef]
- Rajanbabu, V.; Chen, J.Y. Applications of antimicrobial peptides from fish and perspectives for the future. Peptides 2011, 32, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Ennaas, N.; Hammami, R.; Beaulieu, L.; Fliss, I. Purification and characterization of four antibacterial peptides from protamex hydrolysate of Atlantic mackerel (Scomber scombrus) by-products. Biochem. Biophys. Res. Commun. 2015, 462, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Ennaas, N.; Hammami, R.; Gomaa, A.; Bedard, F.; Biron, E.; Subirade, M.; Beaulieu, L.; Fliss, I. Collagencin, an antibacterial peptide from fish collagen: Activity, structure and interaction dynamics with membrane. Biochem. Biophys. Res. Commun. 2016, 473, 642–647. [Google Scholar] [CrossRef]
- Hatab, S.; Chen, M.L.; Miao, W.; Lin, J.; Wu, D.; Wang, C.; Yuan, P.; Deng, S. Protease Hydrolysates of Filefish (Thamnaconus modestus) Byproducts Effectively Inhibit Foodborne Pathogens. Foodborne Pathog. Dis. 2017, 14, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, J.; Ding, S.; Qi, B. Isolation and identification of novel antioxidant and antimicrobial oligopeptides from enzymatically hydrolyzed anchovy fish meal. Process. Biochem. 2018, 74, 148–155. [Google Scholar] [CrossRef]
- Sila, A.; Nedjar-Arroume, N.; Hedhili, K.; Chataigné, G.; Balti, R.; Nasri, M.; Dhulster, P.; Bougatef, A. Antibacterial peptides from barbel muscle protein hydrolysates: Activity against some pathogenic bacteria. LWT—Food Sci. Technol. 2014, 55, 183–188. [Google Scholar] [CrossRef]
- Collins, F.W.J.; O’Connor, P.M.; O’Sullivan, O.; Rea, M.C.; Hill, C.; Ross, R.P. Formicin—A novel broad-spectrum two-component lantibiotic produced by Bacillus paralicheniformis APC 1576. Microbiology 2016, 162, 1662–1671. [Google Scholar] [CrossRef] [PubMed]
- Houyvet, B.; Zanuttini, B.; Corre, E.; Le Corguillé, G.; Henry, J.; Zatylny-Gaudin, C. Design of antimicrobial peptides from a cuttlefish database. Amino Acids 2018, 50, 1573–1582. [Google Scholar] [CrossRef]
- Cantisani, M.; Finamore, E.; Mignogna, E.; Falanga, A.; Nicoletti, G.F.; Pedone, C.; Morelli, G.; Leone, M.; Galdiero, M.; Galdiero, S. Structural insights into and activity analysis of the antimicrobial peptide myxinidin. Antimicrob. Agents Chemother. 2014, 58, 5280–5290. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Mukai, Y.; Niidome, T.; Takashi, C.; Tokunaga, Y.; Hatakeyama, T.; Aoyagi, H. Interaction of pleurocidin and its analogs with phospholipid membrane and their antibacterial activity. J. Pept. Res. 2001, 57, 119–126. [Google Scholar] [CrossRef]
- Fukuoka, Y.; Matsushita, Y.; Furukawa, S.; Niidome, T.; Hatakeyama, T.; Aoyagi, H. Structure-Activity Relationship of Model Peptides Based on Pleurocidin, an Antibacterial Peptide. Bull. Chem. Soc. Jpn. 2003, 76, 1857–1861. [Google Scholar] [CrossRef]
- Lee, J.; Lee, D.G. Influence of the hydrophobic amino acids in the N- and C-terminal regions of pleurocidin on antifungal activity. J. Microbiol. Biotechnol. 2010, 20, 1192–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.K.; Lee, S.A.; Shin, S.; Lee, J.Y.; Jeong, K.W.; Nan, Y.H.; Park, Y.S.; Shin, S.Y.; Kim, Y. Structural flexibility and the positive charges are the key factors in bacterial cell selectivity and membrane penetration of peptoid-substituted analog of Piscidin 1. Biochim. Biophys. Acta 2010, 1798, 1913–1925. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Shin, A.; Jeong, K.W.; Jin, B.; Jnawali, H.N.; Shin, S.; Shin, S.Y.; Kim, Y. Role of phenylalanine and valine10 residues in the antimicrobial activity and cytotoxicity of piscidin-1. PLoS ONE 2014, 9, e114453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Offret, C.; Fliss, I.; Bazinet, L.; Marette, A.; Beaulieu, L. Identification of A Novel Antibacterial Peptide from Atlantic Mackerel belonging to the GAPDH-Related Antimicrobial Family and Its In Vitro Digestibility. Mar. Drugs 2019, 17, 413. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.K.; Lee, M.J.; Go, H.J.; Park, T.H.; Park, N.G. Purification and characterization of YFGAP, a GAPDH-related novel antimicrobial peptide, from the skin of yellowfin tuna, Thunnus albacares. Fish Shellfish Immunol. 2012, 33, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, M.J.; Go, H.J.; Kim, Y.J.; Park, N.G. Antimicrobial function of the GAPDH-related antimicrobial peptide in the skin of skipjack tuna, Katsuwonus pelamis. Fish Shellfish Immunol. 2014, 36, 571–581. [Google Scholar] [CrossRef]
- Branco, P.; Francisco, D.; Chambon, C.; Hebraud, M.; Arneborg, N.; Almeida, M.G.; Caldeira, J.; Albergaria, H. Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 2014, 98, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Wagener, J.; Schneider, J.J.; Baxmann, S.; Kalbacher, H.; Borelli, C.; Nuding, S.; Kuchler, R.; Wehkamp, J.; Kaeser, M.D.; Mailander-Sanchez, D.; et al. A peptide derived from the highly conserved protein GAPDH is involved in tissue protection by different antifungal strategies and epithelial immunomodulation. J. Investig. Dermatol. 2013, 133, 144–153. [Google Scholar] [CrossRef] [Green Version]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Juhl, C.; Beck-Sickinger, A.G. Chapter Two—Molecular Tools to Characterize Adiponectin Activity. In Vitamins & Hormones; Litwack, G., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 90, pp. 31–56. [Google Scholar]
- Shire, S.J. 2—Analytical tools used in the formulation and assessment of stability of monoclonal antibodies (mAbs). In Monoclonal Antibodies; Shire, S.J., Ed.; Woodhead Publishing: Sawston, UK, 2015. [Google Scholar]
- Hammami, R.; Bedard, F.; Gomaa, A.; Subirade, M.; Biron, E.; Fliss, I. Lasso-inspired peptides with distinct antibacterial mechanisms. Amino Acids 2015, 47, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial Peptides from Fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Menif, E.; Offret, C.; Labrie, S.; Beaulieu, L. Identification of Peptides Implicated in Antibacterial Activity of Snow Crab Hepatopancreas Hydrolysates by a Bioassay-Guided Fractionation Approach Combined with Mass Spectrometry. Probiotics Antimicrob. Proteins 2018, 11, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Branco, P.; Kemsawasd, V.; Santos, L.; Diniz, M.; Caldeira, J.; Almeida, M.G.; Arneborg, N.; Albergaria, H. Saccharomyces cerevisiae accumulates GAPDH-derived peptides on its cell surface that induce death of non-Saccharomyces yeasts by cell-to-cell contact. FEMS Microbiol. Ecol. 2017, 93, fix055. [Google Scholar] [CrossRef] [Green Version]
- Gan, B.H.; Gaynord, J.; Rowe, S.M.; Deingruber, T.; Spring, D.R. The multifaceted nature of antimicrobial peptides: Current synthetic chemistry approaches and future directions. Chem. Soc. Rev. 2021, 50, 7820–7880. [Google Scholar] [CrossRef]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Combination Effects of Antimicrobial Peptides. Antimicrob. Agents Chemother. 2016, 60, 1717–1724. [Google Scholar] [CrossRef] [Green Version]
- Massicotte, M.-A.; Vincent, A.T.; Schneider, A.; Paquet, V.r.E.; Frenette, M.; Charette, S.J. One Aeromonas salmonicida subsp. salmonicida isolate with a pAsa5 variant bearing antibiotic resistance and a pRAS3 variant making a link with a swine pathogen. Sci. Total Environ. 2019, 690, 313–320. [Google Scholar] [CrossRef]
- Paquet, V.E.; Vincent, A.T.; Moineau, S.; Charette, S.J. Beyond the A-layer: Adsorption of lipopolysaccharides and characterization of bacteriophage-insensitive mutants of Aeromonas salmonicida subsp. salmonicida. Mol. Microbiol. 2019, 112, 667–677. [Google Scholar] [CrossRef]
- Souza, A.L.; Diaz-Dellavalle, P.; Cabrera, A.; Larranaga, P.; Dalla-Rizza, M.; De-Simone, S.G. Antimicrobial activity of pleurocidin is retained in Plc-2, a C-terminal 12-amino acid fragment. Peptides 2013, 45, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Parente, A.M.S.; Daniele-Silva, A.; Furtado, A.A.; Melo, M.A.; Lacerda, A.F.; Queiroz, M.; Moreno, C.; Santos, E.; Rocha, H.A.O.; Barbosa, E.G.; et al. Analogs of the Scorpion Venom Peptide Stigmurin: Structural Assessment, Toxicity, and Increased Antimicrobial Activity. Toxins 2018, 10, 161. [Google Scholar] [CrossRef] [Green Version]
- Nešuta, O.; Hexnerová, R.; Buděšínský, M.; Slaninová, J.; Bednárová, L.; Hadravová, R.; Straka, J.; Veverka, V.; Čeřovský, V. Antimicrobial Peptide from the Wild Bee Hylaeus signatus Venom and Its Analogues: Structure–Activity Study and Synergistic Effect with Antibiotics. J. Nat. Prod. 2016, 79, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Shagaghi, N.; Palombo, E.A.; Clayton, A.H.A.; Bhave, M. Antimicrobial peptides: Biochemical determinants of activity and biophysical techniques of elucidating their functionality. World J. Microbiol. Biotechnol. 2018, 34, 62. [Google Scholar] [CrossRef] [PubMed]
- Park, N.G.; Silphaduang, U.; Moon, H.S.; Seo, J.K.; Corrales, J.; Noga, E.J. Structure-activity relationships of piscidin 4, a piscine antimicrobial peptide. Biochemistry 2011, 50, 3288–3299. [Google Scholar] [CrossRef]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollmann, A.; Martinez, M.; Noguera, M.E.; Augusto, M.T.; Disalvo, A.; Santos, N.C.; Semorile, L.; Maffia, P.C. Role of amphipathicity and hydrophobicity in the balance between hemolysis and peptide-membrane interactions of three related antimicrobial peptides. Colloids Surf. B Biointerfaces 2016, 141, 528–536. [Google Scholar] [CrossRef]
- Jiang, Z.; Kullberg, B.J.; van der Lee, H.; Vasil, A.I.; Hale, J.D.; Mant, C.T.; Hancock, R.E.; Vasil, M.L.; Netea, M.G.; Hodges, R.S. Effects of hydrophobicity on the antifungal activity of alpha-helical antimicrobial peptides. Chem. Biol. Drug Des. 2008, 72, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Dennison, S.R.; Harris, F.; Bhatt, T.; Singh, J.; Phoenix, D.A. The effect of C-terminal amidation on the efficacy and selectivity of antimicrobial and anticancer peptides. Mol. Cell Biochem. 2009, 332, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Strandberg, E.; Tiltak, D.; Ieronimo, M.; Kanithasen, N.; Wadhwani, P.; Ulrich, A.S. Influence of C-terminal amidation on the antimicrobial and hemolytic activities of cationic α-helical peptides. Pure Appl. Chem. 2007, 79, 717–728. [Google Scholar] [CrossRef]
- Houyvet, B.; Bouchon-Navaro, Y.; Bouchon, C.; Goux, D.; Bernay, B.; Corre, E.; Zatylny-Gaudin, C. Identification of a moronecidin-like antimicrobial peptide in the venomous fish Pterois volitans: Functional and structural study of pteroicidin-alpha. Fish Shellfish Immunol. 2018, 72, 318–324. [Google Scholar] [CrossRef]
- Kuete, V.; Efferth, T. Cameroonian medicinal plants: Pharmacology and derived natural products. Front. Pharmacol. 2010, 1, 123. [Google Scholar] [CrossRef] [Green Version]
- Ríos, J.L.; Recio, M.C. Medicinal plants and antimicrobial activity. J. Ethnopharmacol. 2005, 100, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. BBA—Biomembr. 2016, 1858, 936–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, W.W.; Phipps, B.M.; Ishiguro, E.E.; Olafson, R.W.; Trust, T.J. Surface layer virulence A-proteins from Aeromonas salmonicida strains. Can. J. Biochem. Cell Biol. Rev. Can. Biochim. Biol. Cell. 1984, 62, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Arnesen, K.R.; Mikkelsen, H.; Schroder, M.B.; Lund, V. Impact of reattaching various Aeromonas salmonicida A-layer proteins on vaccine efficacy in Atlantic cod (Gadus morhua). Vaccine 2010, 28, 4703–4708. [Google Scholar] [CrossRef] [PubMed]
- van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. Properties and mechanisms of action of naturally occurring antifungal peptides. Cell Mol. Life Sci. 2013, 70, 3545–3570. [Google Scholar] [CrossRef]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar]
- Travkova, O.G.; Moehwald, H.; Brezesinski, G. The interaction of antimicrobial peptides with membranes. Adv. Colloid Interface Sci. 2017, 247, 521–532. [Google Scholar] [CrossRef]
- Moffat, L.; Jones, D.T. Semi-Supervised Learning of Protein Secondary Structure from Single Sequences. bioRxiv 2020, 1–13. [Google Scholar] [CrossRef]
- Monincova, L.; Veverka, V.; Slaninova, J.; Budesinsky, M.; Fucik, V.; Bednarova, L.; Straka, J.; Cerovsky, V. Structure-activity study of macropin, a novel antimicrobial peptide from the venom of solitary bee Macropis fulvipes (Hymenoptera: Melittidae). J. Pept. Sci. 2014, 20, 375–384. [Google Scholar] [CrossRef]
- Rahmanpour, A.; Ghahremanpour, M.M.; Mehrnejad, F.; Moghaddam, M.E. Interaction of Piscidin-1 with zwitterionic versus anionic membranes: A comparative molecular dynamics study. J. Biomol. Struct. Dyn. 2013, 31, 1393–1403. [Google Scholar] [CrossRef]
- Talandashti, R.; Mahdiuni, H.; Jafari, M.; Mehrnejad, F. Molecular Basis for Membrane Selectivity of Antimicrobial Peptide Pleurocidin in the Presence of Different Eukaryotic and Prokaryotic Model Membranes. J. Chem. Inf. Model. 2019, 59, 3262–3276. [Google Scholar] [CrossRef] [PubMed]
- Farkas, A.; Maroti, G.; Kereszt, A.; Kondorosi, E. Comparative Analysis of the Bacterial Membrane Disruption Effect of Two Natural Plant Antimicrobial Peptides. Front. Microbiol. 2017, 8, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, J.; McGoverin, C.; Vanholsbeeck, F.; Swift, S. Optimisation of the Protocol for the LIVE/DEAD((R)) BacLight(TM) Bacterial Viability Kit for Rapid Determination of Bacterial Load. Front. Microbiol. 2019, 10, 801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.U.; Egli, T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, M.; Azevedo, N.F.; Ivask, A. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep. 2019, 9, 6483. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Xiang, Y.; Xu, M. From red to green: The propidium iodide-permeable membrane of Shewanella decolorationis S12 is repairable. Sci. Rep. 2015, 5, 18583. [Google Scholar] [CrossRef] [Green Version]
- Davey, H.M.; Hexley, P. Red but not dead? Membranes of stressed Saccharomyces cerevisiae are permeable to propidium iodide. Environ. Microbiol. 2011, 13, 163–171. [Google Scholar] [CrossRef]
- Branco, P.; Francisco, D.; Monteiro, M.; Almeida, M.G.; Caldeira, J.; Arneborg, N.; Prista, C.; Albergaria, H. Antimicrobial properties and death-inducing mechanisms of saccharomycin, a biocide secreted by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 159–171. [Google Scholar] [CrossRef]
- Woodburn, K.W.; Jaynes, J.M.; Clemens, L.E. Evaluation of the Antimicrobial Peptide, RP557, for the Broad-Spectrum Treatment of Wound Pathogens and Biofilm. Front. Microbiol. 2019, 10, 1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, ix–xv. [Google Scholar] [CrossRef] [Green Version]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Bo, J.; Yang, Y.; Zheng, R.; Fang, C.; Jiang, Y.; Liu, J.; Chen, M.; Hong, F.; Bailey, C.; Segner, H.; et al. Antimicrobial activity and mechanisms of multiple antimicrobial peptides isolated from rockfish Sebastiscus marmoratus. Fish Shellfish Immunol. 2019, 93, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Cuenca-Estrella, M.; Cantón, E. EUCAST and CLSI: Working together towards a Harmonized Method for Antifungal Susceptibility Testing. Curr. Fungal Infect. Rep. 2013, 7, 59–67. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Cantón, E.; Pemán, J. Antifungal Susceptibility Testing of Filamentous Fungi. Curr. Fungal Infect. Rep. 2012, 6, 41–50. [Google Scholar] [CrossRef]
- Carson, C.F.; Mee, B.J.; Riley, T.V. Mechanism of action of Melaleuca alternifolia (tea tree) oil on Staphylococcus aureus determined by time-kill, lysis, leakage, and salt tolerance assays and electron microscopy. Antimicrob. Agents Chemother. 2002, 46, 1914–1920. [Google Scholar] [CrossRef] [Green Version]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Comparative mode of action of the antimicrobial peptide melimine and its derivative Mel4 against Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 7063. [Google Scholar] [CrossRef] [Green Version]
- Bessalle, R.; Haas, H.; Goria, A.; Shalit, I.; Fridkin, M. Augmentation of the antibacterial activity of magainin by positive-charge chain extension. Antimicrob. Agents Chemother. 1992, 36, 313–317. [Google Scholar] [CrossRef] [Green Version]

| Peptide Analogs | Sequences | Net Charges | Isoelectric Points | Molar Weights (g/mol) | GRAVY Indexes |
|---|---|---|---|---|---|
| 1 (SJGAP) | VKVGINGFGRIGRLVTRAAFHGKKVEIVAIND | +4 | 11.4 | 3436.07 | 0.272 |
| 2 | VKVGINGFGRIG | +2 | 11.4 | 1216.45 | 0.558 |
| 3 | IGRLVTRAAFHG | +2 | 12.1 | 1297.53 | 0.433 |
| 4 | HGKKVEIVAIND | 0 | 7.6 | 1322.53 | −0.225 |
| VKVGINGFGRIGRLVTRAAFHGKKVAIVAINA | +6 | 12.4 | 3334.02 | 0.603 | |
| 6 * | VKVGINGFGRIGRLVTRAAFHGKKVKIVAINK | +8 | 12.5 | 3448.21 | 0.247 |
| 7 * | VKVGINGFGRIGRLVTRLLFHGKKVEIVLIND | +4 | 11.4 | 3562.21 | 0.459 |
| 8 * | VKVGINGFGRIGRLVTRAAFHGKKVEIVAIND-NH2 | +5 | 11.9 | 3435.08 | 0.272 |
| Peptide Analogs | Antibacterial Activity (MIC; MBC) (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| E. coli ATCC 25922 | E. coli ATCC 11229 | P. aeruginosa ATCC 27853 | A. salmonicida 69 R3 | A. salmonicida 69 R5 | M. luteus LMA-272 | L. ivanovii ATCC 19119 | |
| 1 (SJGAP) | n.a. 1 | ||||||
| 2 | n.a. | ||||||
| 3 | n.a. | ||||||
| 4 | n.a. | ||||||
| 5 | 32; 128 | 32; 64 | 16; n.b.a. 2 | 32; 64 | 32; 128 | 64; n.b.a. | 32; n.b.a. |
| 6 | 64; 64 | 16; 32 | 16; 128 | 64; 128 | 64; n.b.a. | 64; n.b.a. | 32; n.b.a. |
| 7 | n.a. | ||||||
| 8 | 64; 64 | 32; 32 | 64; n.b.a. | 128; n.b.a. | n.a. | n.a. | n.a. |
| Peptide Analogs | Antifungal Activity (MIC; MFC) (μg/mL) | |||||
|---|---|---|---|---|---|---|
| A. niger 3071-13 | M. racemosus LMA-722 | Paecilomyces sp. 5332-9a | R. mucilaginosa 27173 | Z. rouxii LL12_088 | S. boulardii 27169 | |
| 1 (SJGAP) | n.a. 1 | n.a. | 128; - | n.a. | 128; n.f.a. 2 | n.a. |
| 2 | n.a. | |||||
| 3 | n.a. | |||||
| 4 | n.a. | n.a. | n.a. | n.a. | 64; n.f.a. | n.a. |
| 5 | n.a. | n.a. | 32; n.f.a. | 32; 64 | 64; n.f.a. | n.a. |
| 6 | n.a. | n.a. | 32; n.f.a. | 32; 64 | 64; n.f.a. | n.a. |
| 7 | n.a. | n.a. | 16; n.f.a. | 32; 64 | 64; n.f.a. | n.a. |
| 8 | n.a. | n.a. | 64; n.f.a. | n.a. | 64; n.f.a. | n.a. |
| Peptide Analogs | Secondary Structure Contents (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PB (10 mM) | TFE 25% | TFE 50% | TFE 75% | |||||||||
| α-Helix | β-Strands | Unrd * | α-Helix | β-Strands | Unrd * | α-Helix | β-Strands | Unrd * | α-Helix | β-Strands | Unrd * | |
| 1 (SJGAP) | 6 | 35 | 37 | 4 | 42 | 33 | 6 | 40 | 32 | 7 | 38 | 33 |
| 2 | 6 | 44 | 35 | 5 | 40 | 33 | 4 | 41 | 34 | 5 | 38 | 35 |
| 3 | 5 | 33 | 39 | 6 | 37 | 35 | 15 | 33 | 30 | 18 | 29 | 31 |
| 4 | 6 | 32 | 39 | 5 | 32 | 37 | 4 | 39 | 35 | 6 | 34 | 33 |
| 5 | 6 | 33 | 39 | 5 | 41 | 32 | 6 | 41 | 32 | 8 | 38 | 32 |
| 6 | 6 | 30 | 41 | 6 | 42 | 31 | 39 | 17 | 24 | 41 | 14 | 28 |
| 7 | 8 | 33 | 36 | 4 | 42 | 33 | 7 | 38 | 33 | 16 | 30 | 33 |
| 8 | 7 | 30 | 40 | 5 | 40 | 34 | 25 | 23 | 31 | 36 | 18 | 26 |
| Peptide Analogs | Sequence | Research Interest/Analog Modification |
|---|---|---|
| 1 (SJGAP) | VKVGINGFGRIGRLVTRAAFHGKKVEIVAIND | Native SJGAP; model for this study |
| 2 | VKVGINGFGRIG | N-terminal segment of SJGAP |
| 3 | IGRLVTRAAFHG | Middle segment of SJGAP |
| 4 | HGKKVEIVAIND | C-terminal segment of SJGAP |
| 5 * | VKVGINGFGRIGRLVTRAAFHGKKVAIVAINA | Substitution of anionic residues with neutral alanine residues |
| 6 * | VKVGINGFGRIGRLVTRAAFHGKKVKIVAINK | Substitution of anionic residues with cationic lysine residues (+4 net charge) |
| 7 * | VKVGINGFGRIGRLVTRLLFHGKKVEIVLIND | Substitution of alanine residues by more hydrophobic leucine residues |
| 8 * | VKVGINGFGRIGRLVTRAAFHGKKVEIVAIND-NH2 | C-terminal amidation |
| Genus | Species | Strain | Type | Study Relevance |
|---|---|---|---|---|
| Escherichia | coli | ATCC 25922 | Gram-negative | Human pathogen; reference strain [87] |
| Escherichia | coli | ATCC 11229 | Gram-negative | Non-pathogenic E. coli strain |
| Pseudomonas | aeruginosa | ATCC 27853 | Gram-negative | Human pathogen |
| Aeromonas | salmonicida | 69 R3 a | Gram-negative | Fish pathogen |
| Aeromonas | salmonicida | 69 R5 a | Gram-negative | Fish pathogen |
| Micrococcus | luteus | LMA-272 b | Gram-positive | Human skin flora; opportunistic pathogen |
| Listeria | ivanovii | ATCC 19119 | Gram-positive | Human pathogen |
| Rhodotorula | mucilaginosa | 27 173 c | Yeast | Human pathogen; food spoilage |
| Saccharomyces | boulardii | 27 169 c | Yeast | Food spoilage |
| Zygosaccharomyces | rouxii | LL12_088 d | Yeast | Food spoilage |
| Aspergillus | niger | 3071-13 e | Filamentous fungi | Food spoilage |
| Mucor | racemosus | LMA-722 b | Filamentous fungi | Food spoilage; opportunistic pathogen |
| Paecilomyces | sp. | 5332-9 e | Filamentous fungi | Food spoilage; opportunistic pathogen |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cashman-Kadri, S.; Lagüe, P.; Fliss, I.; Beaulieu, L. Determination of the Relationships between the Chemical Structure and Antimicrobial Activity of a GAPDH-Related Fish Antimicrobial Peptide and Analogs Thereof. Antibiotics 2022, 11, 297. https://doi.org/10.3390/antibiotics11030297
Cashman-Kadri S, Lagüe P, Fliss I, Beaulieu L. Determination of the Relationships between the Chemical Structure and Antimicrobial Activity of a GAPDH-Related Fish Antimicrobial Peptide and Analogs Thereof. Antibiotics. 2022; 11(3):297. https://doi.org/10.3390/antibiotics11030297
Chicago/Turabian StyleCashman-Kadri, Samuel, Patrick Lagüe, Ismail Fliss, and Lucie Beaulieu. 2022. "Determination of the Relationships between the Chemical Structure and Antimicrobial Activity of a GAPDH-Related Fish Antimicrobial Peptide and Analogs Thereof" Antibiotics 11, no. 3: 297. https://doi.org/10.3390/antibiotics11030297
APA StyleCashman-Kadri, S., Lagüe, P., Fliss, I., & Beaulieu, L. (2022). Determination of the Relationships between the Chemical Structure and Antimicrobial Activity of a GAPDH-Related Fish Antimicrobial Peptide and Analogs Thereof. Antibiotics, 11(3), 297. https://doi.org/10.3390/antibiotics11030297







